Abstract
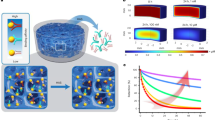
Aptamers are being applied as affinity reagents in analytical applications owing to their high stability, compact size and amenability to chemical modification. Generating aptamers with different binding affinities is desirable, but systematic evolution of ligands by exponential enrichment (SELEX), the standard for aptamer generation, is unable to quantitatively produce aptamers with desired binding affinities and requires multiple rounds of selection to eliminate false-positive hits. Here we introduce Pro-SELEX, an approach for the rapid discovery of aptamers with precisely defined binding affinities that combines efficient particle display, high-performance microfluidic sorting and high-content bioinformatics. Using the Pro-SELEX workflow, we were able to investigate the binding performance of individual aptamer candidates under different selective pressures in a single round of selection. Using human myeloperoxidase as a target, we demonstrate that aptamers with dissociation constants spanning a 20-fold range of affinities can be identified within one round of Pro-SELEX.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information . The sequencing data files are too large to be publicly shared, but they are available from the corresponding author upon reasonable request. The chip design (in the format of STL) and running protocol of the microfluidic chip will be available free of charge from the publisher’s website as a supplementary file. Source data are provided with this paper.
Code availability
The code corresponding to the AptaZ algorithm can be accessed at https://github.com/dwangnu/AptaZ.
References
Drabovich, A. P. et al. Smart aptamers facilitate multi-probe affinity analysis of proteins with ultra-wide dynamic range of measured concentrations. J. Am. Chem. Soc. 129, 7260–7261 (2007).
Wilson, B. D. et al. Re-evaluating the conventional wisdom about binding assays. Trends Biochem. Sci. 45, 639–649 (2020).
Arroyo-Currás, N. et al. Real-time measurement of small molecules directly in awake, ambulatory animals. Proc. Natl Acad. Sci. USA 114, 645–650 (2017).
Fercher, C. et al. Recombinant antibody engineering enables reversible binding for continuous protein biosensing. ACS Sens. 6, 764–776 (2021).
Plaxco, K. W. et al. Seconds-resolved, in situ measurements of plasma phenylalanine disposition kinetics in living rats. Anal. Chem. 93, 4023–4032 (2021).
Xue, L. et al. Solid-state nanopore sensors. Nat. Rev. Mater. 5, 931–951 (2020).
Sevenler, D. et al. Beating the reaction limits of biosensor sensitivity with dynamic tracking of single binding events. Proc. Natl Acad. Sci. USA 116, 4129–4134 (2019).
Wang, T. et al. Three decades of nucleic acid aptamer technologies: lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 37, 28–50 (2019).
Liu, J. et al. Functional nucleic acid sensors. Chem. Rev. 109, 1948–1998 (2009).
Song, S. et al. Aptamer-based biosensors. Trends Anal. Chem. 27, 108–117 (2008).
Wang, B. et al. Wearable aptamer-field-effect transistor sensing system for noninvasive cortisol monitoring. Sci. Adv. 8, eabk0967 (2022).
Nakatsuka, N. et al. Aptamer-field-effect transistors overcome Debye length limitations for small-molecule sensing. Science 362, 319–324 (2018).
Mage, P. L. et al. Closed-loop control of circulating drug levels in live animals. Nat. Biomed. Eng. 1, 0070 (2017).
Ferguson, B. S. et al. Real-time, aptamer-based tracking of circulating therapeutic agents in living animals. Sci. Transl. Med. 5, 213ra165 (2013).
Zhao, C. et al. Implantable aptamer-field-effect transistor neuroprobes for in vivo neurotransmitter monitoring. Sci. Adv. 7, eabj7422 (2021).
Qian, S. et al. Aptamers from random sequence space: accomplishments, gaps and future considerations. Anal. Chim. Acta 1196, 339511 (2022).
Li, F. et al. Aptamers facilitating amplified detection of biomolecules. Anal. Chem. 87, 274–292 (2015).
Ellington, A. D. et al. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. et al. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Mendonsa, S. D. et al. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 126, 20–21 (2004).
Berezovski, M. et al. Non-SELEX selection of aptamers. J. Am. Chem. Soc. 128, 1410–1411 (2006).
Lou, X. et al. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl Acad. Sci. USA 106, 2989–2994 (2009).
Wang, J. et al. Particle display: a quantitative screening method for generating high-affinity aptamers. Angew. Chem. Int. Ed. 53, 4796–4801 (2014).
Zhang, R. et al. Association between myeloperoxidase levels and risk of coronary artery disease. JAMA 286, 2136–2142 (2001).
Baldus, S. et al. Myeloperoxidase serum levels predict risk in patients with acute coronary syndromes. Circulation 108, 1440–1445 (2003).
Labib, M. et al. Single-cell mRNA cytometry via sequence-specific nanoparticle clustering and trapping. Nat. Chem. 10, 489–495 (2018).
