Abstract
Chronic pancreatitis (CP) is characterized by irreversible fibrotic destruction and impaired pancreatic function. CP disrupts lipid metabolism and causes the imbalance of gut microbiota which in turn exacerbates pancreatic fibrosis. Statins alter gut microbiota and exert anti-inflammatory effects, but its role in CP has not been fully elucidated. Here, we found that statins-associated higher abundance of Lactobacillus intestinalis (L.intestinalis) maintained gut homeostasis that restrained bacteria translocation from gut to the pancreas, which eventually aggravated pancreatic fibrosis through inhibiting CD8+T cells-dependent immunity. Fecal microbiota transplantation (FMT) or L.intestinalis administration inhibited the infiltration of CD8+T cells and macrophages that delayed CP progression. L.intestinalis restrained the recruitment of M1 macrophages and limited the release of Ccl2/7 in the colon, which prevented epithelial damage and epithelial barrier dysfunction through blocking Ccl2/7-Ccr1 signaling. Our findings elucidate that the utilization of statin therapy or supplementation of L.intestinalis can be potential approach for the therapies of CP.
Similar content being viewed by others
Introduction
Chronic pancreatitis (CP) refers to a progressive inflammatory disease of the pancreas caused by many factors, including genetics, environment and alcohol consumption. The pathology of CP is characterized by pancreatic atrophy, damage to acinar cells and irreversible fibrosis that lead to permanent pancreatic endocrine and exocrine insufficiency1,2,3. The immune subtypes including macrophages, dendritic cells and CD8+T cells are recruited in fibrotic zones when CP occurs4. Immune cells release inflammatory mediators which can activate pancreatic stellate cells (PSCs) or trigger pro-inflammatory signals that aggravate pancreatic damage and fibrosis progression5,6.
The gut-pancreas axis is considered as crossroads of pancreatic diseases7,8,9. CP causes gut microbiota dysbiosis which impairs pancreatic enzyme secretion and pancreatic endocrine insufficiency10,11,12. Fecal microbiota transplantation (FMT) reconfigures gut microbiota that reprograms immune microenvironment in the gastrointestinal tract and distant organs13,14. Our study has reported that FMT alleviates pancreatic fibrosis through limiting macrophages and CD4+T cell infiltration15. Understanding the mechanism of intestinal microorganisms influencing CP progression and the optional procedures to restore gut microbiota balance will be beneficial for CP treatment.
CP is accompanied by lipid disorders, while oxidative stress molecules produced by abnormal lipid accumulation also accelerate pancreatic injury16. Statins are competitive inhibitors of β-Hydroxy β-methylglutaryl-CoA (HMG-CoA) reductase, rate-limiting enzyme of cholesterol synthesis, which can interfere lipid and fatty acid metabolism17. Statins can suppress collagen expression and expedite fibroblast apoptosis that restrain the progression of fibrotic diseases18,19. The impacts of statin therapy-associated gut microbiota on host metabolism have been described, while its role on suppressing pancreatic fibrosis has not been explored20,21.
In current study, we observed that CP caused abnormal lipid metabolism and disrupted gut microbiota that accelerated pancreatic fibrosis. Targeting lipid metabolism using statin therapy maintained gut homeostasis and intercepted pathogenic bacteria migration from the gut to the pancreas, which attenuated pancreatic fibrosis through CD8+T cells-dependent manner. Manipulation of statin therapy associated Lactobacillus intestinalis (L.intestinalis) using FMT or oral supplementation alleviated epithelium injury and suppressed bacteria migration through influencing macrophage-epithelium cells interactions. Our findings establish the causal evidence of statin therapy-related gut microbiota achieving local and distal immune modulation which suppresses pancreatic fibrosis through gut-pancreas communication. This opens the gate to manage gut microbiota for CP interventions.
Methods
Ethics statement
For human studies, written informed consent was obtained from all subjects before the study protocol and this study was approved by the ethics committee of the First Affiliated Hospital of Harbin Medical University (2024IIT092). Written informed consent was also obtained from all participants before the study protocol began. All animal experimental procedures were carried out according to protocols approved by the Institutional Animal Ethics Committee of the First Hospital of Harbin Medical University (IACUC No. 2022148). Animal experiments were performed in accordance with the Guide for the Care and Use of Medical Laboratory Animals.
Human study design
All patients and healthy volunteers were recruited from the First Affiliated Hospital of Harbin Medical University (Harbin, China). Human pancreatic tissues were collected from patients underwent surgery including eight patients diagnosed with CP and eight patients diagnosed with benign pancreatic disease. For FMT studies, fecal samples were collected from three healthy volunteers, three patients who were administrated with atorvastatin for over three months and another six volunteers without following conditions: chronic metabolic, cardiovascular or gastrointestinal diseases, pregnancy, and medical treatment that may influence gut microbiome. The healthy subjects and patients who had received any antibiotics or probiotics four weeks prior to sample collection were also excluded. The plasma and stool were collected from extra 20 healthy volunteers, and the total cholesterol (TC) levels were detected, and gut microbiota was sequenced (Supplementary Table 1).
Animal experiments and ethical statement
C57BL/6N mice (6–8 weeks old, 20 ± 2 g body weight) were purchased from Vital River Laboratory Animal Technology Co., Ltd (Beijing, China), and homozygous CD8a-deficient mice (referred to as CD8-KO) were purchased from Saiye Biotechnology Co., Ltd. (Suzhou, China). All animals were raised in a specific pathogen-free environment with controlled light conditions (12-h light/dark cycle). Age- and sex-matched mice and littermate controls were used at 8–10 weeks of age, and were randomly assigned into different treatment groups. For the CD8-KO experiment, four males and two females were used in each group, while all subsequent experiments utilized only male mice.
For the CP model, caerulein (Sigma-Aldrich, C9026) was given six intraperitoneal injections (50 μg/kg body weight) every other day at 1-h intervals (3 days a week). Mice were treated three times a week for 3 weeks (at least 2 days separating each dose) and sacrificed the next day after the last injection22. The blood, stool, pancreas, and colon tissues were collected.
For atorvastatin treatment, atorvastatin (MCE, Shanghai, China) or atorvastatin calcium tablet suspension (Pfizer, Shanghai, China) was given by oral gavage (10 mg/kg body weight) from two days after the first dose of caerulein injection. Mice were treated three times a week for 3 weeks.
Fecal microbiota transplantation
Mice were given an Abx cocktail composed of streptomycin (5 mg/ml), clindamycin (0.1 mg/ml), and amoxicillin (0.5 mg/ml) in sterile drinking water for 1 week23. For FMT experiments, fecal samples were collected from volunteers or patients and processed immediately after fresh harvest. In brief, an appropriate amount of fresh stool was dissolved in sterile PBS (0.1 g/ml) and the supernatant was filtered and stored at −80 °C for further use. FMT was administrated with fecal solution (200 μl per dose, three times a week) for 2 weeks by oral gavage after Abx treatment. Then, CP model was built and FMT was performed twice a week for the following 3 weeks.
Microbial strain
L.intestinalis (B293738, same as ATCC 49335) was purchased from Mingzhou Biological Technology (Ningbo, China) and cultured under microaerobic conditions on MRS Broth at 37 °C. Mice were given an Abx cocktail for 1 week as described previously before oral supplementation of L.intestinalis. (1 × 109 colony forming unit (CFU)/ml) was given for 2 weeks (200 μl per dose, three times a week) by oral gavage. Then, CP model was built and L.intestinalis was given twice a week for the following 3 weeks.
Tissue RNA extraction and qRT-PCR
Total RNA was extracted using RNA Mini Kit (Qiagen, 74134), according to the manufacturer’s instructions, and then reverse transcribed using PrimeScript RT Master Mix (Takara, RR036A). Quantitative Real-Time PCR (qRT-PCR) was performed with SYBR Green Real-time PCR Master Mix (Takara, RR820A) and quantified by ABI PRISM 7500 Sequence Detection System (Applied Biosystems). The sequences of oligonucleotide primers were designed according to the cDNA sequences in the GenBank database using Primer Express software (Applied Biosystems). Gapdh was used as the reference gene and the sequences of primers are listed in Supplementary Table 2.
