Abstract
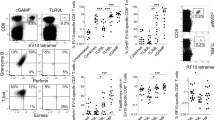
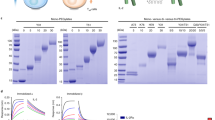
Complexes of extracellular nucleic acids (NAs) with endogenous proteins or peptides, such as LL37, break immune balance and cause autoimmune diseases, whereas NAs with arginine-enriched peptides do not. Inspired by this, we synthesize a polyarginine nanoparticle PEG-TK-NPArg, which effectively inhibits Toll-like receptor-9 (TLR9) activation, in contrast to LL37. To explore the discrepancy effect of PEG-TK-NPArg and LL37, we evaluate the periodic structure of PEG-TK-NPArg-NA and LL37-NA complexes using small-angle X-ray scattering. LL37-NA complexes have a larger inter-NA spacing that accommodates TLR9, while the inter-NA spacing in PEG-TK-NPArg-NA complexes mismatches with the cavity of TLR9, thus inhibiting an interaction with multiple TLR9s, limiting their clustering and damping immune induction. Subsequently, the inhibitory inflammation effect of PEG-TK-NPArg is proved in an animal model of rheumatoid arthritis. This work on how the scavenger-NA complexes inhibit the immune response may facilitate proof-of-concept research translating to clinical application.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the article and Supplementary Information. Source data are provided with this paper.
References
Davidson, A. & Diamond, B. Autoimmune diseases. N. Engl. J. Med. 345, 340–350 (2001).
Steinman, L. Immune therapy for autoimmune diseases. Science 305, 212–216 (2004).
Maecker, H. T. et al. New tools for classification and monitoring of autoimmune diseases. Nat. Rev. Rheumatol. 8, 317–328 (2012).
Wahren-Herlenius, M. & Dörner, T. Immunopathogenic mechanisms of systemic autoimmune disease. Lancet 382, 819–831 (2013).
Tu, Z. et al. Design of therapeutic biomaterials to control inflammation. Nat. Rev. Mater. 7, 557–574 (2022).
Stearns, N. A., Lee, J., Leong, K. W., Sullenger, B. A. & Pisetsky, D. S. The inhibition of anti-DNA binding to DNA by nucleic acid binding polymers. PLoS ONE 7, e40862 (2012).
Jain, S. et al. Nucleic acid scavengers inhibit thrombosis without increasing bleeding. Proc. Natl Acad. Sci. USA 109, 12938–12943 (2012).
Holl, E. K. et al. The nucleic acid scavenger polyamidoamine third-generation dendrimer inhibits fibroblast activation and granulation tissue contraction. Plast. Reconstr. Surg. 134, 420e–433e (2014).
Liu, F. et al. A cationic metal-organic framework to scavenge cell-free DNA for severe sepsis management. Nano Lett. 21, 2461–2469 (2021).
Liu, F. et al. Targeting multiple mediators of sepsis using multifunctional tannic acid-Zn2+-gentamicin nanoparticles. Matter 4, 3677–3695 (2021).
Huang, H. et al. Nanoparticulate cell-free DNA scavenger for treating inflammatory bone loss in periodontitis. Nat. Commun. 13, 5925 (2022).
Shi, C. et al. A nanoparticulate dual scavenger for targeted therapy of inflammatory bowel disease. Sci. Adv. 8, eabj2372 (2022).
Sun, M. et al. Circulating cell-free DNAs as a biomarker and therapeutic target for acetaminophen-induced liver injury. Adv. Sci. 10, 2206789 (2023).
Liang, H. et al. Cationic nanoparticle as an inhibitor of cell-free DNA-induced inflammation. Nat. Commun. 9, 4291 (2018).
Peng, B. et al. Tuned cationic dendronized polymer: molecular scavenger for rheumatoid arthritis treatment. Angew. Chem. Int. Ed. 58, 4254–4258 (2019).
Wu, J. et al. Cationic block copolymer nanoparticles with tunable DNA affinity for treating rheumatoid arthritis. Adv. Funct. Mater. 30, 2000391 (2020).
Tian, J. et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 8, 487–496 (2007).
Yanai, H. et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462, 99–103 (2009).
Li, Y., Berke, I. C. & Modis, Y. DNA binding to proteolytically activated TLR9 is sequence‐independent and enhanced by DNA curvature. EMBO J. 31, 919–931 (2012).