Labib, M. et al. Tracking the expression of therapeutic protein targets in rare cells by antibody-mediated nanoparticle labelling and magnetic sorting. Nat. Biomed. Eng. 5, 41–52 (2021).
Wang, Z. et al. Efficient recovery of potent tumour-infiltrating lymphocytes through quantitative immunomagnetic cell sorting. Nat. Biomed. Eng. 6, 108–117 (2022).
Wang, Z. et al. Ultrasensitive detection and depletion of rare leukemic B cells in T cell populations via immunomagnetic cell ranking. Anal. Chem. 93, 2327–2335 (2021).
Bock, L. C. et al. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355, 564–566 (1992).
Tang, W. H. W. et al. Usefulness of myeloperoxidase levels in healthy elderly subjects to predict risk of developing heart failure. Am. J. Cardiol. 103, 1269–1274 (2009).
Vallée-Bélisle, A. et al. Engineering biosensors with extended, narrowed or arbitrarily edited dynamic range. J. Am. Chem. Soc. 134, 2876–2879 (2012).
Tu, J. et al. The era of digital health: a review of portable and wearable affinity biosensors. Adv. Funct. Mater. 30, 1906713 (2020).
Deng, R. et al. Recognition-enhanced metastably shielded aptamer for digital quantification of small molecules. Anal. Chem. 90, 14347–14354 (2018).
Das, J. et al. Reagentless biomolecular analysis using a molecular pendulum. Nat. Chem. 13, 428–434 (2021).
Wang, L. et al. Rapid and ultrasensitive electromechanical detection of ions, biomolecules and SARS-CoV-2 RNA in unamplified samples. Nat. Biomed. Eng. 6, 276–285 (2022).
Zhang, Z. et al. High-affinity dimeric aptamers enable the rapid electrochemical detection of wild-type and B.1.1.7 SARS-CoV-2 in unprocessed saliva. Angew. Chem. Int. Ed. 60, 24266–24274 (2021).
Li, J. et al. Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res. 49, 7267–7279 (2021).
Yang, G. et al. Identification of SARS-CoV-2-against aptamer with high neutralization activity by blocking the RBD ___domain of spike protein 1. Signal Transduct. Target. Ther. 6, 227 (2021).
Lapa, S. A. et al. The toolbox for modified aptamers. Mol. Biotechnol. 58, 79–92 (2016).
Gawande, B. N. et al. Selection of DNA aptamers with two modified bases. Proc. Natl Acad. Sci. USA 114, 2898–2903 (2017).
Gordon, C. K. L. et al. Click-particle display for base-modified aptamer discovery. ACS Chem. Biol. 14, 2652–2662 (2019).
Pfeiffer, F. et al. Identification and characterization of nucleobase-modified aptamers by click-SELEX. Nat. Protoc. 13, 1153–1180 (2018).
Wang, Z. et al. Ultrasensitive and rapid quantification of rare tumorigenic stem cells in hPSC-derived cardiomyocyte populations. Sci. Adv. 6, eaay7629 (2020).
Edgar, R. C. & Bateman, A. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Acknowledgements
This research was supported in part by the Canadian Institutes of Health Research (grant no. FDN-148415) and the Collaborative Health Research Projects Program (CIHR/NSERC partnered). This research is part of the University of Toronto’s Medicine by Design initiative, which receives funding from the Canada First Research Excellence Fund. This research was supported in part by the McCormick Catalyst Fund at Northwestern University.
Author information
Authors and Affiliations
Contributions
D.C., Z.W. and S.O.K. conceived and designed the experiments. D.C. performed the aptamer selection and validation. Z.W. performed the chip fabrication and wrote the code for AptaZ. All authors discussed the results, analysed the data and contributed to the preparation and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.O.K. is an inventor on a patent entitled ‘Device for capture of particles in a flow’ (US patent US10073079) that is licensed to CTRL Therapeutics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Chunhai Fan, Yingfu Li and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supporting discussion, Tables 1–5 and Figs. 1–12.
Supplementary Data
Source data used to generate Supplementary Figures.
Supplementary Data
Chip design.
Supplementary Data
Fabrication and running protocol of the microfluidic chip.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chang, D., Wang, Z., Flynn, C.D. et al. A high-dimensional microfluidic approach for selection of aptamers with programmable binding affinities. Nat. Chem. 15, 773–780 (2023). https://doi.org/10.1038/s41557-023-01207-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-023-01207-z
This article is cited by
-
Microfluidic technologies for enhancing the potency, predictability and affordability of adoptive cell therapies
Nature Biomedical Engineering (2025)
-
Single-step discovery of high-affinity RNA ligands by UltraSelex
Nature Chemical Biology (2025)
-
One-step screening of myeloperoxidase aptamer using porous PEG hydrogel
Advanced Composites and Hybrid Materials (2025)
-
DNA framework signal amplification platform-based high-throughput systemic immune monitoring
Signal Transduction and Targeted Therapy (2024)
-
Blocker-SELEX: a structure-guided strategy for developing inhibitory aptamers disrupting undruggable transcription factor interactions
Nature Communications (2024)