Western blot
Protocol for protein extraction was used as supplied by MinuteTM Total Protein Extraction Kit for Animal Cultured Cells and Tissues (SD-001, Invent Biotechnologies Inc., USA). Western blot was performed as described previously24. Briefly, extracted pancreatic tissue proteins were mixed with SDS loading buffer and subjected to loading, electrophoresis, and membrane transfer, followed by blocking and immunostaining with primary antibodies. After the incubation with secondary antibodies, the expressions of proteins were analyzed with a molecular imager system (BIO-RAD. Hercules, USA) and an enhanced chemiluminescence method (Thermo Fisher Science, Massachusetts, USA) to visualize the bands. Protein levels were quantified using Image Studio Lite (LI-COR Biosciences).
Histopathology processing immunohistochemistry and immunofluorescence
Pancreatic and colon tissues were fixed in 4% paraformaldehyde for 24 h, paraffin-embedded and sectioned for hematoxylin and eosin (H&E) staining, Trichrome staining, IHC, and IF. The severity of pancreatitis was analyzed using histopathologic scoring and fibrosis area using ImageJ (version 1.4.3). For IHC and IF, paraffin sections were dewaxed to water, microwave heated for antigen repair, incubated overnight at 4 °C with primary antibody, then incubated with biotin-labeled secondary antibody or IF secondary antibodies. The antibodies of anti-CD8α (Cell Signaling Technology, 98941), anti-Ly6G (Cell Signaling Technology, 87048), anti-F4/80 (Cell Signaling Technology, 70076), anti-COL1A1 (Cell Signaling Technology, 72026), anti-α-SMA (Cell Signaling Technology, 19245) and goat anti-Rabbit IgG (Vector, MP-7451) were used for IHC, and anti-ZO-1 (ProteinTech, 21773-1-AP), anti-Occludin (ProteinTech, 27260-1-AP), anti-Claudin-1 (ProteinTech, 28674-1-AP), FITC-conjugated goat anti-rabbit IgG (Servicebio, GB22303) and DAPI (Servicebio, G1012) were used for IF. The number of positive cells or the area of positive cells was quantified using ImageJ.
Fluorescence in situ hybridization (FISH)
FISH was conducted according to the manufacturer’s instructions using BersinBio™ Fluorescent in situ hybridization Kit (BersinBio, Guangzhou, China). Briefly, paraffin sections were baked at 56 °C for 30 min, dewaxed to water in xylene and ethanol, and then incubated in a drop of pre-hybridization solution at 37 °C for 30 min. The hybridization mixture that contained the probe (the primers sequence of 16s RNA are listed in Supplementary Table 2) was dropped onto the slices and incubated overnight in the dark. On the next day, DAPI was incubated, and image was taken using a confocal fluorescence microscope (20×, Olympus, Japan). The quantification wasper formed using ImageJ.
16S rRNA sequencing and analyses
Total bacterial DNA was extracted from samples using the Power Soil DNA Isolation Kit (MO BIO Laboratories). DNA quality and quantity were assessed by the ratios of 260 nm/280 nm and 260 nm/230 nm. The V3-V4 region of the bacterial 16S rRNA gene was amplified with the common primer pair (Forward primer, 5’-ACTCCTACGGGAGGCAGCA-3’, reverse primer, 5’-GGACTACHVGGGTWTCTAAT-3’) combined with adapter sequences and barcode sequences. PCR products were purified with VAHTSTM DNA Clean Beads. All PCR products were quantified using Quant-iTTM dsDNA HS Reagent and pooled. High-throughput sequencing analysis of bacterial rRNA genes was performed on the purified, pooled sample using the Illumina Hiseq 2500 platform (2 × 250 paired ends) provided by Biotree Co. Ltd. (Shanghai, China). Alpha diversity was estimated by calculating the Chao1 richness, observed OTU, ACE, Shannon-Wiener diversity index, and Simpson diversity index from different perspectives, which focus on the microbial community abundance and evenness. Beta diversity evaluates divergence among microbial communities based on measuring the distance matrix of dissimilarity. In this study, principal components analysis was performed to assess variance among different communities. Analysis of similarities (ANOSIM) test was performed to calculate the significance of dissimilarity. The predominance of bacterial communities between groups was analyzed using the linear discriminant analysis (LDA) effect size (LEfSe) (LDA score (log10) = 3 as cutoff value). Bray-Curtis dissimilarity matrices were calculated and used for ordination by PCoA. These matrices were also used to assess differences in β-diversity by ANOSIM. PCoA was carried out for all dimension reduction analyses using the vegan package in R software.
Targeted lipidomic analysis
LC-MS/MS analyses were performed using a UHPLC system (1290, Agilent Technologies) with a Phenomen Kinetex 1.7 u C18 100A Column (100 × 2.1 mm) coupled to Triple TOF 6600 (Q-TOF, AB Sciex). The Triple TOF mass spectrometer was used for its ability to acquire MS/MS spectra on an information-dependent basis (IDA) during an LC/MS experiment. In this mode, the acquisition software (Analyst TF 1.7, AB Sciex) continuously evaluates the full scan survey MS data as it collects and triggers the acquisition of MS/MS spectra depending on preselected criteria. ESI source conditions were set as following: Ion source gas 1 as 60, Ion source gas 2 as 60, Curtain gas as 30, source temperature 550 °C, Ion Spray Voltage Floating 5500 V or −4500 V in positive or negative modes, respectively. For data preprocessing and annotation: An in-house program, namely, LipidAnalyzer, was developed using R for automatic data analysis. The raw data files (.wiff format) were converted to files in mzXML format using the “msconvert” program from ProteoWizard (version 3.0.6150). Then, the mzxML files were loaded into LipidAnalyzer for data processing. Peak detection was first applied to the MS1 data. The CentWave algorithm in XCMS was used for peak detection, With the MS/MS spectrum, lipid identification was achieved through a spectral match using an in-house MS/MS spectral library. the absolute quantitation of lipids can be achieved using the peak area, SIL-IS, and RF information.
RNA sequencing analysis
In our study, pancreas and colon tissues from mice were harvested after inducing CP model. RNAs were isolated using RNeasy kit (Qiagen), and total amounts and integrity of RNA were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The gene expressions of each sample were investigated using the Illumina NovaSeq 6000 according to the manufacturer’s guide (Illumina, San Diego, CA, USA).
Tissue dissociation, cell purification, and single-cell RNA-seq analysis
Colon tissues were washed with 1× DPBS (Thermo Fisher, 14190144) on ice and the digestion buffer (PBS with 5% FBS) which included 0.25% Trypsin (Thermo Fisher, 25200-072) and 10 μg/ml DNase I (Sigma-Aldrich, 11284932001) was added. Colon tissues were dissociated at 37 °C for 40 min with a shaking speed of 50 r.p.m. Cell suspensions were filtered using a 40 μm nylon cell strainer and red blood cells were removed by 1× Red Blood Cell Lysis Solution (Thermo Fisher, 00-4333-57). Dissociated cells were washed with 1х DPBS containing 2% FBS. Cells were stained with 0.4% Trypan blue (Thermo Fisher, 14190144) to check the viability of Countess® II Automated Cell Counter (Thermo Fisher). Next, single-cell suspensions were selected using CD45 microbeads and magnetic bead column separation (Miltenyi Biotec).
For 10× library preparation and sequencing, briefly, all the remaining procedures including the library construction were performed according to the standard manufacturer’s protocol (CG000206 Rev D). Sequencing libraries were quantified using a High Sensitivity DNA Chip (Agilent) on a Bioanalyzer 2100 and the Qubit High Sensitivity DNA Assay (Thermo Fisher). The libraries were sequenced on NovaSeq6000 (Illumina) using 2 × 150 chemistry.
