Abstract
The biolistic delivery system is an essential tool in plant genetic engineering, capable of delivering DNAs, RNAs, and proteins independent of tissue type, genotype, or species. However, its efficiency and consistency remain longstanding challenges despite decades of widespread use. Here, through advanced simulations, we identify gas and particle flow barriers as the root cause of these limitations. We show that a flow guiding barrel (FGB) achieves a 22-fold enhancement in transient transfection efficiency, a 4.5-fold increase in CRISPR-Cas9 ribonucleoprotein editing efficiency in onion epidermis, and a 17-fold improvement in viral infection efficiency in maize seedlings. Furthermore, stable transformation frequency in maize using B104 immature embryos increases over 10-fold, while in planta CRISPR-Cas12a-mediated genome editing efficiency in wheat meristems doubles in both T0 and T1 generations. This study provides insights into the fundamental mechanisms underlying biolistic inefficiency and demonstrates a practical solution that enables broader and more reliable applications in plant genetic engineering.
Similar content being viewed by others
Introduction
Genetic modification enhances crop traits like yield, nutrition, pest resistance, and resilience, significantly contributing to global food security1,2,3,4,5,6,7. Beyond agriculture, it holds promise for biofuels, pharmaceuticals, and biotechnology8,9. While CRISPR-Cas enables precise genome modifications compared to traditional plant breeding10,11,12, substantial challenges remain in delivering genetic materials to plant cells and regenerating transformed tissues13.
Agrobacterium-mediated infection and biolistic delivery are the primary methods for plant transformation. Agrobacterium-mediated transformation offers high efficiency and throughput and reliably generates single-copy DNA insertion events14,15,16. However, its narrow host range limits its use for transforming recalcitrant crops17,18, and its reliance on DNA delivery can lead to off-target effects19,20. In contrast, delivering non-DNA-based CRISPR reagents, such as RNA or ribonucleoproteins (RNPs), minimizes off-target effects and generates transgene-free edited plants, which are highly desirable for plant breeding and regulatory approval19,20.
Biolistic delivery, or particle bombardment, complements Agrobacterium-mediated transformation by enabling the delivery of virtually any biological cargo to nearly any plant species or tissue type21. Biolistics becomes particularly valuable for transforming tissue types or species resistant to Agrobacterium infection. However, biolistic delivery faces notable challenges with efficiency, consistency, and tissue damage caused by high-velocity microprojectiles, which hinder regeneration and transformation22,23,24,25. Additionally, it often leads to fragmented and multiple transgene insertions in the genome, resulting in unpredictable gene expression14,16.
Despite its critical role and widespread use, the gene gun device developed for biolistic delivery has seen little technological advancement over the past three decades. No comprehensive study has yet addressed the fundamental causes of its inefficiency and inconsistency. A limited understanding of particle flow dynamics in this system has hindered its full potential. Modernizing this technology could unlock new opportunities, particularly as biolistic delivery remains the only effective tool for creating DNA-free gene-edited plants using CRISPR-Cas RNPs for plants that cannot be regenerated from protoplasts19,20. This capability directly addresses public concerns and regulatory hurdles associated with Agrobacterium, a disarmed plant pathogen complicating its use in transgenic crop production13,19,20.
In this study, we employ a highly interdisciplinary approach that integrates advanced engineering tools with plant biotechnology to tackle these issues. Through computational simulations, we gain insight of the internal mechanism of the Bio-Rad PDS-1000/He system and identify gas and particle flow barriers as the root cause of inefficiency and inconsistency. To address this issue, we develop the flow guiding barrel (FGB), a 3D-printed device, to systematically optimize particle and gas flow. This design enhances performance by generating a 4-fold larger target area and higher-velocity microprojectiles. In plant cell delivery, the FGB achieves a 2- to 30-fold improvement across various applications and target tissues. Tests included delivery of DNA, protein, and RNP into onion epidermal cells and infectious DNA clones of soybean mosaic virus (SMV) and sugarcane mosaic virus (SCMV) into soybean and sweet corn leaves26. In maize B104 embryos, the FGB increases throughput from 30–40 to 100 embryos per bombardment and improves stable transformation frequency by over 10-fold. FGB also augments in planta transformation of wheat shoot apical meristems (SAM). A 2-fold increase in editing efficacy of CRISPR-Cas12a is achieved in both T0 and T1 generations with a single bombardment per plate, compared to the conventional three-bombardment protocol. The improved germline editing of the polyploid wheat genome by CRISPR-Cas12a effectively addresses the previously reported low-editing efficiency problem27, highlighting the potential application of FGB in plant genome editing.
Results
Flow guiding barrel design and biolistic delivery parameter optimization in onion epidermis
There is limited literature exploring the fundamental causes of inefficiencies and inconsistencies in biolistic delivery systems, although it is recognized that the velocities and quantities of projectiles delivered to the samples are critical to performance21. To bridge this gap, we develop computational fluid dynamic (CFD) simulation models to analyze helium and particle flows, assuming laminar flow and 600 nm gold particles within the Bio-Rad PDS 1000/He system, the most widely used gene gun. These simulations revealed that a major issue lies in the small aperture of the internal barrel (d = 10 mm), which is smaller than the typical sample size and severely restricts particle flow, resulting in significant particle loss (Supplementary Fig. 1). Furthermore, the current design disrupts helium flow, producing an inconsistent diffusive flow pattern that reduces the pressure, decreases particle velocity, and causes uneven distribution on the target tissue (Fig. 1a). These findings pinpoint critical limitations in the gene gun design and led us to hypothesize that engineering the flow dynamics within the gene gun could significantly improve its efficiency and consistency.
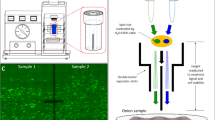
a Comparison between the conventional gene gun and the gene gun equipped with the FGB helium and particle flow velocity profiles using computational fluid dynamics simulation model. b Schematic illustration of biolistic delivery using the conventional gene gun and the gene gun equipped with the FGB system. Images of FITC-loaded gold particles bombarded into agar gel using both delivery methods. The FGB delivers to a larger target area, allowing for higher throughput experiments. Portions of this figure were created in BioRender. c Schematic illustration of FGB placement inside the gene gun, the barrel design in the slicing software prior to 3D printing, and representative images of GFP-DNA delivery into onion epidermis cells with different FGB geometries. The lines through the entire onion samples are autofluorescence from creases in the onion tissue on the agar plate and are excluded from analysis. d Optimization of the FGB length using transient delivery of GFP-DNA into onion epidermis (n = 8 biological replicates). e Optimization of the FGB diameter using transient delivery of GFP-DNA into onion epidermis (n = 23 biological replicates for No Barrel, and n = 10 biological replicates for 10 mm, 15 mm, and 20 mm diameter). f Penetration depth in agar gel using the two systems quantified via fluorescent confocal microscopy. g Bombardment area analysis conducted via fluorescent optical images using both delivery systems (n = 3 replicates). Error bars represent the mean ± s.d. P values were calculated using a two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < .001. h) Simulation particle velocities upon impacting the tissues using the conventional gene gun and the gene gun equipped with the FGB. Source data are provided as a Source Data file.
To test the hypothesis, we developed the FGB device using SolidWorks® and fabricated it via Fused Deposition Modeling (FDM) 3D printing, enabling rapid modifications to optimize performance. This FGB integrates seamlessly with the existing Bio-Rad PDS-1000/He gene gun by replacing the internal spacer rings (Fig. 1b, c). Key design parameters, including the FGB’s diameter and length (Fig. 1c), were fine-tuned to enhance performance. Transient DNA delivery of a green fluorescent protein (GFP) construct into onion epidermis demonstrated that the FGB performs better at longer target distances and reduced pressures (Fig. 1d, e, Supplementary Fig. 2–4).
