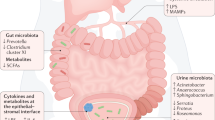
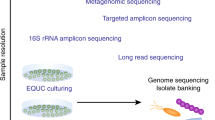
Abstract
Genitourinary cancers account for 20% of cancer instances globally and pose a substantial burden. The microbiome, defined as the ecosystem of organisms that reside within and on the human body, seems to be closely related to multiple cancers. Research on the gut microbiome has yielded substantial insights into the interactions of this entity with the immune system and cancer therapeutic efficacy, whereas the urinary microbiome has been relatively less well-studied. Advances in next-generation sequencing technologies led to new discoveries in the urinary microbiome, which might aid in early detection, risk stratification and personalized treatment strategies in genitourinary cancers. Mechanistic investigations have also suggested a role for the urinary microbiome in modulating the tumour microenvironment and host immune response. For example, distinct urinary microbial signatures have been linked to bladder cancer occurrence and recurrence risk, with specific taxa associated with cytokine production and inflammation. Urinary microbiome signatures have also been explored as potential biomarkers for non-invasive cancer detection. However, challenges remain in standardizing methodologies, validating findings across studies, and establishing causative mechanisms. As investigations into the urinary microbiome continue to evolve, so does the potential for developing microbiome-modulating therapies and enhancing diagnostic capabilities to improve outcomes in patients with genitourinary cancers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Schafer, E. J. et al. Disparities and trends in genitourinary cancer incidence and mortality in the USA. Eur. Urol. 84, 117–126 (2023).
Yafi, F. A. et al. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol. Oncol. 33, 66.e25–31 (2015).
Tan, W. C. & Kanesvaran, R. Current standards and practice changing studies in genitourinary (GU) cancers — a review of studies in localized/early GU cancers. ESMO Open. 7, 100432 (2022).
Gilbert, J. et al. Current understanding of the human microbiome. Nat. Med. 24, 392–400 (2018).
Hou, K. et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 7, 1–28 (2022).
Thursby, E. & Juge, N. Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836 (2017).
Schwabe, R. F. & Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 13, 800–812 (2013).
Nelson, D. E. et al. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. PLoS ONE 5, e14116 (2010).
Dong, Q. et al. The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PLoS ONE 6, e19709 (2011).
Nelson, D. E. et al. Bacterial communities of the coronal sulcus and distal urethra of adolescent males. PLoS ONE 7, e36298 (2012).
Wolfe, A. J. et al. Evidence of uncultivated bacteria in the adult female bladder. J. Clin. Microbiol. 50, 1376–1383 (2012).
Fouts, D. E. et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J. Transl. Med. 10, 174 (2012).
Khasriya, R. et al. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J. Clin. Microbiol. 51, 2054–2062 (2013).
Hilt, E. E. et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J. Clin. Microbiol. 52, 871–876 (2014).
Wolfe, A. J. & Brubaker, L. “Sterile Urine” and the presence of bacteria. Eur. Urol. 68, 173–174 (2015).
Markowski, M. C. et al. The microbiome and genitourinary cancer: a collaborative review. Eur. Urol. 75, 637–646 (2019).
Pohl, H. G. et al. The urine microbiome of healthy men and women differs by urine collection method. Int. Neurourol. J. 24, 41–51 (2020).
Lagier, J.-C. et al. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 16, 540–550 (2018).
Deen, N. S., Ahmed, A., Tasnim, N. T. & Khan, N. Clinical relevance of expanded quantitative urine culture in health and disease. Front. Cell Infect. Microbiol. 13, 1210161 (2023).
Hu, T., Chitnis, N., Monos, D. & Dinh, A. Next-generation sequencing technologies: an overview. Hum. Immunol. 82, 801–811 (2021).
Halderman, A. A. & Lane, A. P. Organism and microbiome analysis: techniques and implications for chronic rhinosinusitis. Otolaryngol. Clin. North. Am. 50, 521–532 (2017).
