Abstract
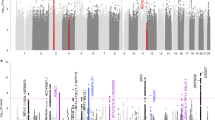
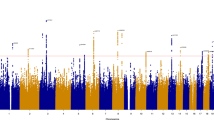
Subjective well-being (SWB) is important for understanding human behaviour and health. Although the connection between SWB and psychiatric disorders has been studied, common genetic mechanisms remain unclear. This study aimed to explore the genetic relationship between SWB and psychiatric disorders. Bivariate causal mixture modelling (MiXeR), polygenic risk score (PRS) and Mendelian randomization (MR) analyses showed substantial polygenic overlap and associations between SWB and the psychiatric disorders. Subsequent replication studies in East Asian populations confirmed the polygenic overlap between schizophrenia and SWB. The conditional and conjunctional false discovery rate analyses identified additional or shared genetic loci associated with SWB or psychiatric disorders. Functional annotation revealed enrichment of specific brain tissues and genes associated with SWB. The identified genetic loci showed cross-ancestry transferability between the European and Korean populations. Our findings provide valuable insights into the common genetic mechanisms underlying SWB and psychiatric disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data from 23andMe Inc. and UKB can be obtained by applying to each respective website (23andMe Inc., https://research.23andme.com/dataset-access/; UKB, https://www.ukbiobank.ac.uk). Summary statistics from ref. 4 are publicly accessible on the Social Science Genetic Association Consortium website (https://www.thessgac.org). Summary statistics for genetic correlation analysis are available from various sources, including the Psychiatric Genomics Consortium (https://www.med.unc.edu/pgc/download-results), GWAS ATLAS (https://atlas.ctglab.nl/traitDB) and GWAS Catalogue (https://www.ebi.ac.uk/gwas/studies). Full summary statistics of the KBA GWAS can be found in the NHGRI-EBI GWAS Catalogue (https://www.ebi.ac.uk/gwas/downloads).
Code availability
In this study, existing pipelines were utilized to obtain the results, and no new code was created. Further information regarding the software and data utilization can be found in Supplementary Information, including the URLs. The protocol and specific details of our MR analyses were not registered in advance.
References
Diener, E., Oishi, S. & Tay, L. Advances in subjective well-being research. Nat. Hum. Behav. 2, 253–260 (2018).
Depression and Physical Illness (Cambridge Univ. Press, 2006).
Vukasović, T. & Bratko, D. Heritability of personality: a meta-analysis of behavior genetic studies. Psychol. Bull. 141, 769–785 (2015).
Okbay, A. et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016).
Kim, S. et al. Shared genetic architectures of subjective well-being in East Asian and European ancestry populations. Nat. Hum. Behav. 6, 1014–1026 (2022).
Andreassen, O. A. et al. New insights from the last decade of research in psychiatric genetics: discoveries, challenges and clinical implications. World Psychiatry 22, 4–24 (2023).
Baselmans, B. M. L. et al. Multivariate genome-wide analyses of the well-being spectrum. Nat. Genet. 51, 445–451 (2019).
Røysamb, E. & Nes, R. B. The role of genetics in subjective well-being. Nat. Hum. Behav. 3, 3 (2019).
Frei, O. et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat. Commun. 10, 2417 (2019).
Hindley, G. et al. Charting the landscape of genetic overlap between mental disorders and related traits beyond genetic correlation. Am. J. Psychiatry 179, 833–843 (2022).
Andreassen, O. A. et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 9, e1003455 (2013).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Watson, H. J. et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat. Genet. 51, 1207–1214 (2019).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Johnson, E. C. et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry 7, 1032–1045 (2020).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Evangelista, J. E. et al. Enrichr-KG: bridging enrichment analysis across multiple libraries. Nucleic Acids Res. 51, W168–w179 (2023).
Klopfenstein, D. V. et al. GOATOOLS: a Python library for Gene Ontology analyses. Sci. Rep. 8, 10872 (2018).