Simulations revealed that the FGB enables more uniform laminar particle flow, unlike the diffusive flow in the conventional gun (Fig. 1a). While the conventional device directs only 21% of loaded particles to the target, the FGB achieves nearly 100% delivery. FITC-labeled gold particles bombarded into a 1.5% agarose gel confirmed these findings. The FGB produced a maximum depth of 35 µm, with peaks at 5–7.5 µm and 22.5–25 µm over a 7.07 cm² target area (Fig. 1b, f, g), compared to the conventional device’s 15 µm depth, single peak at 7.5 µm, and 1.77 cm² area (Fig. 1b, f, g). These results are consistent with the simulation, which showed that the FGB achieved twice the particle velocity and four times the coverage area compared to the conventional device (Fig. 1a, h), thus validating our simulation model. These results demonstrate the FGB’s ability to modulate flow dynamics, significantly improving projectile coverage, velocity, and penetration. Moreover, this study provides insights into the fundamental mechanisms that drive gene gun performance.
Improved delivery of GFP-DNA, FITC-BSA protein, and CRISPR-Cas9 RNP in onion epidermal cells
Transient expression of GFP from a plasmid DNA construct (pLMNC95; Supplementary Fig. 5) in onion epidermal cells was first used to benchmark the FGB. With 22 ng of pLMNC95 DNA, the FGB demonstrated a 22-fold improvement, yielding an average of 3,351 green fluorescent cells compared to 153 cells with the original device (Fig. 2a). Even with reduced DNA (2.2 ng) and spermidine (1 μL), the FGB still achieved 1031 green fluorescent cells per bombardment (Fig. 2b), a 7-fold improvement over the conventional device with full DNA loading (22 ng). The high efficacy with a small DNA quantity suggests the potential to reduce transgene copy number in stable transformation28.
a 22 ng of GFP-DNA and 10 μL of spermidine were bombarded into onion epidermis comparing the two delivery systems in transient cell transfection (n = 21 biological replicates for No Barrel, n = 8 for FGB). b 2.2 ng of GFP-DNA and 1 μL of spermidine were delivered with the FGB and compared with 22 ng of gfp-DNA and 10 μL of spermidine with conventional device (n = 23 biological replicates for No Barrel, n = 6 for FGB). c 25 μg of FITC-BSA and 1 μL of TransIT-2020 were delivered using both devices and compared by counting the number of FITC-BSA-positive onion cells (n = 5 biological replicates). d Transient Cas9-RNP editing in onion epidermis using both delivery systems verified by NGS (n = 9 biological replicates). The tracrRNA on the RNP-complex has red fluorescence, hence the greater red fluorescence is due to a more effective delivery of the RNP to the tissue. Box plots in (a) and (b) extend from the 25th to the 75th percentiles, with a box in the center representing the mean and whiskers extending from the minimum to the maximum values. Error bars represent the mean ± s.d. P values were calculated using a two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < .001. Scale bar = 1 cm. Source data are provided as a Source Data file.
To validate the FGB’s capability to improve protein delivery, we tested the delivery of FITC-labeled bovine serum albumin (FITC-BSA). The FGB showed a 4-fold increase of internalization of FITC-BSA (Fig. 2c), confirmed via microscopy (Supplementary Fig. 6). It also improved Cas9-RNP delivery, achieving a 4.5-fold increase in editing of the flavonoid 3’-hydroxylase (F3’H) gene in onion epidermal cells, with an average of 6.6% editing validated by next-generation sequencing (NGS, Fig. 2d). This enhancement highlights the FGB’s advantage in DNA-free delivery, as Agrobacterium cannot deliver RNPs.
Improved delivery of virus-infectious clones in soybean and maize seedlings
Biolistics is an important method for introducing infectious DNA clones of viruses into plant species29. To evaluate the FGB for this purpose, two-week-old sweet corn and soybean seedlings were biolistically inoculated with infectious clones of GFP-expressing sugarcane mosaic virus (SCMV-CS1-GFP, Supplementary Fig. 7) and soybean mosaic virus (SMV-GFP, Supplementary Fig. 8) respectively (Fig. 3a, b). The FGB achieved an 83.5% infection rate for SCMV-CS1-GFP to sweet corn seedlings with enhanced GFP expression, compared to 5% with the conventional device (Fig. 3c). Similarly, delivery of SMV-GFP to soybean seedlings reached a 100% infection rate versus 66% with the original system (Fig. 3d). These results highlight the FGB’s effectiveness in delivering higher titer of infectious viruses to target tissues, demonstrating its potential to improve the efficiency of plant-virus interaction and virus-based gene function studies30,31,32.
a Schematic showing the bombardment procedure. This figure was created in BioRender. b Images of juvenile sweet corn, the bombardment setup with the FGB, and infected sweet corn at 4 weeks post-bombardment. c SCMV-GFP DNA was bombarded into leaves of sweet corn seedlings and GFP fluorescence was imaged at 4 weeks post-bombardment. d SMV-GFP DNA was bombarded into leaves of soybean seedlings and GFP fluorescence was imaged at 4 weeks post-bombardment. Error bars represent the mean ± s.d. (n = 3 independent experiments with 8 replicates per independent experiment). P values were calculated using a two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. Source data are provided as a Source Data file.
Maize B104 embryo transient and stable transformation improvements
To evaluate the FGB’s potential for enhancing stable transformation, we biolistically delivered pCBL101-mCherry (Supplementary Fig. 9) into immature maize B104 embryos. Leveraging the FGB’s ability to distribute particles over a larger area, we targeted 100 maize embryos per bombardment plate, a significant increase from the 30–40 embryos per plate used in the conventional methods (Fig. 4a; Supplementary Fig. 10a). Based on transient expression data, FGB biolistic parameters were optimized to 425 psi with a 12 cm target distance (Supplementary Fig. 10b). A comparison of transient mCherry expression in embryos bombarded with the FGB versus the conventional system showed a 2.5-fold improvement, with transfection rates increasing from 37.67% to 92.16% (Fig. 4b, c). Qualitative assessment of mCherry expression in embryos further confirmed the superior transient expression achieved with the FGB (Supplementary Fig. 11a, b). Embryos bombarded with the FGB consistently showed higher expression levels.
a Schematic showing the number of embryos bombarded and the spread of embryos on the plate using the new protocol vs the conventional protocol. This figure was created in BioRender. b Optical fluorescent images of transiently expressing embryos using both delivery systems. c Transient expression analysis on maize embryos using the two delivery systems. Efficiency was quantified as the number of embryos transiently expressing/number of embryos bombarded. d Embryo stable transformation frequency using both delivery systems. Transformation frequency is defined as the number of rooted plants per bombardment of 100 embryos. Box plot in (c) extends from the 25th to the 75th percentiles, with a box in the center representing the mean and whiskers extending from the minimum to the maximum values. Error bars represent the mean ± s.d. (n = 6 independent experiments). P values were calculated using a two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. Source data are provided as a Source Data file.
Next, embryos were evaluated for stable transformation following a published QuickCorn protocol for Agrobacterium-mediated transformation33 (Supplementary Table 1). The QuickCorn protocol uses an antibiotic-resistance gene neomycin phosphotransferase II (NptII) for selection and bypasses the callus bulking phase33. For the biolistic delivery experiments, we further optimized the QuickCorn protocol by adjusting the antibiotic concentration used in the plant growth media and the duration of tissue recovery (Supplementary Table 2 and Supplementary Fig. 12a and 12b). Compared to the conventional maize B104 biolistic transformation protocol using bialaphos herbicide for selection34, the modified QuickCorn protocol drastically reduced the transformation timeline from 165 days to 72 days (Supplementary Table 2 and Supplementary Fig. 12c).