For single-cell RNA-seq data processing, reads were processed using the Cell Ranger 2.1.0 pipeline with default and recommended parameters. FASTQs generated from Illumina sequencing output were aligned to the mouse genome, version GRCm38, using the STAR algorithm. Next, Gene-Barcode matrices were generated for each individual sample by counting UMIs and filtering non-cell associated barcodes. Finally, we generate a gene-barcode matrix containing the barcoded cells and gene expression counts. This output was then imported into the Seurat (v2.3.0) R toolkit for quality control and downstream analysis of our single-cell RNA-seq data. The expression of mitochondria genes was calculated using the Percentage Feature Set function of the Seurat package. To remove low-activity cells, cells with more than 10 percent expression of mitochondria genes were excluded. The normalized data (NormalizeData function in Seurat package) was performed for extracting a subset of variable genes. Variable genes were identified while controlling for the strong relationship between variability and average expression. Next, we integrated data from different samples after identifying “anchors” between datasets using FindIntegrationAnchors and IntegrateData in the Seurat package. The clusters were visualized by Uniform Manifold Approximation and Projection (UMAP).
Differentially expressed genes (DEGs) analysis
To identify differentially expressed genes (DEGs), we used the Seurat FindMarkers function based on Wilcoxon rank sum test with default parameters, and selected the genes expressed in more than 10% of the cells in both groups of cells and with an average log (Fold Change) value greater than 0.25 as DEGs. The p value less than 0.05 was used as the criterion to evaluate the statistical significance.
Pathway enrichment analysis
To investigate the potential functions of immune-relevant genes, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were used with the “clusterProfiler” R package v 3.16.1. Pathways with p value less than 0.05 were considered as significantly enriched. Selected significant pathways were plotted as bubble plots. Gene Set Enrichment Analysis (GSEA) was conducted using clusterProfiler package (version 3.5). The fold change of gene expression between two groups was calculated, and the gene list was generated according to the change of |log2FC|.
Establishment and treatment of mouse colon organoids
The protocol of isolating crypt was based on previous reports and modified25,26. Briefly, the entire colon from the cecum to the anus were completely removed. Colon was longitudinally incised, and feces and mucus were gently scraped off by using a microscope slide. Colon was cut into 2 mm pieces, followed by extensive washes with PBS to remove contaminants. Crypts were released from colon pieces by incubation for 30–60 min at 4 °C in PBS containing 10 mM EDTA (Solarbio, E1170) and antibiotics. Crypts were then filtered through a 100 μm strainer to remove villus material and tissue fragments. Isolated crypts were counted and every 300–500 crypts were mixed with 50 μl of Matrigel (VivoMatter Biotech Co. Ltd, VM002-PRF-10) and plated in 24-well plates. Crypts were cultured in Mouse Colon Organoid Culture Medium (Shanghai JFKR Organoid Biotechnology Co. Ltd, JFKR-MNC-100) at 37 °C in humidified 5% CO2 atmosphere. Culture medium was replaced every 2–3 days. The crypts are cultured for 7–14 days to form colon organoids with a diameter of 300–500 μm.
L. intestinalis was cultured in MRS medium overnight and the supernatant was spun for filtered through a 0.22 μm filter to remove the bacteria body. Colon organoids were stimulated by LPS (200 ng/ml, MCE, HY-D1056), in combination with 5% L.intestinalis supernatant or 5% MRS medium. After 24 h treatment, the organoids were harvested for total RNA extraction and qRT-PCR.
Flow cytometry
Fresh pancreatic tissue was cut up with scissors, incubated for 10 min at 37 °C in digestion buffer (1 mg/ml collagenase P dissolved in HBSS), terminated, and filtered through a 70 μm filter to obtain a single cell suspension. Samples were incubated with LIVE/DEAD fixable Violet dead cell stain kit (Invitrogen, L34955) for 15 min to stain dead cells. Next, single cells were incubated with primary antibodies including anti-CD45 (Biolegend, 103132), anti-CD3 (Biolegend, 100234), anti-CD8a (Biolegend, 100714), anti-CD4 (Biolegend, 100555), anti-CD11b (Biolegend, 101217), anti-Ly6G (Biolegend, 127607), anti-F4/80 (Biolegend, 123141) and anti-CD86 (Biolegend, 105029) for 30 min at 4 °C. For intracellular staining such as anti-CD206 (Biolegend, 141719), cells were fixed and permeabilized using fixative solution (Invitrogen, 00-8222-49) and Permeabilization Buffer (Invitrogen, 00-8333-56). Cells were analyzed by flow cytometry (BD FACS Calibur Flow Cytometer) with FACS Diva software. Data were analyzed using FlowJo (Flow Jo V10.8.1, LLC, Ashland, OR, USA).
Statistical and correlation analysis
All error bars represent the standard error of the mean (SEM). Statistical significance was assigned at P values of <0.05 and detected by GraphPad Prism 8.0 (GraphPad Software, San Diego, USA). The Shapiro–Wilk test was used to determine the sample distribution type. A two-tailed Student’s t-test was used to evaluate statistical significance between two groups for normal distribution. For the nonparametric tests, the two-tailed Mann-Whitney test was used to evaluate statistical significance between two groups. For more than two groups, the significance was calculated by the Kruskal–Wallis test or ordinary one-way ANOVA depending on the sample distribution type; a post hoc Tukey test was used to conduct multiple comparisons. Correlations between enriched species and circulating metabolites were tested with Spearman’s correlation. P values of <0.05 were considered statistically significant. *P < 0.05, **P < 0.01 and ***P < 0.001. The linear correlation between differentially enriched bacteria species and metabolites was calculated by Pearson correlation. Heatmaps were hierarchically clustered to represent the species-metabolite-associated patterns based on the correlation distance. The analyses and visualizations were implemented in python (v2.7.9) with the numpy (v1.9.2), scipy (v0.15.1) and matplotlib (v1.4.3) packages.
Results
Statins alter disorders of lipid metabolism and alleviate CP
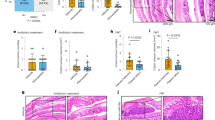
CP disrupted host lipid homeostasis27, we performed lipid profiles test in mice to investigate the alterations of plasma lipids in CP. We observed that the plasma levels of total cholesterol (TC) and low-density lipoprotein cholesterol (LDL-C) in the CP group were significantly elevated compared to control group, while the level of high-density lipoprotein cholesterol (HDL-C) was decreased. (Fig. 1a). Dietary intakes are key determinants of lipid metabolism28, we then investigated whether disruption of lipid metabolism using high-fat diet (HFD) can affect pancreatic fibrosis (Fig. 1b). We found that HFD aggravated pancreatic fibrosis and local inflammation compared to standard diet (SD) in CP model (Fig. 1c, d, Supplementary Fig. 1a). Statins have been demonstrated to possess anti-inflammatory properties in addition to their lipid-lowering effects29, we then explored whether atorvastatin (ATOR) therapy can alleviate pancreatic fibrosis (Fig. 1e). We found that the expression of α-smooth muscle actin (α-SMA) was increased in CP tissues, while reduced by ATOR treatment (Fig. 1f, g). In parallel, ATOR decreased the interstitial fibrosis and inflammation scores, showing a lesser number of infiltrated inflammatory cells and reduced area of acinar atrophy and interstitial fibrosis in CP mice (Fig. 1h, i, Supplementary Fig. 1b–e). Consistent with these, ATOR reduced the levels of Fibronectin and α-SMA in CP tissues (Fig. 1j). Notably, we found that ATOR can inhibit neutrophil infiltration in pancreatic tissue during the early stage of pancreatic inflammation (24 h after cerulein induction), thereby reducing pancreatic necrosis and the severity of inflammation (Supplementary Fig. 2a–h). These suggest that alteration of abnormal lipid metabolism through statin therapy can alleviate pancreatic fibrosis.