Spencer, T. J. et al. Impact of tic disorders on ADHD outcome across the life cycle: findings from a large group of adults with and without ADHD. Am. J. Psychiatry 158, 611–617 (2001).
Lowe, T. L., Capriottim, M. R. & McBurnett, K. Long-term follow-up of patients with Tourette’s syndrome. Mov. Disord. Clin. Pract. 6, 40–45 (2019).
Coleman, J. R. I. et al. The genetics of the mood disorder spectrum: genome-wide association analyses of more than 185,000 cases and 439,000 controls. Biol. Psychiatry 88, 169–184 (2022).
Kichaev, G. et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 104, 65–75 (2019).
Jansen, P. R. et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat. Genet. 51, 394–403 (2019).
Dashti, H. S. et al. Genetic determinants of daytime napping and effects on cardiometabolic health. Nat. Commun. 12, 900 (2021).
Mei, L. et al. Overlapping common genetic architecture between major depressive disorders and anxiety and stress-related disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 113, 110450 (2022).
Wen, X. et al. Unbalanced amygdala communication in major depressive disorder. J. Affect. Disord. 329, 192–206 (2023).
Wang, C. et al. Quantitative EEG abnormalities in major depressive disorder with basal ganglia stroke with lesions in different hemispheres. J. Affect. Disord. 215, 172–178 (2017).
Zhang, X. et al. Severity related neuroanatomical and spontaneous functional activity alteration in adolescents with major depressive disorder. Front. Psychiatry 14, 1157587 (2023).
Wang, X. et al. Disrupted functional connectivity of the cerebellum with default mode and frontoparietal networks in young adults with major depressive disorder. Psychiatry Res. 324, 115192 (2023).
Sankar, A. et al. Altered frontal cortex functioning in emotion regulation and hopelessness in bipolar disorder. Bipolar Disord. 23, 152–164 (2021).
Jones, H. J. et al. Associations between plasma fatty acid concentrations and schizophrenia: a two-sample Mendelian randomisation study. Lancet Psychiatry 8, 1062–1070 (2021).
Bloch, M. H. & Qawasmi, A. Omega-3 fatty acid supplementation for the treatment of children with attention-deficit/hyperactivity disorder symptomatology: systematic review and meta-analysis. J. Am. Acad. Child Adolesc. Psychiatry 50, 991–1000 (2011).
Vezini, A., Balosso, S. & Ravizza, T. Neuroinflammatory pathways as treatment targets and biomarkers in epilepsy. Nat. Rev. Neurol. 15, 459–472 (2019).
Yuan, N. Y. et al. Arachidonic acid cascade and eicosanoid production are elevated while LTC4 synthase modulates the lipidomics profile in the brain of the HIVgp120-transgenic mouse model of neuroHIV. Cells 11, 2123 (2022).
Bradberry, M. M. et al. N-glycoproteomics of brain synapses and synaptic vesicles. Cell Rep. 42, 112368 (2023).
Horowitz, A. M. et al. Blood factors transfer beneficial effects of exercise on neurogenesis and cognition to the aged brain. Science 369, 167–173 (2020).
Waszkiewicz, N. et al. Salivary exoglycosidases as markers of alcohol dependence. Alcohol Alcohol. 49, 409–416 (2014).
Dalenberg, J. R. et al. Short-term consumption of sucralose with, but not without, carbohydrate impairs neural and metabolic sensitivity to sugar in humans. Cell Metab. 31, 493–502.e7 (2020).
Belsky, D. W. et al. Genetics and the geography of health, behaviour and attainment. Nat. Hum. Behav. 3, 576–586 (2019).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Kim, Y. & Han, B. G. Cohort profile: the Korean Genome and Epidemiology Study (KoGES) Consortium. Int. J. Epidemiol. 46, 1350 (2017).
Kim, J.-H. The reliability and validity test of Psychosocial Well-being Index (PWI). J. Korean Acad. Nurs. 29, 304–313 (1999).
Goldberg, D. P. et al. The validity of two versions of the GHQ in the WHO study of mental illness in general health care. Psychol. Med. 27, 191–197 (1997).