Embryos bombarded with the FGB device achieved a stable transformation frequency of 11.33%, compared to only 0.33% with the conventional device (Fig. 4d), representing a significant improvement of more than 30-fold. Most transgenic plants recovered from FGB-bombarded embryos were located in the central and middle regions of the petri dish (Supplementary Fig. 10a). This distribution aligns with areas of the strongest particle flow, as indicated by simulation and gel penetration tests (Fig. 1d–h).
We also directly compared the FGB performance against the Agrobacterium-mediated maize transformation system used in our labs at Iowa State University.31 The experiments assessed transformation using the Agrobacterium-mediated method with or without a vir gene ternary helper plasmid33 and the biolistic delivery system with or without the FBG device (Supplementary Table 3). The FGB significantly outperformed Agrobacterium-mediated methods without a vir gene ternary helper plasmid, although its transformation frequency was lower than that of the Agrobacterium-mediated method with the helper plasmid. Transgenic plants generated using the FGB exhibited comparable transgene integration copy numbers to those produced via Agrobacterium (3.2 vs. 3.7 copies of the mCherry gene, Supplementary Table 3, Supplementary Fig. 13a). In contrast, a single transgenic plant produced by the conventional biolistic system exhibited a high copy number, exceeding 24 copies for the NptII gene and 32 copies for mCherry. We attribute the lower copy number observed with the FGB, compared to the conventional device, to its more uniform delivery of projectiles. This uniformity results in fewer particles reaching each cell, thereby reducing the DNA copy numbers per cell. These findings demonstrate the substantial improvements of the FGB over the original biolistic device, enhancing transformation efficiency and reducing tissue culture time. Additionally, it can also improve the quality of transgenic events, as low copy number transgene integration reduces genome disruptions.
Enhanced SAM-based CRISPR Cas12a genome editing efficiency in wheat using the FGB
SAM-based delivery of genetic materials is particularly promising for crops recalcitrant to tissue culture35,36. To explore its potential in wheat, we adapted a protocol to deliver DNA vectors into SAMs, aiming to generate transgenic or genome-edited wheat plants (Fig. 5a). Building on earlier observations of the FGB’s ability to achieve deeper tissue penetration (Fig. 1f), we hypothesized that it would deliver a higher number of particles to the SAM, particularly targeting the underlying L2 layer cells, a layer of cells in SAM that is responsible for the development of germline tissues. This enhanced penetration is expected to improve SAM-specific delivery and germline transmission (Fig. 5b).
a Schematic illustration of the procedure for delivering DNA constructs to wheat SAM using biolistic delivery. Portions of this figure were created in BioRender. b Comparison between the conventional gene gun and the gene gun equipped with the FGB system. The FGB assembly features a modified stop screen, which differs in size from the conventional version. c Visualization of GFP expression under the two different conditions. The SAM region is outlined, with red arrows indicating GFP expression within SAM cells. d Quantification of GFP delivery efficiency using the SAM-based biolistic method and (e) Percentage of SAM cells exhibiting GFP fluorescence after bombardment. Error bars represent the mean ± s.d. (n = 3 independent experiments). P values were calculated using a two-tailed Student’s t test. *P < 0.05, **P < 0.01. The GFP fluorescence was detected 12 h after bombardment and embryos carrying 10 or more GFP signal spots were considered GFP-positive (d). The GFP fluorescence was detected 12 h after bombardment and embryos whose SAM (L1/L2/L3 layer) cells showing GFP signals were counted as positive (e).
We first varied conditions to compare delivery efficiency between the conventional device and the FGB using a GFP reporter plasmid (Supplementary Fig. 14). Conventional bombardment (Condition 1) used the previously reported settings of 1350 psi, 6 cm flight distance, and 3 bombardments27,35,37. This condition resulted in an overall GFP delivery efficiency of 93% (based on GFP signal in any cell), but SAM-specific delivery (based on GFP signal in SAM-specific region) was limited to only 17% (Fig. 5d, e, and Supplementary Table 4). Reducing to 1 bombardment (Condition 2) dramatically reduced overall GFP delivery to just 3%, with no expression in SAM tissue. In contrast, the FGB (conditions 3, 4, and 5) achieved 100% GFP delivery efficiency, with SAM-specific delivery of 33%, 53%, and 17%, respectively (Fig. 5d, e, and Supplementary Table 4). Conditions 3 and 4 demonstrated a 2 to 3-fold improvement in SAM-specific delivery compared to the conventional method (Fig. 5c). These findings suggest that even at relatively low gas pressures with a single bombardment, the FGB enhances wheat SAM transformation efficiency, likely due to its ability to generate higher-velocity projectiles for deeper tissue penetration.
Based on the improved SAM-specific GFP delivery, we hypothesized that the FGB would enhance the efficiency of CRISPR-based genome editing in wheat. We focused on CRISPR-Cas12a-mediated genome editing, as previous SAM-based genome editing efforts focused on CRISPR-Cas9 while Agrobacterium-mediated CRISPR-Cas12a delivery showed low editing efficiency in wheat27,35,38,39,40. We targeted the TaPDS gene, present in the A, B, and D sub-genomes, using a construct containing ttLbCas12a and the crRNA41, both driven by the ZmUbi promoter (Supplementary Fig. 15a, b).
We tested all experimental conditions with three independent replicates and analyzed editing efficiency by NGS (Table 1). Condition 1 (conventional gene gun, 3 bombardments) resulted in an average genome editing efficiency of 4.0% in the T0 generation, while Condition 2 (conventional gene gun, 1 bombardment) did not generate positive plants. In contrast, Conditions 3 and 4 with FGB achieved genome editing efficiencies of 8.7% and 9.3%, respectively, representing a 2-fold or more improvement in the T0 generation.
To assess the inheritance of edits, the positive T0 plants were self-pollinated, and the resulting seeds were germinated to analyze the T1 generation. Under Condition 1, the editing efficiency in the T1 generation was 27.8%. In comparison, Conditions 3 and 4 achieved significantly higher efficiencies of 45.0% and 55.6% respectively, while Condition 5 also reached 44.4% (Table 1). Genotyping analysis confirmed that the mutations in T0 plants (Table 2) were successfully inherited to the T1 progeny, which carried homozygous or heterozygous mutations at the target gene homeologs (Table 3). These results validate our hypothesis that the FGB substantially enhances SAM-based delivery, improving the L2 layer cell targeting and increasing the inheritance of mutations from T0 to T1.
Discussion
In conventional gene guns, restrictive geometry creates diffusive flow dynamics, causing random deflection of projectiles and inconsistent delivery beyond biological sample variations. To enhance biolistic efficiency, the Hepta adaptor was developed to simultaneously load and deliver seven samples, resulting in a 4- to 5-fold increase in transfected cells42. However, this design consumed seven times more resources and retained the restrictive geometry of conventional gene guns. Consequently, it did not improve particle velocity or penetration depth, both essential for effective delivery. The bulky structure and increased complexity further limited its practicality. Ultimately, the drawbacks of the Hepta adaptor outweighed the advantages, leading to its discontinuation. Guided by computational simulations, we developed the FGB to eliminate these limitations in the original gene gun system and the Hepta adaptor. The FGB collimates nearly all projectiles into a laminar flow, ensuring uniform impact, higher velocity, and consistent delivery, even at lower pressures. Across various tissue types, genotypes, and plant species, the FGB consistently achieved a 2- to 30-fold improvement in delivery efficiency. Further simulations and gel-trapping studies confirmed its ability to deliver substantially more microcarriers at higher velocities with even distribution over a larger target area. The innovation allows for using smaller DNA quantities, thereby reducing transgene copy numbers and enhancing the utility of the biolistic delivery systems28.