a Comparison of lipid levels in the plasma between CP and control mice (n = 6 per group). b Schematic illustration of experimental mouse models induced by high-fat diet (HFD), followed by caerulein injection. c Representative images of H&E staining, Trichrome staining, and IHC for Collagen-1 in SD-NC, SD-CP, HFD-NC, and HFD-CP groups. d Pathology scores, quantification of fibrotic area and Collagen-1 positive area in SD-NC, SD-CP, HFD-NC, and HFD-CP groups (n = 8 per group). e Schematic illustration of CP mouse models, including PBS and atorvastatin (ATOR) treatment groups. f Western blot showing the expression of α-SMA in pancreatic tissues from NC, CP, and CP + ATOR mice, GAPDH was used as an endogenous control. g The quantification of α-SMA expression in pancreatic tissues (n = 3 per group). h Representative images of H&E staining, Trichrome staining, and IHC of α-SMA in NC, CP, and CP + ATOR groups. i Pathology scores, quantification of fibrotic area, and α-SMA positive area in each group (n = 7 per group). j qRT-PCR detecting the expressions of α-SMA and fibronectin in CP tissues (n = 5 per group), Gapdh was used as an endogenous control. Data are presented as mean ± SEM. ns, no significance; *p < 0.05; **p < 0.01; ***p < 0.001. Figure (b, e) is from Figdraw 2.0 of HOME for Researchers (Permissions for use have already been obtained).
Statin therapy alleviates pancreatic fibrosis through CD8+T-dependent immunity
Immune cells and secreted inflammatory factors play crucial roles in accelerating pancreatic fibrosis. To dissect the immunoregulatory mechanism of statin therapy alleviating pancreatic fibrosis, differential genes in the pancreas between CP and control group were compared (Fig. 2a). GO analysis revealed that the inflammatory-related processes, such as leukocyte migration involved in inflammatory response, positive regulation of T cell proliferation activation were activated by upregulated genes in CP group (Fig. 2b). Genes which were involved in those processes included S100a8, S100a9, Ccl5, Cd8a and Ccl7 (Fig. 2c). CP causes local and systemic immune activations, we then investigated the changes of infiltrated immune cells in CP tissues which could reflect the anti-fibrotic effects of statin therapy. We found that ATOR restrained the infiltration of CD8+T cells, macrophages in CP tissues, but had no effect on neutrophils (Fig. 2d, e). Our findings also revealed that HFD exacerbated pancreatic fibrosis through increasing the infiltration of CD8+T cells and macrophages (Fig. 2f, g). The roles of CD8+T cells in CP development have been documented30. To dissect whether ATOR limited CP progression by affecting CD8+T cells dependent immune phenotypes, we explored the anti-fibrotic effects of statin therapy in CD8 knockout mice vs wild type (WT) mice (Fig. 2h). We observed that pancreatic inflammation and interstitial fibrosis were significantly downregulated and the number of infiltrated macrophages was decreased in CD8 knockout mice, while ATOR treatment lost the anti-fibrotic effect in the CD8-deficient mice (Fig. 2i, j, Supplementary Fig. 3a, b). These suggest that management of lipid metabolism by statin therapy reduces pancreatic fibrosis through CD8+T cells specific manner.
a Volcano plots presenting the differential genes in pancreatic tissues between NC and CP mice (log2FC ≥ 1.5, p < 0.05). b GO analysis indicating upregulated pathways including positive regulation of T cell proliferation activation and lymphocyte proliferation in CP group compared to NC group. c GSEA analysis enriching upregulated expression of inflammation-related factors by different genes between CP and NC mice. d Representative images of CD8, F4/80, and Ly6G positive cells using IHC in NC, CP, and CP + ATOR groups. e The quantification of CD8, F4/80 and Ly6G positive cells in each group (n = 7 per group). f Representative images of CD8, F4/80, and Ly6G positive cells in CP mice. g The comparison of CD8, F4/80, and Ly6G positive cells between SD and HFD groups in CP mice (n = 8 per group). h Schematic illustration of CP mouse (CD8-KO) models, including PBS and ATOR treatment groups. i Representative images of H&E staining, Trichrome staining, and F4/80 positive cells by IHC in CP (WT mice), CP (CD8-KO mice), and CP (CD8-KO mice)+ATOR groups. j Pathology scores, quantification of fibrotic area and F4/80 positive cells between groups (n = 6 per group). Data are presented as mean ± SEM. ns, no significance; ns, no significance; *p < 0.05; **p < 0.01; ***p < 0.001. h is from Figdraw 2.0 of HOME for Researchers (Permissions for use have already been obtained).
Statin therapy suppresses pancreatic fibrosis through altering gut microbiota and limiting its migration
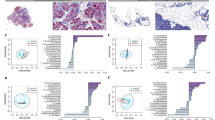
CP caused the dysbiosis of gut microbiota, to investigate whether ATOR suppresses pancreatic fibrosis through influencing gut microbiota, the gut microbial profiles of mice were assessed using 16S rRNA sequencing. We found decreased α-diversity of gut microbiota in CP mice compared with the control mice, while it was increased after ATOR intervention (Fig. 3a). β-diversity was shown by principal coordinates analysis (PCoA) indicating that the microbial communities were significantly changed after ATOR intervention (Fig. 3b). We then compared different taxa of gut microbiota in three groups and found that the abundances of Firmicutes and Bacteroidetes were decreased in CP mice, while they were increased by ATOR treatment. The abundance of Verrucomicrobiota was upregulated in CP mice and was decreased by ATOR treatment (Supplementary Fig. 4a, b). At the genus level, the increased abundances of Streptococcus and Akkermansia were found in CP mice, while both genus were decreased after ATOR intervention (Fig. 3c, d). In addition, Lactobacillus, Lachnospiraceae NK4A136 group, Alistipes, and Bacillus were downregulated in CP group, while upregulated by ATOR intervention (Fig. 3c, d, Supplementary Fig. 4c-e). To further understand the mechanism of statins-altered gut microbiota delaying pancreatic fibrosis, differential genes between CP in combination with ATOR vs CP alone were identified (Supplementary Fig. 4f). GO analysis indicated that response to bacterium was upregulated after ATOR intervention (Fig. 3e). Imbalance of gut microbiota and local inflammation cause bacteria migration from the gut to the distal organs31. We found increased bacteria signals in CP tissues compared to normal pancreas, while the bacteria loads were significantly reduced after ATOR intervention (Fig. 3f, g). To further explore whether the similar phenotype can be found in CP patients, eight CP tissues and eight normal pancreatic tissues were collected, and the microorganisms were profiled. The increased number of OTU and α-diversity of tissue microbiome in CP tissues were observed compared with normal pancreas (Fig. 3h). These suggest that bacteria are translocated from the gut to the pancreas when CP occurs. ATOR affects gut microbiota composition and harnesses gut bacteria migration which may limit CP development.
a The Chao1 and Shannon analyses determined the α-diversity of gut microbiota in NC, CP, and CP + ATOR groups (n = 7 per group). b PCoA analysis indicating the β-diversity of gut microbiota (n = 7 per group). c Heatmap visualizing differential abundance of bacterial taxa at the genus level (n = 7 per group). d The comparison of relative abundances of Streptococcus, Akkermansia, and Lactobacillus in fecal samples (n = 7 per group). e GO analysis indicating the biological processes enriched by upregulated genes between CP + ATOR and CP mice. f Representative images of 16S rRNA staining by FISH. g The comparable number of 16S rRNA positive area between groups (n = 7 per group). h Venn map reveals the disparities in species numbers and the Chao1 analyses comparing the α-diversity of microorganisms in pancreatic tissues between NP and CP groups (n = 8 per group). i, j GO analysis mapping activated pathways including focal adhesion and ECM receptor interaction between CP and NC mice. k Heatmap visualizing differential genes in colon tissues (n = 3 per group). l Representative images for ZO-1, Occludin, and Claudin-1 by IF. m The quantification of ZO-1, Occludin, and Claudin-1 positive area in the colon (n = 7 per group). Data are presented as mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001.