Tringe, S. G. & Hugenholtz, P. A renaissance for the pioneering 16S rRNA gene. Curr. Opin. Microbiol. 11, 442–446 (2008).
Schloss, P. D. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLOS Comput. Biol. 6, e1000844 (2010).
Clarridge, J. E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17, 840–862 (2004).
Brubaker, L., Putonti, C., Dong, Q. & Wolfe, A. J. The human urobiome. Mamm. Genome 32, 232–238 (2021).
Logsdon, G. A., Vollger, M. R. & Eichler, E. E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 21, 597 (2020).
Gao, B. et al. An introduction to next generation sequencing bioinformatic analysis in gut microbiome studies. Biomolecules 11, 530 (2021).
Mostafa, M. H., Sheweita, S. A. & O’Connor, P. J. Relationship between schistosomiasis and bladder cancer. Clin. Microbiol. Rev. 12, 97–111 (1999).
Adebayo, A. S. et al. The microbiome in urogenital schistosomiasis and induced bladder pathologies. PLoS Negl. Trop. Dis. 11, e0005826 (2017).
Chorbińska, J. et al. Is the urinary and gut microbiome associated with bladder cancer? Clin. Med. Insights Oncol. 17, 11795549231206796 (2023).
Bučević Popović, V. et al. The urinary microbiome associated with bladder cancer. Sci. Rep. 8, 12157 (2018).
Hrbáček, J., Tláskal, V., Čermák, P., Hanáček, V. & Zachoval, R. Bladder cancer is associated with decreased urinary microbiota diversity and alterations in microbial community composition. Urol. Oncol. 41, 107.e15–107.e22 (2023).
Uzelac, M. et al. Urinary microbiome dysbiosis and immune dysregulations as potential diagnostic indicators of bladder cancer. Cancers 16, 394 (2024).
Parra-Grande, M. et al. Profiling the bladder microbiota in patients with bladder cancer. Front Microbiol 12, 718776 (2021).
Pederzoli, F. et al. Sex-specific alterations in the urinary and tissue microbiome in therapy-naïve urothelial bladder cancer patients. Eur. Urol. Oncol. 3, 784–788 (2020).
Qiu, Y. et al. Deciphering the influence of urinary microbiota on FoxP3+ regulatory T cell infiltration and prognosis in Chinese patients with non-muscle-invasive bladder cancer. Hum. Cell 35, 511–521 (2022).
Mai, G. et al. Common core bacterial biomarkers of bladder cancer based on multiple datasets. Biomed. Res. Int. 2019, 4824909 (2019).
Gedefie, A. et al. Acinetobacter baumannii biofilm formation and its role in disease pathogenesis: a review. Infect. Drug Resist. 14, 3711–3719 (2021).
Brossard, K. A. & Campagnari, A. A. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect. Immun. 80, 228–233 (2012).
Zhang, Y., Wang, W., Zhou, H. & Cui, Y. Urinary Eubacterium sp. CAG:581 promotes non-muscle invasive bladder cancer (NMIBC) development through the ECM1/MMP9 pathway. Cancers 15, 809 (2023).
Zeng, J. et al. Alterations in urobiome in patients with bladder cancer and implications for clinical outcome: a single-institution study. Front. Cell Infect. Microbiol. 10, 555508 (2020).
Ślusarczyk, A. et al. Changes in the urinary microbiome after transurethral resection of non-muscle-invasive bladder cancer: insights from a prospective observational study. Ann. Surg. Oncol. 31, 4773–4786 (2024).
Bajic, P., Wolfe, A. J. & Gupta, G. N. Old instillations and new implications for bladder cancer: the urinary microbiome and intravesical BCG. BJU Int. 124, 7–8 (2019).
Ingersoll, M. A. & Albert, M. L. From infection to immunotherapy: host immune responses to bacteria at the bladder mucosa. Mucosal Immunol. 6, 1041–1053 (2013).