The FGB greatly improved the delivery of viral infectious DNA clones, achieving nearly 100% infection rates in corn and soybean seedlings, which will enhance applications such as in planta protein production43, gRNA evaluation44, and virus-induced genome editing44,45. It also addresses a critical bottleneck in delivering CRISPR-Cas RNPs for precise, DNA-free editing. The flexibility of the FGB-aided delivery was further demonstrated when combined with the newly developed QuickCorn method for maize transformation33. Using the adapted protocol with the FGB device, we achieved an 11.33% stable transformation efficiency with 100 embryos per bombardment in 72 days compared to 0.33% with the conventional gene gun. Furthermore, the DNA copy numbers of transgenic lines produced by the FGB (3.2 copies) were comparable to those generated via Agrobacterium (3.7 copies), which is likely attributed to the even distribution of DNA projected over the larger targeted sample areas.
In direct delivery to wheat SAMs, the FGB demonstrated a unique capability of delivering the particles deeper into the L2 layer cells, which gives rise to germline transformation and genome editing. The FGB achieved up to 53% SAM-specific GFP delivery with only 1 bombardment per plate, which represents a marked increase compared to the conventional method requiring 3 bombardments per plate. These improvements translate into higher efficiency and probability of heritable mutations, which are crucial for creating genome-edited wheat plants. Indeed, in our experiments of testing CRISPR-Cas12a, the FGB conferred a 2-fold increase in genome editing efficiency in both T0 and T1 generations. With rapid advances in SAM-based genome editing27,35,36,39, the FGB’s ability to target more L2 layer cells marks a significant step toward enhancing genotype-independent transformation and genome editing across crops.
A distinctive aspect of the FGB, is the innovation is rooted in physics and engineering design rather than biology. Therefore, we believe this technology is readily adaptable to diverse biological systems. Our demonstrated applications include transient gene expression, stable transformation, and genome editing using CRISPR-Cas9-RNP and CRISPR-Cas12a plasmid systems. The potential of FGB extends well beyond our case studies in wheat, maize, soybean, and onion presented here. The FGB is expected to help achieve transgene-free genome editing in crops via delivering CRISPR-Cas systems such as RNPs20,46,47,48 or RNA47,49. The digitized design process facilitated by simulation models allows rapid prototyping through 3D printing. This approach offers precise control, full customization, quick turnaround times, and flexibility for future applications. Such adaptability is crucial for developing transformation methods tailored for new species, genotypes, or explants. For example, it is possible to tailor FGB geometries for specific applications, such as further improvement of SAM-based transformation or delivery to intact seedlings. In summary, the FGB not only represents a major innovative upgrade to the standard gene gun system commercialized since 1988 but also lays the groundwork for next-generation biolistic delivery methods. The technology revitalizes biolistic delivery, transforming it into a powerful platform for advancing genetic engineering.
Methods
COMSOL simulations
In this study, COMSOL Multiphysics was utilized to investigate the particle velocities, helium gas velocity, and particle distribution area for both the conventional gene gun system and the flow-guiding barrel (FGB) system. The simulations were conducted by integrating and solving the laminar flow and particle tracing modules of COMSOL, which allowed for detailed analysis of the fluid-particle interactions (Supplementary Method 1).
COMSOL geometry and model setup
The chamber geometry was set to a radius of 30 mm with a height of 125 mm. For simulating the FGB, the outer barrel had a radius of 14 mm and a height of 45 mm, while the inner barrel had a radius of 10 mm with a height of 45 mm. The target distance was set at 9 cm. For simulating the conventional gene gun system, the outer barrel had a radius of 13.5 mm and a height of 5 mm, while the inner barrel had a radius of 5 mm and a height of 5 mm. Two spacer rings were placed directly under the barrel with a radius of 13.5 mm and a total height of 10 mm. The target distance was set at 6 cm. A physics-controlled mesh with fine element size was used. To accurately capture the fluid dynamics and particle motion, the gene gun chamber geometry was constructed to reflect real-world dimensions, with particular attention to the detailed features of the FGB. The mesh density was optimized to provide high resolution near the barrel exit, where gas velocity gradients and particle acceleration are most significant. Finer meshing was applied near the barrel walls and the particle release surface to accurately simulate wall effects and particle interactions.
COMSOL boundary conditions and initial conditions
The helium gas flow was modeled as a velocity inlet with a 5 mm radius and a 20 mm height for the inlet boundary using compressible flow. The outlet boundary was modeled as a zero pressure to simulate the vacuum in the chamber during bombardment. A deformed macrocarrier with a 4 mm radius was positioned 5 mm above the barrel and set as the particle release surface. At 0.07 s into the simulation, 1000 gold particles were input into the simulation model and released from the particle release surface, and their velocities and spatial distribution were tracked as they traveled through the barrel and impacted the target.
COMSOL simulation and particle tracking
The simulations were run as time-dependent studies, with the particles’ velocities and distribution being recorded at the bottom of the chamber (target ___location). Motion of particles tracked using the Lagrangian method for particle tracing. This allowed the capture of individual particle trajectories under the influence of external forces, including drag, gravity, and gas flow. Particle velocities were integrated over time to capture transient behaviors, while particle and helium gas interactions evaluated at each timestep. The helium gas was modeled as a compressible fluid, while particles were treated as discrete entities interacting with the surrounding flow. The Stokes drag model was applied to describe particle motion, appropriate for the particle size and gas flow velocity. The simulations accounted for both the initial velocity and acceleration of particles induced by the expanding gas, as well as the opposing drag force exerted by the helium flow. While we do not believe gravitational force has a substantial impact, it is included in the simulation to accurately reflect real-life experimental conditions. The particle release mechanism was modeled as a discrete event at t = 0.07 s, corresponding to the expected activation time of the rupture disk and the subsequent macrocarrier impact.
We calculated the Reynolds numbers for both the conventional gene gun and the FGB using the estimated inlet velocity of 200 m/s50 (Supplementary Method 1), confirming that the flow regime is laminar. The particle distribution was assumed to be uniform. While we acknowledge this limitation, as particles aggregate upon drying on the macrocarrier, both the FGB and the conventional gene gun follow identical sample preparation protocols. As a result, the particle size distribution is expected to remain consistent across both systems, and we do not anticipate that aggregation would qualitatively affect the comparative outcomes of the simulation.
COMSOL simulation results and analysis
Results were extracted based on the time-dependent simulation, with particle velocities and distribution patterns recorded. The final particle distribution on the target was analyzed for both the conventional and FGB systems, focusing on factors such as the uniformity of particle deposition, particle distribution, and the velocity profiles of the particles under both barrel configurations. In both cases, particle velocities were tracked throughout their trajectories, from the release surface (macrocarrier) to their final position on the target, providing detailed data on the effectiveness of each gene gun system barrel configuration in terms of particle delivery and distribution precision.
Flow guiding barrel design
The flow guiding barrel (FGB) and its various design iterations were digitally created using SolidWorks (Dassault Systèmes, Waltham, MA, USA), a computer-aided design (CAD) software. The design process involved simulating fluid flow and particle dynamics to optimize the geometry for efficient particle acceleration and distribution. Once the designs were finalized, STL files were generated and then sliced into G-code using FlashPrint, a slicing software that converts the 3D models into a format compatible with the 3D printer. This process allows for fine-tuning of parameters such as layer height, print speed, and infill pattern, all of which were optimized to balance print quality and material usage.