To explore whether the migration of gut bacteria is caused by injured gut barrier function, the differentially expressed genes between CP vs control mice in the colon tissues were compared (Supplementary Fig. 4g). GO analysis indicated that Focal adhesion (Fig. 3i) and ECM receptor interaction (Fig. 3j) functional panels were downregulated in CP mice. Genes which were contributed to the above two processes were visualized and we found that Ccl2, Ccl7, Ccl12, Ccl17, and other pro-inflammatory factors such as Il17d, Mapk3 and Mapk13 in CP mice were increased. In addition, expressions of genes (Tjp1, Ocln, Cldn2, Egfr) that contribute to intestinal wall integrity were decreased in CP mice (Fig. 3k). Next, the expressions of zonula occludens-1 (ZO-1), Occludin (Ocln) and Claudin-1 (Cldn1) that can be used to assess gut permeability were detected. We found that their expressions were downregulated in CP group compared to control group, while increased by ATOR intervention (Fig. 3l, m). These suggest that CP impairs gut barrier function and promotes pathogenic bacteria migration that promotes pancreatic fibrosis. ATOR remodels gut microbial composition and improves gut barrier function which restrains bacteria translocation and CP development.
Statin therapy-associated gut microbiota in human alleviates pancreatic fibrosis
To investigate whether the alteration of gut microbiota by statin therapy in human exhibits anti-fibrotic effects, fecal samples from three healthy volunteers and three patients who have been administrated with atorvastatin over three months were collected, and the effects of FMT from human into mice on pancreatic fibrosis were explored (Fig. 4a). Our results showed that FMT from patients who have received long-term statin therapy (FMT-ATOR) suppressed pancreatic fibrosis compared with FMT from healthy volunteers (FMT-HC) (Fig. 4b, c, Supplementary Fig. 5a, b). FMT reshapes the gut microbiota community that affects host immunity13,14,32. The immune profiles in CP tissues were then compared between FMT-HC and FMT-ATOR groups, and we found that FMT-ATOR decreased the number of infiltrated CD8+T cells, macrophages, and neutrophils (Fig. 4d, e). In parallel, decreased proportion of CD8+T cells, CD4+T cells, macrophages and neutrophils were observed in FMT-ATOR group (Fig. 4f, g, Supplementary Fig. 5c). We then tested the levels of ZO-1, Occludin, and Claudin-1 in the colon tissues and found that their expressions were upregulated in FMT-ATOR group compared to FMT-HC group (Fig. 4h, i). In addition, FMT-ATOR reduced the expressions of inflammatory factors, such as Ccl2 and Ccl7 in the colon tissue compared to FMT-HC (Fig. 4j).
a Schematic illustration of CP mouse models induced by caerulein injection, including PBS, Abx treatment, and FMT groups. b Representative images of H&E staining, Trichrome staining, and IHC for Collagen-1 in FMT-HC and FMT-ATOR groups. c Pathology scores, quantification of fibrotic area and Collagen-1 positive area between FMT-HC and FMT-ATOR groups (n = 8 per group). d Representative image of CD8, F4/80 and Ly6G positive cells. e The quantification of CD8, F4/80 and Ly6G positive cells by IHC (n = 8 per group). f, g The proportion of CD8+T cells (CD8+CD3+CD45+), CD4+T cells (CD4+CD3+CD45+), macrophages (F4/80+CD11b+CD45+), and neutrophils (Ly6G+CD11b+CD45+) was compared by flow cytometry (n = 7 per group). h Representative IF staining for ZO-1, Occludin and Claudin-1 in the colon. i The quantification of ZO-1, Occludin and Claudin-1 positive area in FMT-HC and FMT-ATOR groups (n = 8 per group). j qRT-PCR testing the expressions of Ccl2 and Ccl7 in colon tissues between FMT-HC and FMT-ATOR groups (n = 6 per group), Gapdh was used as an endogenous control. k The linear discriminant analysis effect size (LEfSe) showing different bacterial taxa between FMT-HC and FMT-ATOR groups. Data are presented as mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001. a is from Figdraw 2.0 of HOME for Researchers (Permissions for use have already been obtained).
To investigate the changes of gut microbiota manipulated by statin therapy that may affect pancreatic fibrosis, the fecal samples of donors and receipt mice were sequenced. We found an increased abundance of Bacteroides coprocola, Ruminococcus callidus, and L.intestinalis in ATOR group compared to HC group (Supplementary Fig. 5d). In the FMT mouse model, FMT-ATOR increased α-diversity of gut microbiota compared to FMT-HC (Supplementary Fig. 5e). Further analysis indicated that Firmicutes and Bacteroidetes were the dominant taxa at the phylum level, and the abundance of Actinobacteriota was increased in FMT-ATOR group, while the abundance of Proteobacteria was decreased (Supplementary Fig. 5f). LEfSe showed that Bifidobacterium pseudolongum, Butyricimonas virosa, Alistipes finegoldii, and L.intestinalis were upregulated in FMT-ATOR group (Fig. 4k). Furthermore, we found that the levels of plasma TC and LDL-C in FMT-ATOR group were decreased compared with FMT-HC group (Supplementary Fig. 5g). Taken together, the alteration of gut microbiota by statin therapy suppresses pancreatic fibrosis through limiting the infiltration of immune cells.
Oral supplementation of L. intestinalis relieves pancreatic fibrosis
To investigate the bacterial species that may contribute to anti-fibrotic role after statin therapy, the intersection between upregulated species by CP + ATOR vs CP groups, ATOR vs HC groups, as well as FMT-ATOR vs FMT-HC groups, were visualized (Fig. 5a). L.intestinalis was found as a beneficial bacterium that may attenuate pancreatic fibrosis (Supplementary Fig. 6a). Further study indicated that oral gavage of L.intestinalis (Fig. 5b) suppressed pancreatic fibrosis (Fig. 5c, d, Supplementary Fig. 6b). Simultaneously, supplementation of L.intestinalis inhibited the infiltration of CD8+T cells and macrophages (Fig. 5e–h, Supplementary Fig. 6c). We then explored whether downregulated genes in the colon tissues between ATOR vs control groups were changed by L.intestinalis supplementation and found that Ccl2, Ccl7, Ccl12, and Ccl17 were decreased between L.intestinalis vs PBS (Fig. 5i). Next, markers that can reflect gut permeability were assessed in the colon tissues, and the protein levels of ZO-1, Occludin, and Claudin-1 and the gene expressions of Tjp1 and Ocln were upregulated after L.intestinalis supplementation (Fig. 5j–l). In addition, we found that L.intestinalis treatment reduced plasma TC and LDL-C levels in CP mice (Supplementary Fig. 6d). These suggest that L.intestinalis restores gut homeostasis and improves intestinal barrier function that limits pancreatic fibrosis development.
a The Venn diagram shows the screening scheme for L.intestinalis. b Schematic illustration of CP mouse models including L.int and PBS treatment groups. c Representative images of H&E staining, Trichrome staining and IHC for Collagen-1 in CP + PBS group and CP + L. int group. d Pathology scores, quantification of fibrotic area and Collagen-1 positive area (n = 6 per group). e Representative images of CD8, F4/80, and Ly6G positive cells by IHC. f The comparison of CD8, F4/80 and Ly6G positive cells (n = 6 per group). g, h The proportion of CD8+T cells (CD8+CD3+CD45+) and macrophages (F4/80+CD11b+CD45+) were compared using flow cytometry (n = 6 per group). i qRT-PCR testing the expressions of Ccl2, Ccl7, Ccl12, and Ccl17 in colon tissues between CP + PBS group and CP + L.int group (n = 5 per group), Gapdh was used as an endogenous control. j Representative IF staining for ZO-1, Occludin, and Claudin-1 in the colon. k The quantification of ZO-1, Occludin, and Claudin-1 positive area between CP + PBS group and CP + L.int groups (n = 6 per group). l qRT-PCR testing the expressions of Tjp1 and Ocln in the colon tissues (n = 5 per group), Gapdh was used as an endogenous control. m Heatmap displaying the different levels of Lactobacillus and Bifidobacterium between post- and pre-statins administration. n The comparison of relative abundance of Lactobacillus between L-TC and H-TC individuals (n = 10 per group). o Correlation analysis between the relative abundance of Lactobacillus and plasma TC level. Data are presented as mean ± SEM. ns, no significance; *p < 0.05; **p < 0.01. b is from Figdraw 2.0 of HOME for Researchers (Permissions for use have already been obtained).