Redelman-Sidi, G., Glickman, M. S. & Bochner, B. H. The mechanism of action of BCG therapy for bladder cancer — a current perspective. Nat. Rev. Urol. 11, 153–162 (2014).
McMillan, A., Macklaim, J. M., Burton, J. P. & Reid, G. Adhesion of Lactobacillus iners AB-1 to human fibronectin: a key mediator for persistence in the vagina? Reprod. Sci. 20, 791–796 (2013).
Seow, S. W. et al. Lactobacillus species is more cytotoxic to human bladder cancer cells than Mycobacterium bovis (bacillus Calmette-Guerin). J. Urol. 168, 2236–2239 (2002).
James, C. et al. Impact of intravesical Bacillus Calmette-Guérin and chemotherapy on the bladder microbiome in patients with non-muscle invasive bladder cancer. Front. Cell Infect. Microbiol. 13, 1125809 (2023).
Heidrich, V. et al. The bladder microbiota is not significantly altered by intravesical BCG therapy. Urol Oncol 42, 22.e13–22.e21 (2024).
Pederzoli, F. et al. Is there a detrimental effect of antibiotic therapy in patients with muscle-invasive bladder cancer treated with neoadjuvant pembrolizumab? Eur. Urol. 80, 319–322 (2021).
Febriyanto, T., Muhammad, F., Wijaya, W., Oey, O. & Simadibrata, D. M. Antibiotic use reduces the efficacy of immune checkpoint inhibitors in patients with urothelial carcinoma: a systematic review and meta-analysis. Urol. Oncol. 42, 160.e11–160.e23 (2024).
Ishiyama, Y. et al. Antibiotic use and survival of patients receiving pembrolizumab for chemotherapy-resistant metastatic urothelial carcinoma. Urol. Oncol. 39, 834.e21–834.e28 (2021).
Wu, P. et al. Profiling the urinary microbiota in male patients with bladder cancer in China. Front. Cell Infect. Microbiol. 8, 167 (2018).
Yu, E. Y.-W. et al. Integrative multi-omics analysis for the determination of non-muscle invasive vs. muscle invasive bladder cancer: a pilot study. Curr. Oncol. 29, 5442–5456 (2022).
Roje, B. et al. Gut microbiota carcinogen metabolism causes distal tissue tumours. Nature 632, 1137–1144 (2024).
Pederzoli, F. et al. Stool microbiome signature associated with response to neoadjuvant pembrolizumab in patients with muscle-invasive bladder cancer. European Urology 85, 417–421 (2024).
Bandini, M. et al. Does the administration of preoperative pembrolizumab lead to sustained remission post-cystectomy? First survival outcomes from the PURE-01 study☆. Ann. Oncol. 31, 1755–1763 (2020).
Basile, G. et al. Neoadjuvant pembrolizumab and radical cystectomy in patients with muscle-invasive urothelial bladder cancer: 3-year median follow-up update of PURE-01 trial. Clin. Cancer Res. 28, 5107–5114 (2022).
Bukavina, L. et al. Role of gut microbiome in neoadjuvant chemotherapy response in urothelial carcinoma: a multi-institutional prospective cohort evaluation. Cancer Res. Commun. 4, 1505 (2024).
Chen, C. et al. Urogenital microbiota: potentially important determinant of PD-L1 expression in male patients with non-muscle invasive bladder cancer. BMC Microbiol. 22, 7 (2022).
Wu, C. et al. Urinary microbiome dysbiosis is associated with an inflammatory environment and perturbed fatty acids metabolism in the pathogenesis of bladder cancer. J. Transl. Med. 22, 628 (2024).
Ratajczak, W. et al. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim. Pol. 66, 1–12 (2019).
Yu, H. et al. Urinary microbiota in patients with prostate cancer and benign prostatic hyperplasia. Arch. Med. Sci. 11, 385–394 (2015).