3D printing process
The FGB device was 3D printed using a Flashforge Creator 3 Pro FDM printer (FlashForge, Jinhua, Zhejiang, China), using Polylactic Acid (PLA) filament as the printing material. PLA was selected for its ease of use, relatively low extrusion temperature, and good mechanical properties. The extrusion temperature was set to 200 °C, consistent with the manufacturer’s recommendations for PLA. The heated print bed was maintained at 60 °C to promote good adhesion and reduce warping during the printing process. The infill pattern was set to hexagonal with 100% infill for optimal strength and durability, ensuring the structural integrity of the printed FGB. The layer height was set to 0.2 mm for a balance of detail and print speed. The print speed was set to 60 mm/s, with a travel speed of 80 mm/s, to optimize printing time while minimizing defects. Additionally, rafts were enabled in the slicing software to ensure good adhesion of the printed object to the print bed and to minimize the risk of print failure, particularly for the larger and more complex geometries of the FGB designs. The printing times for the FGB designs varied depending on the complexity and size of the iteration. Print times ranged from 1 to 2 h per design, with more complex iterations taking longer due to the increased number of layers and detail required for accurate representation of the design.
Gel penetration experiment
For gel penetration experiment, a 1.5% agarose gel was made to mimic the plant tissue. A Zeiss 780 confocal microscope (Carl Zeiss, Inc., Jena, Germany) was used to track the FITC-labeled particles and determine penetration depths via z-stacking. The confocal was equipped with a FITC filter (excitation: 460–500, emission: 512–542 nm). ImageJ was used to count the particles.
Microscopy using an inverted fluorescence microscope
Images were taken using a Leica DMi8 inverted microscope with an automated stage and digital camera. Low magnification was used in conjunction with the tile scan feature to merge multiple images into a mosaic. A typical image is shown in Fig. 1C. Fluorescent images were captured with a FITC filter (excitation: 460–500, emission: 512–542 nm) or a Texas Red filter (excitation 542–582, emission: 604–644 nm).
Onion epidermis GFP-DNA biolistic bombardment
Sections of onion epidermis (≥ 3 × 4 cm, with the long axis perpendicular to the scale veins) were removed from the inner surface of onion scales and placed on agar plates. The conventional device for the BioRad PDS-1000/He (Bio-Rad Life Science, Hercules, CA, USA) was assembled as described in the Bio-Rad manual, available from the company website, with the stopping screen holder placed between the spacer rings. The flow guiding barrel replaced the conventional device with a stopping screen placed between the barrel and the stopping screen holder. To ensure a fair comparison between the FGB and the original device, the amounts of gold, DNA, and delivery agent are kept consistent for both the No Barrel and FGB experiments. However, the rupture pressure and target distance are individually optimized for each plant tissue and for each device. As a result, the pressures and distances may vary between experiments. Gold particles (0.6 µm, Bio-Rad Life Science) were washed with isopropanol24, followed by washes with water and final resuspension in water to give the indicated concentration. Gold aliquots were transferred to 1.5 mL microfuge tubes and stored frozen until use. To precipitate DNA onto gold, 25 µL of sonicated 12 mg/mL gold was combined with 0.2 µg plasmid DNA pLMNC95, encoding a gene for an endoplasmic reticulum-localized GFP (ER-GFP)51. This was followed by the addition, while vortexing, of 25 µL 2.5 M CaCl2 and 10 µL 0.1 M spermidine (Thermo Fisher Scientific, Waltham, MA, USA). Vortexing was then continued for an additional 1 min. The gold/DNA complex was collected with brief centrifugation, washed with 70% ethanol, and resuspended in 80 µL of 100% ethanol. Tubes of the DNA/gold precipitate were briefly sonicated to achieve a uniform suspension and then 10 µL aliquots were removed during vortexing and spread on the center 1 cm of macrocarriers (Bio-Rad Life Science) which were then allowed to dry in the laminar flow hood. Bombardments with the conventional device were performed with 650 psi rupture disks (Analytical Scientific Instruments, Richmond, CA, USA) and a 6 cm target distance24. Bombardment with the FGB was optimized and performed with same rupture disks and a 9 cm target distance. Bombardments with both devices were performed with a vacuum level of −28 in. Hg.
Onion epidermis FITC-BSA biolistic bombardment
Onion epidermis and gold particles were prepared, as discussed above. To precipitate FITC-BSA (Sigma-Aldrich, St. Louis Missouri, USA) onto gold, 25 µL of sonicated 12 mg/mL gold was combined with 300 µg FITC-BSA protein. This was followed by the addition, while vortexing, of 2 µL TransIT-2020 (Mirus Bio, Madison, WI, USA). Vortexing was then continued for an additional 1 min. The gold/DNA complex was collected with brief centrifugation and resuspended in 128 µL of 100% DI water. Tubes of the gold/FITC-BSA precipitate were briefly sonicated to achieve a uniform suspension, and then 16 µL aliquots were removed during vortexing and spread on the center 1 cm of macrocarriers, which were then allowed to dry in the laminar flow hood. Bombardments with the conventional device were optimized and performed with 650 psi rupture disks and a 6 cm target distance. Bombardments with the FGB were optimized and performed with 650 psi rupture disks and a 9 cm target distance. Bombardments with both devices were performed with a vacuum level of −28 in. Hg.
Onion epidermis CRISPR-Cas9 RNP biolistic bombardment
Custom-synthesized crRNA targeting the F3’H gene (10 nmol), 5’ ATTO 550 tracrRNA (5 nmol), and SpCas9 (62 µmol) were purchased from IDT (Integrated DNA Technologies, Coralville, IA) and stored at −20 °C until use. Briefly, on the day of Cas9-RNP delivery to plant cells, 1 µL of crRNA and 1 uL of tracrRNA were dissolved in 9 µL of nuclease free-IDTE buffer (10 mM Tris, 0.1 mM EDTA, pH 7.5) each. To form the gRNA complex, 8.4 µL of each 10 µM crRNA and 10 µM tracrRNA were mixed with an additional 23.2 µL nuclease free-IDTE buffer (1 × -TE buffer, pH 7.5) in 1.5 mL centrifuge tubes and placed in a 95 °C heat block for 5 min. After 5 min, the tubes were centrifuged at 16,000 x g for 5 s and placed at RT (22 °C) for 10 min. 62 µM SpCas9 was diluted 10x with 1X PBS buffer (pH 7.4) to a 1 µg/µL concentration. To form the RNP complex, 13.2 µL of diluted SpCas9 was added to this tube. The solution was mixed by pipetting up and down, and the tube was incubated at RT for 10 min. This RNP complex was used for biolistic delivery into the onion epidermis.
Onion epidermis and gold particles were prepared as described in the “Onion epidermis GFP-DNA biolistic bombardment” section. To precipitate Cas9-RNP onto gold, 21 µL of sonicated 12 mg/mL gold was combined with 53.2 µL Cas9-RNP targeting the F3’H gene. This was followed by the addition, while vortexing, of 1 µL TransIT-2020 (Mirus Bio, Madison, WI, USA). Vortexing was then continued for an additional 1 min. The gold/RNP complex was collected with brief centrifugation and resuspended in 96 µL of 100% DI water. Tubes of the gold/RNP precipitate were briefly sonicated to achieve a uniform suspension, and then 16 µL aliquots were removed during vortexing and spread on the center 1 cm of macrocarriers, which were then allowed to dry in the laminar flow hood. Bombardments with the conventional device were optimized and performed with 650 psi rupture disks and a 6 cm target distance. Bombardments with the FGB were optimized and performed with 650 psi rupture disks and a 9 cm target distance. Bombardments with both devices were performed with a vacuum level of −28 in. Hg.
Cell counting analysis
Cell analysis was done using CellProfiler 3.1.9 for Windows and was run on a Windows 10 PC. The modules included in the pipelines were used without customization beyond the available options included in the software24.