Bartels, M. & Boomsma, D. I. Born to be happy? The etiology of subjective well-being. Behav. Genet. 39, 605–615 (2009).
Fat, L. N. et al. Evaluating and establishing national norms for mental wellbeing using the short Warwick–Edinburgh Mental Well-being Scale (SWEMWBS): findings from the Health Survey for England. Qual. Life Res. 26, 1129–1144 (2017).
Wood, A. R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Guerreiro, R. et al. Genome-wide analysis of genetic correlation in dementia with Lewy bodies, Parkinson’s and Alzheimer’s diseases. Neurobiol. Aging 38, 214.e7–214.e10 (2016).
Hartwig, F. P., Davey Smith, G. & Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998 (2017).
Bowden, J. et al. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Verbanck, M. et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Nazarzadeh, M. et al. Plasma lipids and risk of aortic valve stenosis: a Mendelian randomization study. Eur. Heart J. 41, 3913–3920 (2020).
Brion, M. J., Shakhbazov, K. & Visscher, P. M. Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol. 42, 1497–1501 (2013).
Sanderson, E., Spiller, W. & Bowden, J. Testing and correcting for weak and pleiotropic instruments in two-sample multivariable Mendelian randomization. Stat. Med. 40, 5434–5452 (2021).
Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 7, e34408 (2018).
Yavorska, O. O. & Burgess, S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 46, 1734–1739 (2017).
Watanabe, K. et al. Author correction: genetic mapping of cell type specificity for complex traits. Nat. Commun. 11, 1718 (2020).
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Shim, I. et al. Clinical utility of polygenic scores for cardiometabolic disease in Arabs. Nat. Commun. 14, 6535 (2023).
Hao, L. et al. Development of a clinical polygenic risk score assay and reporting workflow. Nat. Med. 28, 1006–1013 (2022).
Shim, H. et al. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS ONE 10, e0120758 (2015).
Acknowledgements
This study was conducted using bioresources from the National Biobank of Korea at the Korea Disease Control and Prevention Agency, South Korea (KBN-2021-031). This study was supported by grants from the National Research Foundation of Korea funded by the Ministry of Science and Information and Communication Technologies, South Korea (Grant Nos. NRF-2021R1A2C4001779 and RS-2024-00335261 to W.M. and NRF-2022R1A2C2009998 to H.-H.W.); the NAVER Digital Bio Innovation Research Fund, funded by NAVER Corporation (Grant No. 37-2023-0140); and by an MD-PhD/Medical Scientist Training Program grant from the Korea Health Industry Development Institute (KHIDI), which is funded by the Ministry of Health and Welfare, Republic of Korea (Grant No. HC20C0005).
Author information
Authors and Affiliations
Contributions
W.M. and H.-H.W. had full access to all data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis. J.Y.J., W.M. and H.-H.W. conceived and designed the study. J.Y.J., Y.A. and J.-W.P. performed the statistical analyses. J.Y.J., Y.A., J.-W.P. and S.L. drafted the manuscript. K.S.O., O.A.A., W.M. and H.-H.W. supervised the study and critically revised the manuscript. All authors including K.J., S.K., S.-H.J., H.K., B.K., M.Y.H., Y.J.K., W.-Y.P. and A.O. contributed to the data interpretation, writing of the manuscript, and reading and approval of the final draft for submission.
Corresponding authors
Ethics declarations
Competing interests
W.-Y.P. is employed by the commercial company GENINUS. O.A.A. is a consultant for HealthLytix and cortecs.ai; he has received speaker’s honoraria from Janssen, Sunovion, and Lundbeck. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Human Behaviour thanks Shitao Rao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11.
Supplementary Tables
Supplementary Tables 1–29.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jung, J.Y., Ahn, Y., Park, JW. et al. Polygenic overlap between subjective well-being and psychiatric disorders and cross-ancestry validation. Nat Hum Behav 9, 1272–1282 (2025). https://doi.org/10.1038/s41562-025-02155-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41562-025-02155-z