To further understand the impacts of statin therapy on human gut microbiota composition, six published studies that have compared different gut microbiota between pre- and post-statins therapy were collected (Fig. 5m). The positive correlations between increased abundances of Lactobacillus and Bifidobacterium with better lipid-lowering effects were found and higher abundance of Firmicutes in fecal samples, as well as increased ratio of Lactobacillus to Bifidobacterium can predict better response to statin therapy33,34. Patients with high cholesterol level indicates lower proportion of Lactobacillus and Bifidobacterium in the gut, which are associated with increased systemic inflammation21,35,36,37. We then investigated the association between the level of cholesterol and the abundance of Lactobacillus in human. Plasma and fecal samples from 20 healthy volunteers were collected, the total cholesterol (TC) level was measured, and gut microbiome was sequenced (Supplementary Fig. 6e, Supplementary Table 1). Based on the median TC value (4.5 mmol/L), individuals were divided into low TC (L-TC, n = 10) and high TC group (H-TC, n = 10). We found that volunteers in L-TC group showed higher abundance of Lactobacillus, and the abundance of Lactobacillus was negatively correlated with plasma TC level (Fig. 5n, o). These suggest that statin therapy may modulate total cholesterol metabolism through enhancing the abundance of Lactobacillus.
L.intestinalis maintains intestinal homeostasis by inhibiting Ccl2/7-Ccr1 signaling between macrophages and epithelium cells
To investigate the mechanism by which the augmentation of L.intestinalis ameliorates intestinal injury, six donors’ stool samples were divided into high-L. intestinalis group (High L.i donor) and low-L.intestinalis group (Low L.i donor), as determined by the level of L.intestinalis, and FMT mouse model was then built (Figs. 6a, 7). We observed a differential abundance of L.intestinalis in CP mice that confirmed the successful colonization of L.intestinalis following FMT (Fig. 6b). Compared with FMT from High L.i donors (FMT-H L.i), an increased percentage of pancreatic fibrosis were found in mice received FMT from Low L.i donor (FMT-L L.i) (Fig. 6c, Supplementary Fig. 7a, b). To further assess the alterations of FMT on immune populations in the colon, CD45+ cells from FMT-H L.i and FMT-L L.i mice were sorted and single-cell RNA sequencing was conducted (Supplementary Fig. 7c). After quality control of raw data, a total of 18,724 cells were obtained for subsequent analysis. Using UMAP plot to visualize cell annotation and the markers of each cell type were listed (Fig. 6d, Supplementary Fig. 7d). We found that the proportion of M1 macrophage was increased in FMT-L L.i group compared with FMT-H L.i group (Fig. 6e). To explore the effect of L.intestinalis on macrophage polarization, mouse splenocytes were stimulated with lipopolysaccharides (LPS) and then co-cultured with L.intestinalis supernatants (Fig. 6f). The supernatants of L.intestinalis decreased the proportion of M1 macrophages (CD86+F4/80+CD11b+), while it increased the proportion of M2 macrophages (CD206+F4/80+CD11b+) (Fig. 6g). Given the links between macrophages recruitment and pro-inflammatory chemokines secretion, we further investigated the differences between FMT-L L.i and FMT-H L.i groups within M1 macrophages (Fig. 6h). GO analysis indicated that the activations of inflammatory processes including regulation of leukocyte differentiation and regulation of inflammatory response were upregulated by FMT-L L.i (Fig. 6i). Genes which were involved in those processes included Tnf, Tlr6, Ccl2, Ccl7 and Cxcl2 (Fig. 6j). KEGG analysis suggested that pro-inflammatory pathways such as chemokine signaling pathway and cytokine-cytokine receptor interaction were upregulated in the FMT-L L.i group (Supplementary Fig. 7e). Simultaneously, decreased levels of Ccl2 and Ccl7 were observed within M1 macrophage subpopulation in FMT-H L.i group (Fig. 6k). Next, bone marrow-derived macrophages (BMDM) of mice was induced into M1 macrophages and stimulated by L.intestinalis supernatants. We found that L.intestinalis inhibited the secretion of Ccl2 and Ccl7 by M1 macrophages (Fig. 6l). To investigate whether L.intestinalis can recruit M1 macrophages, we employed a transwell assay and found that the addition of L.intestinalis supernatant led to a significant reduction in the capacity of M1 macrophages migration (Fig. 6m, n). As epithelial cells are key components of forming the intestinal barrier, we then compared the different genes between two groups in the epithelial cells (Supplementary Fig. 7f). GO analysis showed that leukocyte migration, leukocyte proliferation and leukocyte chemotaxis were upregulated by FMT-L L.i (Supplementary Fig. 7g). The upregulations of Ccl2 and Ccl7 are involved in the pro-inflammatory signaling pathways through binding with their receptors38, we then co-cultured mouse colonic organoid (Fig. 6o, Supplementary Fig. 7h) with L.intestinalis supernatants and found that the level of Ccr1 was decreased, while the level of Ccr2 was unchanged (Fig. 6p). These suggest that increased abundance of L.intestinalis attenuates ameliorate epithelial injury by blocking Ccl2/7-Ccr1 signaling between M1 macrophage and epithelial cells.
a qRT-PCR detecting the expressions of L. intestinalis in volunteers’ stool samples (n = 3 per group). b The comparison of relative abundance of L.intestinalis in fecal samples between FMT-H L.i group and FMT-L L.i group (n = 6 per group). c Representative images of H&E staining, Trichrome staining, pathology scores, and quantification of fibrotic area in both groups (n = 6 per group). d UMAP plot displaying distinct clusters of 18,724 cells obtained from FMT-H L.i and FMT-L L.i mice (n = 1 per group). e Proportion of cell types. f Flow chart depicting the co-culture of immune cells derived from the spleen with LPS and L.i supernatants. g The proportion of M1 macrophages (CD86+F4/80+CD11b+) and M2 macrophages (CD206+F4/80+CD11b+) between LPS and LPS + L.i supernatants groups were compared by flow cytometry (n = 5 per group). h Volcano plots presenting the differential genes in M1 macrophages between FMT-L L.i and FMT-H L.i mice (log2FC ≥ 0.25, p < 0.05). i, j GO analysis showing the enriched pathways by upregulated genes in M1 macrophages between FMT-L L.i and FMT-H L.i groups. k The levels of Ccl2 and Ccl7 in M1 macrophages between FMT-L L.i and FMT-H L.i groups. l qRT-PCR detecting the expressions of Ccr1 and Ccr2 in M1 macrophages after L. intestinalis supernatants or MRS medium intervention (n = 5 per group), Gapdh was used as an endogenous control. m Flow chart depicting the migration of M1 macrophages under L. intestinalis supernatants. n The number of migrated macrophages was compared between the two groups (n = 3 per group). o Flow chart depicting the establishment of colon organoids in mice and co-culture with LPS and L. intestinalis supernatants. p qRT-PCR detecting the expressions of Ccr1 and Ccr2 in colon organoids after co-culture (n = 3 per group), Gapdh was used as an endogenous control. Data are presented as mean ± SEM. ns, no significance; *p < 0.05; **p < 0.01; ***p < 0.001. f, m, o is from Figdraw 2.0 of HOME for Researchers (Permissions for use have already been obtained).

The upregulation of L. intestinalis by statin therapy suppresses the infiltration of CD8+T cells and macrophages that attenuate pancreatic fibrosis. Figure 7 is from Figdraw 2.0 of HOME for Researchers (Permissions for use have already been obtained).