Onion epidermis DNA extraction, PCR, and NGS analysis
DNA was extracted from onion epidermis using QuickExtract DNA Extraction Solution (Lucigen) following the manufacturer’s protocol, except that the incubation time was extended to 1–3 h. An initial PCR was performed using primers flanking the target site in the flavonoid 3’-hydroxylase gene (AY541035.1). This was followed by a nested PCR using primers that incorporated index and Illumina partial adaptor sequences. Indexed samples were combined and submitted to Azenta U.S., Inc. for Amplicon-EZ next-generation sequencing (NGS). Sequencing data was demultiplexed using the Galaxy Barcode Splitter software (www.usegalaxy.org). Editing was analyzed using the CRISPR RGEN Tools Cas-Analyzer software (www.rgenome.net).
Seedling soybean and sweet corn SMV and SCMV biolistic bombardment
A similar DNA binding procedure was performed to precipitate SMV-GFP and SCMV-GFP DNA onto gold, 50 µL of sonicated 10 mg/mL gold was combined with 1 µg plasmid. This was followed by the addition, while vortexing, of 1 µL TransIT-2020. Vortexing was then continued for an additional 1 min. The gold/DNA complex was collected with brief centrifugation and resuspended in 100 µL of 100% ethanol. Tubes of the DNA/gold precipitate were briefly sonicated to achieve a uniform suspension, and then 10 µL aliquots were removed during vortexing and spread on the center 1 cm of macrocarriers, which were then allowed to dry in a laminar flow hood.30,31 Bombardments with the FGB were optimized and performed with 425 psi rupture disks and a 6 cm target distance. It was impossible to use a distance greater than 6 cm, because entire seedlings were placed in the chamber. The use of pressures higher than 425 psi was not possible due to extensive leaf damage. Bombardments with both devices were performed in a vacuum of −28 in. Hg.
Maize pCBL101-mCherry vector construction
pCBL101-mCherry (Supplementary Fig. 8) was constructed via Gibson assembly of SacI and XmaI digested pCBL10152 (Addgene #199722) with the mCherry fragment amplified from pKL201353 (Addgene #172182) using primers P35S-CBL-F1 (5’- atgacatgattacgaattcgagctcGCCCTTAGTATTCCCTACCCC-3’) and Tvsp-CBL-R1 (5’- gtcgactctagaggatccccgggCCCTGGAAAATGCACTCCCT-3’) and Q5 master mix (NEB).
Maize biolistic and Agrobacterium-mediated transformation
Immature embryos of maize inbred B104, harvested from ears at 12–14 days post-pollination, were used for either Agrobacterium-mediated transformation33 or the biolistic delivery of pCBL101-mCherry experiments. For device comparison, the pretreatment was modified from Raji et al.34 by skipping the preculture step. A total of 100 embryos were placed evenly in a circle at the center of the plate filled with osmotic medium and incubated at 28 °C for 4 h before being subjected to bombardment.
DNA coating on gold particles used a published procedure with modifications24. Briefly, to precipitate pCBL101 DNA onto gold, 50 µL of sonicated 10 mg/mL gold was combined with 1 µg plasmid. This was followed by the addition, while vortexing, of 1 µL TransIT-2020. Vortexing was then continued for an additional 1 min. The gold/DNA complex was collected with brief centrifugation and resuspended in 100 µL of 100% ethanol. Tubes of the DNA/gold precipitate were briefly sonicated to achieve a uniform suspension, and then 10 µL aliquots were removed during vortexing and spread on the center 1 cm of macrocarriers, which were then allowed to dry in the laminar flow hood. Bombardments with the conventional device were performed as previously optimized with 650 psi rupture disks and a 6 cm target distance34. Bombardment with the FGB was optimized and performed with 425 psi rupture disks and a 12 cm target distance (Supplementary Fig. 9b). Bombardments with both devices were performed with a vacuum level of −28 in. Hg.
Maize tissue culture and regeneration follow the published Agrobacterium-mediated QuickCorn procedure33 with modifications (Supplementary Data 1). After bombardment, the embryos were incubated at 28 °C for 20 h before moving to the resting medium for 9–12 days and then to another round of resting medium with 37.5 mg/L G418 selection for another 9–12 days. To monitor the development of the embryos located on the outside, middle, and center parts of the osmotic medium, embryos from the osmotic medium plates were placed on three separate resting medium plates based on their original ___location. The regenerated callus pieces were then transferred to the maturation medium amended with 37.5 mg/L G418 and incubated in a dark chamber at 28 °C. After two weeks, the maturation medium was refreshed and incubated for another two weeks. Callus pieces with regenerating shoots were transferred to a rooting medium amended with 75 mg/L (for Agrobacterium experiments) or 50 mg/L (for biolistic experiments) G418 and kept in a light chamber with a 16/8 (light/dark) photoperiod at 28 °C for 14 days. Rooted plantlets were then checked for red fluorescence in the roots using a NIGHTSEA BlueStar flashlight and filter glasses (NIGHTSEA, MA, USA), and leaf samples were harvested for the DNA copy number analysis.
Maize transgene copy number estimation
Transgene copy number analysis was performed using quantitative PCR (qPCR) and Taqman probes54. Total genomic DNA was extracted from maize leaf tissues using the rapid plant genomic DNA preparation method developed by Edwards et al.55. A single copy gene ZmMEK1 was used as a reference for the transgene copy number estimation56.
Briefly, a reference plasmid, pKL2556 (4416 bp; Supplementary Fig. 13a and Supplementary Table 5), containing three fragments from the reference gene (ZmMEK1; 255 bp) and two transgenes (NptII, 556 bp; mCherry, 631 bp) was constructed by PCR amplification using the primers listed in Fig. S12 and Gibson assembly using NEBuilder Hifi DNA assembly master mix (New England Biolabs, Ipswich, MA, USA). qPCR primer and Taqman probes were designed using the PrimerQuestTM Tool (IDT, Coralville, IA, USA) and synthesized by IDT (Coralville, IA, USA). qPCR reactions were carried out in a 20 µL reaction volume with two primer/probe sets for ZmMEK1 (HEX) with either NptII (FAM) or mCherry (FAM). Each reaction contained 0.25 µM probes, 0.5 µM primers, 0.2 pg–2 ng pKL2556 or 5–10 ng of maize genomic DNA, and 1X PrimeTimeTM Gene Expression Master Mix (IDT, Coralville, IA, USA). qPCR reactions were carried out in a Mx3005p qPCR system (Agilent, Santa Clara, CA) using the fast-cycling condition: 3 min at 95 °C for polymerase activation followed by 40 cycles of amplification with 5 s at 95 °C for denaturation and 30 s at 60 °C for annealing and extension. Using the serially diluted pKL2556 DNA, qPCR cycle threshold values (Ct) for the reference gene ZmMEK1 (HEX) and the transgenes (NptII and mCherry) were obtained, and the standard curves were generated to estimate the Ct values (CtR) for the transgenes assuming the equal copy number in the genomic DNA samples. All three primer pairs had similar efficiencies, and the estimated CtR values were 1.008 × Ct (ZmMEK1; r2 = 0.998) for NptII and 1.018 × Ct (ZmMEK1; r2 = 0.999) for mCherry. Because the single copy reference gene ZmMEK1 has two alleles in the genome, an equal transgene copy number would indicate two copies per genome. Therefore, transgene copy numbers were estimated using the following equations:
Wheat SAM preparation and plant growth conditions
Mature seeds of wheat (Triticum aestivum L. cv. “Fielder”) were used in this study. Seeds were sterilized in a 50% bleach solution for 10 min, rinsed with sterile distilled water, and placed in Petri dishes lined with autoclaved filter paper for overnight germination at room temperature. Using a needle (32 G 4 mm; Medt, USA) under a microscope, the coleoptile and leaf primordia covering the SAM were carefully removed. Embryos were excised from the endosperms and cultured on osmotic Murashige and Skoog (MS) medium containing 20 g/L sucrose, 0.98 g/L MES (pH 5.8), 36.4 g/L D-sorbitol, 36.4 g/L D-mannitol, 3% plant preservative mixture (Nacalai Tesque, Japan), and 7.0 g/L phytagel (Sigma-Aldrich, USA). Fifty embryos were placed per plate for bombardment. GFP fluorescence was assessed 12 h post-bombardment using a fluorescence microscope (Zeiss) with a GFP filter.