Tug, S. et al. Correlation between cell free DNA levels and medical evaluation of disease progression in systemic lupus erythematosus patients. Cell. Immunol. 292, 32–39 (2014).
Schmidt, N. W. et al. Liquid-crystalline ordering of antimicrobial peptide-DNA complexes controls TLR9 activation. Nat. Mater. 14, 696–700 (2015).
Holl, E. K. et al. Scavenging nucleic acid debris to combat autoimmunity and infectious disease. Proc. Natl Acad. Sci. USA 113, 9728–9733 (2016).
Lou, H. & Pickering, M. C. Extracellular DNA and autoimmune diseases. Cell. Mol. Immunol. 15, 746–755 (2018).
Duvvuri, B. & Lood, C. Cell-free DNA as a biomarker in autoimmune rheumatic diseases. Front. Immunol. 10, 502 (2019).
Lande, R. et al. CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-α production in systemic sclerosis. Nat. Commun. 10, 1731 (2019).
Lee, E. Y., Lee, M. W. & Wong, G. C. L. Modulation of Toll-like receptor signaling by antimicrobial peptides. Semin. Cell Dev. Biol. 88, 173–184 (2019).
Herster, F. et al. Neutrophil extracellular trap-associated RNA and LL37 enable self-amplifying inflammation in psoriasis. Nat. Commun. 11, 105 (2020).
Michailidou, D. et al. Immune complex-mediated neutrophil activation in patients with polymyalgia rheumatica. Rheumatology 62, 2880–2886 (2023).
Gilliet, M. & Lande, R. Antimicrobial peptides and self-DNA in autoimmune skin inflammation. Curr. Opin. Immunol. 20, 401–407 (2008).
Brooks, H., Lebleu, B. & Vivès, E. Tat peptide-mediated cellular delivery: back to basics. Adv. Drug Deliv. Rev. 57, 559–577 (2005).
Lee, E. Y. et al. A review of immune amplification via ligand clustering by self-assembled liquid-crystalline DNA complexes. Adv. Colloid Interface Sci. 232, 17–24 (2016).
Lee, E. Y. et al. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat. Commun. 10, 1012 (2019).
Zielke, C., Nielsen, J. E., Lin, J. S. & Barron, A. E. Between good and evil: complexation of the human cathelicidin LL-37 with nucleic acids. Biophys. J. 123, 1316–1328 (2024).
Tursi, S. A. et al. Bacterial amyloid curli acts as a carrier for DNA to elicit an autoimmune response via TLR2 and TLR9. PLoS Pathog. 13, e1006315 (2017).
Lee, E. Y. et al. Crystallinity of double-stranded RNA-antimicrobial peptide complexes modulates Toll-like receptor 3-mediated inflammation. ACS Nano 11, 12145–12155 (2017).
Engelberg, Y. & Landau, M. The human LL-37(17-29) antimicrobial peptide reveals a functional supramolecular structure. Nat. Commun. 11, 3894 (2020).
Prell, J. S., O’Brien, J. T., Steill, J. D., Oomens, J. & Williams, E. R. Structures of protonated dipeptides: the role of arginine in stabilizing salt bridges. J. Am. Chem. Soc. 131, 11442–11449 (2009).
Hao, L. T. et al. Strong, multifaceted guanidinium-based adhesion of bioorganic nanoparticles to wet biological tissue. JACS Au. 1, 1399–1411 (2021).
Prevette, L. E., Mullen, D. G. & Banaszak Holl, M. M. Polycation-induced cell membrane permeability does not enhance cellular uptake or expression efficiency of delivered DNA. Mol. Pharm. 7, 870–883 (2010).
Bernkop-Schnürch, A. Strategies to overcome the polycation dilemma in drug delivery. Adv. Drug Deliv. Rev. 136–137, 62–72 (2018).
Guo, P. et al. Dual functionalized amino poly(glycerol methacrylate) with guanidine and Schiff-base linked imidazole for enhanced gene transfection and minimized cytotoxicity. J. Mater. Chem. B 3, 6911–6918 (2015).
Zheng, M. et al. ROS-responsive polymeric siRNA nanomedicine stabilized by triple interactions for the robust glioblastoma combinational RNAi therapy. Adv. Mater. 31, 1903277 (2019).
Lande, R. et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449, 564–569 (2007).