Gonçalves, M. F. M. et al. Microbiota of urine, glans and prostate biopsies in patients with prostate cancer reveals a dysbiosis in the genitourinary system. Cancers 15, 1423 (2023).
Williams, G. D. Two cases of urinary tract infection caused by Propionimicrobium lymphophilum. J. Clin. Microbiol. 53, 3077–3080 (2015).
Shrestha, E. et al. Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. J. Urol. 199, 161–171 (2018).
Pan, S.-Y., Chen, W.-C., Huang, C.-P., Hsu, C. Y. & Chang, Y.-H. The association of prostate cancer and urinary tract infections: a new perspective of prostate cancer pathogenesis. Medicina 59, 483 (2023).
De Marzo, A. M. et al. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 7, 256–269 (2007).
Chagneau, C. V. et al. Uropathogenic E. coli induces DNA damage in the bladder. PLoS Pathog 17, e1009310 (2021).
Wilson, M. R. et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, eaar7785 (2019).
Nougayrède, J.-P. et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006).
Shrestha, E. et al. Oncogenic gene fusions in nonneoplastic precursors as evidence that bacterial infection can initiate prostate cancer. Proc. Natl Acad. Sci. USA 118, e2018976118 (2021).
Olsson, J. et al. Chronic prostatic infection and inflammation by Propionibacterium acnes in a rat prostate infection model. PLoS ONE 7, e51434 (2012).
Shinohara, D. B. et al. A mouse model of chronic prostatic inflammation using a human prostate cancer-derived isolate of Propionibacterium acnes. Prostate 73, 1007–1015 (2013).
Fassi Fehri, L. et al. Prevalence of Propionibacterium acnes in diseased prostates and its inflammatory and transforming activity on prostate epithelial cells. Int. J. Med. Microbiol. 301, 69–78 (2011).
Nickel, J. C. et al. Search for microorganisms in men with urologic chronic pelvic pain syndrome: a culture-independent analysis in the MAPP research network. J. Urol. 194, 127–135 (2015).
Sfanos, K. S., Yegnasubramanian, S., Nelson, W. G. & De Marzo, A. M. The inflammatory microenvironment and microbiome in prostate cancer development. Nat. Rev. Urol. 15, 11–24 (2018).
Ly, L. K. et al. Bacterial steroid-17,20-desmolase is a taxonomically rare enzymatic pathway that converts prednisone to 1,4-androstanediene-3,11,17-trione, a metabolite that causes proliferation of prostate cancer cells. J. Steroid. Biochem. Mol. Biol. 199, 105567 (2020).
Wang, T. et al. An expanded metabolic pathway for androgen production by host-associated bacteria. bioRxiv https://doi.org/10.1101/2024.06.09.598130 (2024).
Maślak, E. et al. A new approach to imaging and rapid microbiome identification for prostate cancer patients undergoing radiotherapy. Biomedicines 10, 1806 (2022).
Helissey, C. et al. Correlation between serum and urine biomarkers and the intensity of acute radiation cystitis in patients treated with radiation therapy for localized prostate cancer: protocol for the radiotoxicity bladder biomarkers (RABBIO) study. JMIR Res. Protoc. 12, e38362 (2023).
Alanee, S. et al. Prospective examination of the changes in the urinary microbiome induced by transrectal biopsy of the prostate using 16S rRNA gene analysis. Prostate Cancer Prostatic Dis. 22, 446–452 (2019).
Chen, V. S. et al. A prospective evaluation of the prostate microbiome in malignant and benign tissue using transperineal biopsy. Prostate 84, 1251–1261 (2024).
Sfanos, K. S. et al. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate 68, 306–320 (2008).
Ahn, H. K., Kim, K., Park, J. & Kim, K. H. Urinary microbiome profile in men with genitourinary malignancies. Invest. Clin. Urol. 63, 569–576 (2022).