For genome editing experiments, the embryos were transferred to normal MS medium (without D-sorbitol and D-mannitol) and incubated at 30 °C for 5 days, followed by transfer to a growth chamber (16 h light/8 h darkness, 22 °C). After three weeks, plants were transferred to soil and grown under the same long-day conditions (16 h light/8 h darkness, 22 °C).
Wheat SAM biolistic delivery of plasmid
Conventional bombardment methods (Condition 1) were adapted from previous reports38,39,41. Briefly, Condition 1 used 0.6 μm gold particles, with a 6.0 cm target distance (from stopping screen to plate), 1,350 psi pressure, and a 6 mm macrocarrier travel distance. Each Petri dish was bombarded three times. Each bombardment delivered 1.5 μg of plasmids (1.5 μL, 1 μg/μL) and 300 μg of gold particles (3 μL, 100 μg/μL). When using the FGB, only a single bombardment per Petri dish was performed.
Wheat T0 and T1 mutation analysis by NGS
NGS of PCR amplicons was utilized to analyze genome editing outcomes in this study. For positive plant screening, the targeted genomic regions were directly amplified from 5th leaf tissue of T0 wheat plants following the instructions of Phire Plant Direct PCR Kit (Thermo Fisher) and barcoded using the Hi-TOM primers57. PCR products were verified by gel electrophoresis. PCR products were pooled together and purified with QIAQuick PCR Purification Kit (QIAGEN) and then quantified by Nanodrop (Thermo Fisher). The purified PCR products were used for Illumina HiSeq2500 sequencing. For targeted mutagenesis, the NGS data were analyzed with CRISPRMatch58 and CRISPResso259. Given the chimeric nature of the SAM-based bombardment, the mutation ratio over 2% was treated as positive plants in the T0 generation. The NGS data was also used to detect the mutations of genome type by using specific sequence of A, B and D genome in hexaploidy wheat. NGS was also used to detect the genotype of positive T1 wheat plants. The genotypes of edited T1 wheat plants were classified based on next-generation sequencing (NGS) data according to the following criteria: homozygous mutants were defined as those in which a single type of mutation accounted for more than 75% of the total sequencing reads; heterozygous mutants each exhibited a single mutation type comprising between 35% and 75% of the reads; and wild-type plants were defined by a total mutation frequency of less than 35%. All primers, crRNA sequences, PCR amplicon sequences, and specific sequences used in this study are provided in the Supplementary Data 2 and 3.
Statistical analysis
Data are presented as the mean and standard deviation. No statistical methods were used to predetermine the sample size. P values were calculated using two-tailed Student’s t tests. Sample sizes are described in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All sequencing data are unavailable at NCBI under accession PRJNA1246628 and PRJNA1246654. Simulation files are available at Figshare [https://doi.org/10.6084/m9.figshare.29218688 and https://doi.org/10.6084/m9.figshare.29218682]. Source data are provided with this paper.
References
Gao, H. et al. Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat. Biotechnol. 38, 579–581 (2020).
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. & Schroeder, J. I. Genetic strategies for improving crop yields. Nature 575, 109–118 (2019).
Gao, C. Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635 (2021).
Li, T., Liu, B., Spalding, M. H., Weeks, D. P. & Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392 (2012).
Napier, J. A., Haslam, R. P., Tsalavouta, M. & Sayanova, O. The challenges of delivering genetically modified crops with nutritional enhancement traits. Nat. Plants 5, 563–567 (2019).
Tuncel, A. et al. Genome-edited foods. Nat. Rev. Bioeng. 1, 799–816 (2023).
Zhu, T., Mettenburg, K., Peterson, D. J., Tagliani, L. & Baszczynski, C. L. Engineering herbicide-resistant maize using chimeric RNA/DNA oligonucleotides. Nat. Biotechnol. 18, 555–558 (2000).
Eidenberger, L., Kogelmann, B. & Steinkellner, H. Plant-based biopharmaceutical engineering. Nat. Rev. Bioeng. 1, 426–439 (2023).
Torney, F., Moeller, L., Scarpa, A. & Wang, K. Genetic engineering approaches to improve bioethanol production from maize. Curr. Opin. Biotechnol. 18, 193–199 (2007).
Molla, K. A., Sretenovic, S., Bansal, K. C. & Qi, Y. Precise plant genome editing using base editors and prime editors. Nat. Plants 7, 1166–1187 (2021).
Shan, Q. et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688 (2013).
Zong, Y. et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440 (2017).
Lee, K. & Wang, K. Strategies for genotype-flexible plant transformation. Curr. Opin. Biotechnol. 79, 102848 (2023).
Liu, J. et al. Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell 31, 368–383 (2019).
Oltmanns, H. et al. Generation of backbone-free, low transgene copy plants by launching T-DNA from the Agrobacterium chromosome. Plant Physiol. 152, 1158–1166 (2010).
Thomson, G., Dickinson, L. & Jacob, Y. Genomic consequences associated with Agrobacterium-mediated transformation of plants. Plant J. 117, 342–363 (2024).
Gelvin, S. B. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37 (2003).
Raman, V. et al. Agrobacterium expressing a type III secretion system delivers Pseudomonas effectors into plant cells to enhance transformation. Nat. Commun. 13, 2581 (2022).
Liang, Z. et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261 (2017).
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K. & Mark Cigan, A. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 1–7 (2016).
Klein, T. M., Wolf, E. D., Wu, R. & Sanford, J. C. High-velocity microprojectiles for delivering nucleic-acids into living cells. Nature 327, 70–73 (1987).
Harkess, A. Smashing barriers in biolistic plant transformation. Plant Cell 31, 273–274 (2019).
Sanford, J. C. The biolistic process. Trends Biotechnol. 6, 299–302 (1988).
Miller, K. et al. An improved biolistic delivery and analysis method for evaluation of DNA and CRISPR-Cas delivery efficacy in plant tissue. Sci. Rep. 11, 7695 (2021).
Miller, K. J. et al. Identifying factors that determine effectiveness of delivery agents in biolistic delivery using a library of amine-containing molecules. ACS Appl. Bio Mater. 5, 4972–4980 (2022).
Jiang, S., Miller, K., Thorpe, C. P., Eggenberger, A. & Wang, K. Flow guiding barrel for improving gene transformation efficiency for biolistic delivery. US Patent Application 2024/0010964 A1 (2024).
Liu, Y. et al. In planta genome editing in commercial wheat varieties. Front. Plant Sci. 12, 648841 (2021).
Brant, E. J., Eid, A., Kannan, B., Baloglu, M. C. & Altpeter, F. The extent of multiallelic, co-editing of LIGULELESS1 in highly polyploid sugarcane tunes leaf inclination angle and enables selection of the ideotype for biomass yield. Plant Biotechnol. J. 22, 2660–2671 (2024).
Lacroix, B. & Citovsky, V. Biolistic approach for transient gene expression studies in plants. Methods Mol. Biol.124, 125-139 (2020).
Hajimorad, M., Domier, L., Tolin, S., Whitham, S. & Saghai Maroof, M. Soybean mosaic virus: a successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 19, 1563–1579 (2018).
Mei, Y., Liu, G., Zhang, C., Hill, J. H. & Whitham, S. A. A sugarcane mosaic virus vector for gene expression in maize. Plant Direct 3, e00158 (2019).