Discussion
CP is characterized by pancreatic inflammation and fibrosis, leading to impaired exocrine function of the pancreas. This pathological condition results in aberrant lipid synthesis and breakdown, often accompanied by disorders in lipid metabolism and gut microbiota dysbiosis16,39. Here, we discover that statin therapy restores gut microbiota relieves the damage of gut barrier and limits pathogenic bacteria translocation in CP mice. Statin therapy-associated gut microbiota, particularly L.intestinalis, downregulates the expressions of pro-inflammatory factors such as Ccl2 and Ccl7 that maintain gut homeostasis, which suppresses pancreatic fibrosis through inhibiting the infiltrations of CD8+T cells and macrophages. Our study underlines the potential roles of statin therapy on gut microbiota re-stabilization that maintains gut barrier integrity and manipulates immune responses in CP.
Increased immune activations have been observed in CP patients, as well as CP mouse models40. The polarization of macrophages towards from M1 phenotype to M2 phenotype, along with cytokines release such as IL-6, TNF-α, and IL-1β damage acinar cells and activate PSCs that cause extremely dense of stroma deposition41. CD8+T cells are increasingly interspersed in CP tissues, while the pro-fibrotic characteristics of CD8+T cells still need to be fully elucidated42,43. Our findings revealed that pancreatic fibrosis causes the re-localization of lymphocytes including CD8+T cells and macrophages that can be exacerbated by a high-fat diet or improved by statins-mediated lipid-lowering effects. The deficiency of CD8 restrains the recruitment of macrophages and impairs anti-fibrotic phenotype, suggesting that statin therapy is involved in CD8+T cells-dependent immune regulatory mechanisms that restrains CP progression. Furthermore, we found that atorvastatin facilitates the protective effect on the early stage of pancreatitis by inhibiting neutrophils recruitment, which adds more evidence of understanding the anti-fibrotic roles of atorvastatin at the phase of CP initiation and progression.
Gut microbiota affects local immune responses and gut barrier functions8,9, which contributes to CP progression12,15. The upregulated abundances of Enterococcus, Escherichia and Streptococcus are associated with pancreatic insufficiency and disease severity in CP patients12. Statin therapy decreases pro-inflammatory bacterial consortia and ameliorates microbial-associated disorders44. Our findings suggest that statin therapy increases the abundance of Lactobacillus that delays pancreatic fibrosis progression. Lactobacillus confers beneficial roles of anti-inflammatory and anti-fibrosis propersities45,46, and maintains intestinal epithelial regeneration and homeostasis47. We found that statin therapy increases the abundance of L. intestinalis which prevents epithelium injury by downregulating the expressions of Ccl2, Ccl7, and Ccl12. Alternatively, L. intestinalis increased the levels of Tjp1, Ocln, Cldn2, Egf, and Lamc1 that activated the ECM receptor interaction and focal adhesion, which enhanced the integrity of intestinal wall and restrained pathogenic bacteria migration48.
Recent studies have confirmed that there are bacteria in normal pancreatic tissue49,50. These bacteria are usually resident bacteria or called commensal bacteria, which can promote the formation of immune tolerance through interaction with immune cells. This tolerance helps prevent the immune system from overreacting to harmless resident bacteria51. An abnormal overgrowth of gut microbiota travels to the pancreas remodels local immune compartments and causes inflammatory mediators’ release52. We found upregulated microbes load in human and mouse CP tissues. Simultaneously, statin therapy reduced bacteria translocation and colonization which causes downregulated anti-microbial response in the pancreas. Microbes are recognized by innate immune cells and then direct adaptive immune responses8. Our findings suggest that microbes in the pancreas are associated with the infiltration of CD8+T cells and macrophages that expedite inflammatory response and pancreatic fibrosis.
Macrophages play a key role in host defense against bacterial pathogens and maintain epithelial barrier integrity in the gut53. However, excessive activation of macrophages can damage mucosal barrier and disrupt intestinal homeostasis through secreting pro-inflammatory factors38,54. Our data revealed that FMT from patients with high abundance of L. intestinalis after statin therapy or oral delivery of L. intestinalis can restrain the infiltration of CD8+T cells and macrophages that alleviated pancreatic fibrosis by decreasing the expressions of Ccl2, Ccl7, Ccl12, and Ccl17 in the colon. Simultaneously, we observed that higher abundance of L. intestinalis exerts inhibitory effects on M1 macrophage polarization and intercepts the release of Ccl2 and Ccl7 in the gut. These suggest that L. intestinalis can alleviate intestinal injury by blocking the signals between M1 macrophage and epithelial cells and impairing the Ccl2/7-induced chemokine cascades, which restores intestinal barrier function and subsequently confines bacterial translocation.
In conclusion, statin therapy associated L. intestinalis maintains intestinal barrier and reduces bacteria translocation through inhibiting Ccl2/7-Ccr1 signaling between macrophages and epithelium cells. This blocks the pathogenic bacteria migration from the gut to the pancreas and decreases CD8+T cells-dependent immune activation which eventually attenuates pancreatic fibrosis. Our study provides new clues about statin therapy and associated manipulation of gut microbiota as compelling approaches for CP treatment.
Data availability
All data presented in this work are available within the article and the supplementary files. Further information or requests for data can be directed to the corresponding authors.
References
Vege, S. S. & Chari, S. T. Chronic pancreatitis. N. Engl. J. Med. 386, 869–878 (2022).
Beyer, G., Habtezion, A., Werner, J., Lerch, M. M. & Mayerle, J. Chronic pancreatitis. Lancet 396, 499–512 (2020).
Gardner, T. B. et al. ACG clinical guideline: chronic pancreatitis. Am. J. Gastroenterol. 115, 322–339 (2020).
Li, L. et al. RB1CC1-enhanced autophagy facilitates PSCs activation and pancreatic fibrogenesis in chronic pancreatitis. Cell Death Dis. 9, 952 (2018).
Zhao, Q., Manohar, M., Wei, Y., Pandol, S. J. & Habtezion, A. STING signalling protects against chronic pancreatitis by modulating Th17 response. Gut 68, 1827–1837 (2019).
Glaubitz, J. et al. In mouse chronic pancreatitis CD25+FOXP3+ regulatory T cells control pancreatic fibrosis by suppression of the type 2 immune response. Nat. Commun. 13, 4502 (2022).
Paik, D. et al. Human gut bacteria produce ΤΗ17-modulating bile acid metabolites. Nature 603, 907–912 (2022).
Michaudel, C. & Sokol, H. The gut microbiota at the service of immunometabolism. Cell Metab. 32, 514–523 (2020).
Thomas, R. M. & Jobin, C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat. Rev. Gastroenterol. Hepatol. 17, 53–64 (2020).
Zhou, C. H. et al. Altered diversity and composition of gut microbiota in Chinese patients with chronic pancreatitis. Pancreatology 20, 16–24 (2020).
Frost, F. et al. The gut microbiome in patients with chronic pancreatitis is characterized by significant dysbiosis and overgrowth by opportunistic pathogens. Clin. Transl. Gastroenterol. 11, e00232 (2020).
Han, M. M. et al. The alterations of gut microbiota in mice with chronic pancreatitis. Ann. Transl. Med. 7, 464 (2019).
Baruch, E. N. et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 371, 602–609 (2021).
Hanssen, N. M. J., de Vos, W. M. & Nieuwdorp, M. Fecal microbiota transplantation in human metabolic diseases: from a murky past to a bright future? Cell Metab. 33, 1098–1110 (2021).
Liu, L. et al. Gut microbiota affects pancreatic fibrotic progression through immune modulation in chronic pancreatitis. Micro. Pathog. 177, 106035 (2023).
Yang, X. et al. Very-low-density lipoprotein receptor-enhanced lipid metabolism in pancreatic stellate cells promotes pancreatic fibrosis. Immunity 55, 1185–1199.e8 (2022).
Trub, A. G. et al. Statin therapy inhibits fatty acid synthase via dynamic protein modifications. Nat. Commun. 13, 2542 (2022).