Park, S. & Lippard, S. J. Redox state-dependent interaction of HMGB1 and cisplatin-modified DNA. Biochemistry 50, 2567–2574 (2011).
DeRouchey, J., Netz, R. R. & Rädler, J. O. Structural investigations of DNA-polycation complexes. Eur. Phys. J. E 16, 17–28 (2005).
Lee, J. et al. Nucleic acid-binding polymers as anti-inflammatory agents. Proc. Natl Acad. Sci. USA 108, 14055–14060 (2011).
Klinman, D. M. Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol. 4, 249–259 (2004).
Zuniga, E. I., McGavern, D. B., Pruneda-Paz, J. L., Teng, C. & Oldstone, M. B. A. Bone marrow plasmacytoid dendritic cells can differentiate into myeloid dendritic cells upon virus infection. Nat. Immunol. 5, 1227–1234 (2004).
Trentham, D. E., Townes, A. S. & Kang, A. H. Autoimmunity to type II collagen: an experimental model of arthritis. J. Exp. Med. 146, 857–868 (1977).
Lee, S.-M. et al. Targeted chemo-photothermal treatments of rheumatoid arthritis using gold half-shell multifunctional nanoparticles. ACS Nano 7, 50–57 (2013).
Liu, X., Chen, S., Yan, Y., Liu, L. & Chen, Y. Nanoparticulate DNA scavenger loading methotrexate targets articular inflammation to enhance rheumatoid arthritis treatment. Biomaterials 286, 121594 (2022).
Acknowledgements
We thank the financial support from the following funding agencies: the National Natural Science Foundation of China (82341037 to Y.C., 22275213 to L.L., 21875290 to L.L. and 92356310 to Y.Z.), the China Postdoctoral Science Foundation (2023M743133 to X.L. and 2023M743140 to J.H.), the Zhejiang Natural Science Foundation (XHD24E1301 to Y.Z.) and support of Sun Yat-sen University (19lgjc01 to L.L.). We thank S. Cheng (University of Chinese Academy of Sciences (UCAS)) for the support in statistical analysis of data and thank the discussion with D. Yu (Westlake University).
Author information
Authors and Affiliations
Contributions
X.L. conducted the main experiments and wrote the paper. S.C. and Y.D. assisted the partial animal studies. J.H. assisted the polypeptide synthesis. Z.L. conducted the SAXS measurement. Y.Z. proposed the multivalent binding mechanism and conducted the SAXS results analysis. L.L. designed and supervised all the experiments and wrote the paper. Y.C. designed and supervised all the experiments and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks Stefan Bauer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Biocompatibility of cationic polypeptide in CIA rats.
Systemic toxicity of cationic polypeptide. (a) H&E staining of heart, liver, spleen, lung and kidney samples extracted from normal rats, PBS, PEG-TK-LArg and PEG-TK-NPArg treated CIA rats with established-stage arthritis (the scale bar is 200 μm). (b) ALP, ALT, AST, creatinine, and BUN analysis of established-stage CIA rats at day 29 (n = 3 biologically independent samples). The dash line area is the normal range. The normal ranges of ALP, ALT, and AST are 84 ~ 162, 34.6 ~ 64.0 and 96 ~ 200 U/L, respectively. The normal ranges of creatinine and BUN levels in rat serum are 44.2 ~ 114.9 μmol/L and 9.73 ~ 14.53 mmol/L, respectively. All the quantitative data are presented as mean ± s.d.
Supplementary information
Supplementary Information
Supplementary Experiment Procedures, Table 1 and Figs. 1–23.
Supplementary Data 1
Source data for supplementary figures.
Source data
Source Data Fig. 2
Statistical source data for Fig. 2.
Source Data Fig. 3
Statistical source data for Fig. 3.
Source Data Fig. 4
Statistical source data for Fig. 4.
Source Data Fig. 5
Statistical source data for Fig. 5.
Source Data Fig. 6
Statistical source data for Fig. 6.
Source Data Extended Data Fig. 1
Statistical source data for Extended Data Fig. 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, X., Chen, S., Huang, J. et al. Synthetic polypeptides inhibit nucleic acid-induced inflammation in autoimmune diseases by disrupting multivalent TLR9 binding to LL37-DNA bundles. Nat. Nanotechnol. 19, 1745–1756 (2024). https://doi.org/10.1038/s41565-024-01759-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41565-024-01759-2