Mei, Y., Zhang, C., Kernodle, B. M., Hill, J. H. & Whitham, S. A. A Foxtail mosaic virus vector for virus-induced gene silencing in maize. Plant Physiol. 171, 760–772 (2016).
Kang, M. et al. An improved Agrobacterium-mediated transformation and genome-editing method for maize inbred B104 using a ternary vector system and immature embryos. Front. Plant Sci. 13, 860971 (2022).
Raji, J. A., Frame, B., Little, D., Santoso, T. J. & Wang, K. Agrobacterium-and biolistic-mediated transformation of maize B104 inbred. Methods Mol. Biol. 1676, 15-40 (2018).
Hamada, H. et al. Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Sci. Rep. 8, 14422 (2018).
Kuwabara, C. et al. A DNA-free and genotype-independent CRISPR/Cas9 system in soybean. Plant Physiol. 196, 2320–2329 (2024).
Hamada, H. et al. An in planta biolistic method for stable wheat transformation. Sci. Rep. 7, 11443 (2017).
Kumagai, Y. et al. Introduction of a second “Green Revolution” mutation into wheat via in planta CRISPR/Cas9 delivery. Plant Physiol. 188, 1838–1842 (2022).
Luo, W., Suzuki, R. & Imai, R. Precise in planta genome editing via homology-directed repair in wheat. Plant Biotechnol. J. 21, 668 (2022).
Wang, W. et al. Expanding the range of editable targets in the wheat genome using the variants of the Cas12a and Cas9 nucleases. Plant Biotechnol. J. 19, 2428–2441 (2021).
Schindele, P. & Puchta, H. Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnol. J. 18, 1118–1120 (2020).
Maliga, P. & Tungsuchat-Huang, T. Plastid transformation in Nicotiana tabacum and Nicotiana sylvestris by biolistic DNA delivery to leaves. Methods Mol Biol. 1132, 147–163 (2014).
Shi, X. et al. Efficient production of antifungal proteins in plants using a new transient expression vector derived from tobacco mosaic virus. Plant Biotechnol. J. 17, 1069–1080 (2019).
Liu, D. et al. Heritable gene editing in tomato through viral delivery of isopentenyl transferase and single-guide RNAs to latent axillary meristematic cells. Proc. Natl Acad. Sci. 121, e2406486121 (2024).
Ellison, E. E. et al. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. plants 6, 620–624 (2020).
Banakar, R. et al. High-frequency random DNA insertions upon co-delivery of CRISPR-Cas9 ribonucleoprotein and selectable marker plasmid in rice. Sci. Rep. 9, 19902 (2019).
Liang, Z. et al. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13, 413–430 (2018).
Zhang, Y., Iaffaldano, B. & Qi, Y. CRISPR ribonucleoprotein-mediated genetic engineering in plants. Plant Commun. 2 (2021).
Zhang, Y. et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617 (2016).
Pirmoradi, F. N. et al. A microarray MEMS device for biolistic delivery of vaccine and drug powders. Hum. Vaccines Immunotherapeutics 11, 1936–1944 (2015).
Luke Mankin, S. & Thompson, W. F. New green fluorescent protein genes for plant transformation: Intron-containing, ER-localized, and soluble-modified. Plant Mol. Biol. Report. 19, 13–26 (2001).
Lee, K., Kang, M., Ji, Q., Grosic, S. & Wang, K. New T-DNA binary vectors with NptII selection and RUBY reporter for efficient maize transformation and targeted mutagenesis. Plant Physiol. 192, 2598–2603 (2023).
McCaw, M. E. et al. Development of a transformable fast-flowering mini-maize as a tool for maize gene editing. Front. Genome Editing 2, 622227 (2021).
Zobrist, J. D. et al. Transformation of teosinte (Zea mays ssp. parviglumis) via biolistic bombardment of seedling-derived callus tissues. Front. Plant Sci. 12, 773419 (2021).
Edwards, K., Johnstone, C. & Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349 (1991).
Hufford, M. B. et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 373, 655–662 (2021).
Liu, Q. et al. Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7 (2019).
You, Q. et al. CRISPRMatch: an automatic calculation and visualization tool for high-throughput CRISPR genome-editing data analysis. Int. J. Biol. Sci. 14, 858 (2018).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Acknowledgements
The authors thank Jacob Zobrist and Minjeong Kang from the K.W. lab for initial help with the biolistic maize B104 transformation using the immature embryo tissue culture protocols, and Kyle Miller, Madison Drent, Riley Behan, and Fei Liu from S.J. lab for their early work on biolistic delivery. We thank Yanhao Cheng in the Y.Q. Lab for providing the ttLbCas12a all-in-one cloning vector for genome editing in wheat. S.J. acknowledges support from the 3 M Non-Tenured Faculty Award, the Digital and Precision Agriculture Award, the Iowa State Startup Fund, the seed grant fund from the Crop Bioengineering Center of Iowa State University, and the Agriculture and Food Research Initiative Grant No. 2019-67013-29016 from the USDA National Institute of Food and Agriculture (NIFA). K.W. acknowledges the support of the Agriculture and Food Research Initiative (Grant No. 2019-67013-29016), Hatch projects #IOW04714 and #IOW04308 from USDA NIFA; K.W.’s contribution to this work is partially supported by (while serving at) the NSF. Y.Q. acknowledges the support of the NSF Plant Genome Research Program (Grant No. IOS-2132693) and the Foundation for Food & Agriculture Research Program on Genotype-Independent Regeneration of Fertile Plants (Grant No. 21010111). C.T. acknowledges the support provided by the NSF DataFEWSion Graduate Traineeship Program (DGE-1828942). The commercialization efforts are supported by the Digital and Precision Agriculture Award from Iowa State University, an NSF I-Corps award (Grant No. 2346805) and DOE STTR awards (Grant No. DE-SC0024762 & DE-SC0025775), all awarded to S.J.
Author information
Authors and Affiliations
Contributions
C.T. conceived the original FGB concept, designed and refined the device, designed and conducted simulation and biolistic delivery experiments for onion, maize, and soybean, analyzed data, and wrote and revised the manuscript. W.L. constructed vectors for wheat, conducted wheat SAM delivery experiment, performed transgenic plants and genome editing analyses, and wrote and revised the manuscript. Q.J. conducted maize transient and stable transformation experiments, analyzed data and helped revise the manuscript. A.E. constructed the vectors for onion, performed genome editing analysis in onion, and helped revise the manuscript. A.C. helped design and conduct SMV-GFP delivery to soybean seedlings and analyzed data. W.X. helped design and conduct SCMV-GFP delivery to corn seedlings and analyzed data. R.S. helped conduct experiments and analyzed data. K.L. constructed vectors for maize, performed transgenic plants and transgene copy number analyses, and helped revise the manuscript. S.W. designed viral delivery experiments for soybean and maize, analyzed data and helped revise the manuscript. Y.Q. designed experiments for wheat, analyzed data and wrote and revised the manuscript. K.W. conceived the concept, designed experiments for onion and maize, analyzed data, and wrote and revised the manuscript. S.J. conceived the original FGB concept, coordinated and oversaw the project, designed biolistic experiments and simulations, analyzed data, and wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.T., K.L., K.W., and S.J. are affiliated with Hermes Biomaterials Inc., which has licensed the technology described in this paper. The method for utilizing the flow guiding barrel for biolistic delivery is currently under patent review (co-authors of this paper S.J., C.T., A.E., and K.W. as well as Kyle Miller of Iowa State University are inventors of the filed patent; Application #18348689). These interests have been reviewed and managed by Iowa State University in compliance with its Conflict of Interest policies. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Guoquan Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Thorpe, C., Luo, W., Ji, Q. et al. Enhancing biolistic plant transformation and genome editing with a flow guiding barrel. Nat Commun 16, 5624 (2025). https://doi.org/10.1038/s41467-025-60761-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60761-x