Cumpanas, A. D. et al. MP64-02 A pilot study on the dark matter of clear cell renal cell carcinoma: the role of the urinary microbiome. J. Urol. 209, e880 (2023).
Dizman, N. et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: a randomized phase 1 trial. Nat. Med. 28, 704–712 (2022).
Cremonesi, E. et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 67, 1984–1994 (2018).
Heidler, S., Lusuardi, L., Madersbacher, S. & Freibauer, C. The microbiome in benign renal tissue and in renal cell carcinoma. Urol. Int. 104, 247–252 (2019).
Wang, J. et al. Uncovering the microbiota in renal cell carcinoma tissue using 16S rRNA gene sequencing. J. Cancer Res. Clin. Oncol. 147, 481–491 (2021).
Liss, M. A. et al. Microbiome within primary tumor tissue from renal cell carcinoma may be associated with PD-L1 expression of the venous tumor thrombus. Adv Urol 2020, 9068068 (2020).
Kovaleva, O. V. et al. Macrophage phenotype in combination with tumor microbiome composition predicts RCC patients’ survival: a pilot study. Biomedicines 10, 1516 (2022).
Jung, C. E. et al. Benchmarking urine storage and collection conditions for evaluating the female urinary microbiome. Sci Rep 9, 13409 (2019).
Jones, C. B., White, J. R., Ernst, S. E., Sfanos, K. S. & Peiffer, L. B. Incorporation of data from multiple hypervariable regions when analyzing bacterial 16S rRNA gene sequencing data. Front Genet 13, 799615 (2022).
Brubaker, L. et al. Forming consensus to advance urobiome research. mSystems 6, e0137120 (2021).
Author information
Authors and Affiliations
Contributions
N.S. and S.L. researched data for the article. All authors contributed substantially to discussion of the content. N.S. and S.L. wrote the article. All authors reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Urology thanks Alan Wolfe and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- ACE index
-
Estimates species richness with greater sensitivity to rare operational taxonomic units.
- Amplicon sequence variants richness
-
Counts the number of unique amplicon sequence variants richness observed in a sample.
- Bray–Curtis distances
-
Non-phylogenetic distance metric that quantifies dissimilarity between two samples based on species abundance and proportion of shared species.
- Chao1 index
-
Estimates total species richness in a sample by accounting for the presence of rare taxa.
- Faith’s Phylogenetic Diversity test
-
Estimates biodiversity by calculating the total branch length of a phylogenetic tree for species in a sample.
- Gini–Simpson index
-
Calculates the probability that two randomly selected organisms in a sample are different.
- Jaccard
-
Measures the proportion of shared species between two samples, ranging from 0 (completely different) to 1 (identical).
- Linear discriminant analysis effect size
-
Statistical method that identifies and ranks differentially abundant features (for example, taxa) between groups using linear discriminant analysis.
- Observed species index
-
Counts the total number of unique species identified in a sample.
- Operational taxonomic units
-
Groups of closely related microbial sequences used to classify microorganisms based on genetic similarity.
- Pielou’s evenness
-
Measures how evenly species are distributed in a sample, ranging from 0 (uneven) to 1 (perfectly even).
- Shannon index
-
Measures microbial diversity by incorporating species richness and evenness.
- Simpson index
-
Calculates the probability that two randomly selected organisms in a sample are the same.
- Unweighted Unifrac
-
Phylogenetic distance metric that compares microbial communities using the presence or absence of taxa, without abundance data.
- Weighted Unifrac
-
Phylogenetic distance metric that compares microbial communities using the presence and relative abundance of taxa.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, S., Sfanos, K. & Singla, N. The role of the urinary microbiome in genitourinary cancers. Nat Rev Urol (2025). https://doi.org/10.1038/s41585-025-01011-z
Accepted:
Published:
DOI: https://doi.org/10.1038/s41585-025-01011-z