Zhu, B., Ma, A. Q., Yang, L. & Dang, X. M. Atorvastatin attenuates bleomycin-induced pulmonary fibrosis via suppressing iNOS expression and the CTGF (CCN2)/ERK signaling pathway. Int. J. Mol. Sci. 14, 24476–24491 (2013).
Trebicka, J. et al. Atorvastatin attenuates hepatic fibrosis in rats after bile duct ligation via decreased turnover of hepatic stellate cells. J. Hepatol. 53, 702–712 (2010).
She, J. et al. Statins aggravate insulin resistance through reduced blood glucagon-like peptide-1 levels in a microbiota-dependent manner. Cell Metab. 36, 408–421.e5 (2024).
Han, J. X. et al. Microbiota-derived tryptophan catabolites mediate the chemopreventive effects of statins on colorectal cancer. Nat. Microbiol. 8, 919–933 (2023).
Sendler, M. et al. Complement component 5 mediates development of fibrosis, via activation of stellate cells, in 2 mouse models of chronic pancreatitis. Gastroenterology 149, 765–776.e10 (2015).
Riquelme, E. et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 178, 795–806.e12 (2019).
Li, L. et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol. Cancer Ther. 15, 2232–2243 (2016).
Wei, W. et al. Psychological stress-induced microbial metabolite indole-3-acetate disrupts intestinal cell lineage commitment. Cell Metab. 36, 466–483.e7 (2024).
Baghdadi, M. B. & Kim, T. H. Analysis of mouse intestinal organoid culture with conditioned media isolated from mucosal enteric glial cells. STAR Protoc. 3, 101351 (2022).
Yang, W. J. et al. Acinar ATP8b1/LPC pathway promotes macrophage efferocytosis and clearance of inflammation during chronic pancreatitis development. Cell Death Dis. 13, 893 (2022).
Lien, E. C. et al. Low glycaemic diets alter lipid metabolism to influence tumor growth. Nature 599, 302–307 (2021).
Bahrami, A., Parsamanesh, N., Atkin, S. L., Banach, M. & Sahebkar, A. Effect of statins on toll-like receptors: a new insight to pleiotropic effects. Pharm. Res 135, 230–238 (2018).
Schmitz-Winnenthal, H. et al. Chronic pancreatitis is associated with disease-specific regulatory T-cell responses. Gastroenterology 138, 1178–1188. (2010).
Van Looveren, K. et al. Glucocorticoids limit lipopolysaccharide-induced lethal inflammation by a double control system. EMBO Rep. 21, e49762 (2020).
Li, G. et al. Gut microbiota aggravates neutrophil extracellular traps-induced pancreatic injury in hypertriglyceridemic pancreatitis. Nat. Commun. 14, 6179 (2023).
Sun, B., Li, L. & Zhou, X. Comparative analysis of the gut microbiota in distinct statin response patients in East China. J. Microbiol. 56, 886–892 (2018).
Liu, Y. et al. Gut microbiome associates with lipid-lowering effect of rosuvastatin in vivo. Front Microbiol 9, 530 (2018).
Vieira-Silva, S. et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 581, 310–315 (2020).
Wang, L. et al. The gut microbiota is associated with clinical response to statin treatment in patients with coronary artery disease. Atherosclerosis 325, 16–23 (2021).
Hu, X. et al. Multi-omics study reveals that statin therapy is associated with restoration of gut microbiota homeostasis and improvement in outcomes in patients with acute coronary syndrome. Theranostics 11, 5778–5793 (2021).
He, J. et al. Fbxw7 increases CCL2/7 in CX3CR1hi macrophages to promote intestinal inflammation. J. Clin. Investig. 129, 3877–3893 (2019).
Wang, D. et al. Pancreatic acinar cells-derived sphingosine-1-phosphate contributes to fibrosis of chronic pancreatitis via inducing autophagy and activation of pancreatic stellate cells. Gastroenterology 165, 1488–1504.e20 (2023).
Xue, J. et al. Alternatively activated macrophages promote pancreatic fibrosis in chronic pancreatitis. Nat. Commun. 6, 7158 (2015).
Reding, T. et al. A selective COX-2 inhibitor suppresses chronic pancreatitis in an animal model (WBN/Kob rats): significant reduction of macrophage infiltration and fibrosis. Gut 55, 1165–1173 (2006).
Schmitz-Winnenthal, H. et al. Chronic pancreatitis is associated with disease-specific regulatory T-cell responses. Gastroenterology 138, 1178–1188 (2010).
Sparmann, G. et al. Cytokine mRNA levels and lymphocyte infiltration in pancreatic tissue during experimental chronic pancreatitis induced by dibutyltin dichloride. Dig. Dis. Sci. 46, 1647–1656 (2001).
Khan, T. J. et al. Effect of atorvastatin on the gut microbiota of high fat diet-induced hypercholesterolemic rats. Sci. Rep. 8, 662 (2018).
Kim, W. et al. Lactobacillus plantarum-derived extracellular vesicles induce anti-inflammatory M2 macrophage polarization in vitro. J. Extracell. Vesicles 9, 1793514 (2020).
Ma, J., Chen, S., Li, Y., Wu, X. & Song, Z. Arbutin improves gut development and serum lipids via Lactobacillus intestinalis. Front. Nutr. 9, 948573 (2022).
Wu, H. et al. Lactobacillus reuteri maintains intestinal epithelial regeneration and repairs damaged intestinal mucosa. Gut Microbes 11, 997–1014 (2020).
Lee, S. H. Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intest. Res. 13, 11–18 (2015).
Geller, L. T. et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160 (2017).
Gao, H. et al. Accumulation of microbial DNAs promotes to islet inflammation and β cell abnormalities in obesity in mice. Nat. Commun. 13, 565 (2022).
Kamada, N. & Núñez, G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology 146, 1477–1488 (2014).
Pöysti, S. et al. Infection with the enteric pathogen C. rodentium promotes islet-specific autoimmunity by activating a lymphatic route from the gut to pancreatic lymph node. Mucosal Immunol. 15, 471–479 (2022).
Chikina, A. S. et al. Macrophages maintain epithelium integrity by limiting fungal product absorption. Cell 183, 411–428.e16 (2020).
Zhang, Y. et al. Gastrointestinal dysmotility predisposes to colitis through regulation of gut microbial composition and linoleic acid metabolism. Adv. Sci. 11, e2306297 (2024).
Acknowledgements
We show our full respect and gratitude to all the participants in the study. This work was supported by the National Natural Science Foundation of China (82270666,82270665,82070658), The Young Elite Scientists Sponsorship Program by CAST (2023QNRC001), The Natural Science Foundation of Heilongjiang Province for Excellent Youths (YQ2023H007) and The Science Fund for Excellent Young Scholars of First Affiliated Hospital of Harbin Medical University (HYD2020YQ0009).
Author information
Authors and Affiliations
Contributions
Y.H.S., S.W.O., and L.W.L. performed animal experiments. Y.H.S., T.Z., S.W.O., and T.Q.L. performed histological evaluation. Y.H.S. and T.Z. performed formal analysis including bioinformatics and statistical analyses. S.W.O., C.Z., and G.Q.L. performed the cell culture and western blot experiments. G.Q.L., Y.K.C., R.B., H.X.Z., X.B.Z., Y.Y., G.W., R.K., and H.C. were involved in collection, preparation, interpretation, validation, and critical review of the data. Y.H.S. and S.W.O. created the manuscript figures and supplementary materials. Y.H.S. and L.L. drafted the manuscript. L.L. and Y.Y. edited and reviewed the manuscript text. B.S. and L.L. acquired funding for the study. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sui, Y., Zhang, T., Ou, S. et al. Statin therapy associated Lactobacillus intestinalis attenuates pancreatic fibrosis through remodeling intestinal homeostasis. npj Biofilms Microbiomes 11, 59 (2025). https://doi.org/10.1038/s41522-025-00695-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-025-00695-w