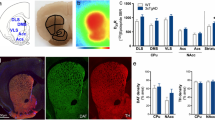
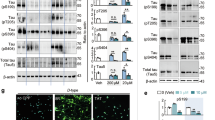
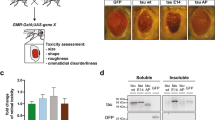
Abstract
Dopamine (DA) is one of the most important neurotransmitters. Its oxidation leads to electrophilic quinone, which covalently modifies nucleophilic residues in proteins, resulting in ‘dopamination’. Individual dopaminated proteins have been studied, most of which were functionally damaged by dopamination. Here, we developed a quantitative chemoproteomic strategy to site-specifically measure proteins’ dopamination. More than 6,000 dopamination sites were quantified. Half-maximal inhibitory concentration values for 63 hypersensitive sites were measured. Among them, hypersensitive dopamination of two cysteines in microtubule-associated protein Tau was biochemically validated and functionally characterized to prevent Tau’s amyloid fibrillation and promote Tau-mediated assembly of microtubules. In addition, endogenous dopamination of Tau in mouse brain was detected through targeted mass spectrometry analysis. Our study not only provides a global portrait of dopamination but also discovers a protective role of DA in regulating the function of Tau, which will enhance our understanding of the physiological and pathological functions of DA in human brain.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The proteomics data (PXD056638) were deposited to the ProteomeXchange. Source data are provided with this paper.
Code availability
The customized code and mathematical algorithm for DIA data analysis and curve fitting were deposited to Zenodo (https://doi.org/10.5281/zenodo.14511272)57.
References
Ehringer, H. & Hornykiewicz, O. Distribution of noradrenaline and dopamine (3-hydroxytyramine) in the human brain and their behavior in diseases of the extrapyramidal system. Klin. Wochenschr. 38, 1236–1239 (1960).
Macedo-Lima, M. & Remage-Healey, L. Dopamine modulation of motor and sensory cortical plasticity among vertebrates. Integr. Comp. Biol. 61, 316–336 (2021).
Sulzer, D., Cragg, S. J. & Rice, M. E. Striatal dopamine neurotransmission: regulation of release and uptake. Basal Ganglia 6, 123–148 (2016).
Segura-Aguilar, J. et al. Protective and toxic roles of dopamine in Parkinson’s disease. J. Neurochem. 129, 898–915 (2014).
Monzani, E. et al. Dopamine, oxidative stress and protein–quinone modifications in Parkinson’s and other neurodegenerative diseases. Angew. Chem. Int. Ed. Engl. 58, 6512–6527 (2019).
Zhou, Z. D. & Lim, T. M. Glutathione conjugates with dopamine-derived quinones to form reactive or non-reactive glutathione-conjugates. Neurochem. Res. 35, 1805–1818 (2010).
Hastings, T. G. The role of dopamine oxidation in mitochondrial dysfunction: implications for Parkinson’s disease. J. Bioenerg. Biomembr. 41, 469–472 (2009).
Zhang, S., Wang, R. & Wang, G. Impact of dopamine oxidation on dopaminergic neurodegeneration. ACS Chem. Neurosci. 10, 945–953 (2019).
Bruning, J. M. et al. Covalent modification and regulation of the nuclear receptor Nurr1 by a dopamine metabolite. Cell. Chem. Biol. 26, 674–685 (2019).
Van Laar, V. S., Mishizen, A. J., Cascio, M. & Hastings, T. G. Proteomic identification of dopamine-conjugated proteins from isolated rat brain mitochondria and SH-SY5Y cells. Neurobiol. Dis. 34, 487–500 (2009).
LaVoie, M. J., Ostaszewski, B. L., Weihofen, A., Schlossmacher, M. G. & Selkoe, D. J. Dopamine covalently modifies and functionally inactivates Parkin. Nat. Med. 11, 1214–1221 (2005).
Yu, G. et al. In vivo protein targets for increased quinoprotein adduct formation in aged substantia nigra. Exp. Neurol. 271, 13–24 (2015).
Hurben, A. K., Erber, L. N., Tretyakova, N. Y. & Doran, T. M. Proteome-wide profiling of cellular targets modified by dopamine metabolites using a bio-orthogonally functionalized catecholamine. ACS Chem. Biol. 16, 2581–2594 (2021).
Bisaglia, M. et al. Dopamine quinones interact with α-synuclein to form unstructured adducts. Biochem. Biophys. Res. Commun. 394, 424–428 (2010).
Girotto, S. et al. Dopamine-derived quinones affect the structure of the redox sensor DJ-1 through modifications at Cys-106 and Cys-53. J. Biol. Chem. 287, 18738–18749 (2012).
Hauser, D. N., Dukes, A. A., Mortimer, A. D. & Hastings, T. G. Dopamine quinone modifies and decreases the abundance of the mitochondrial selenoprotein glutathione peroxidase 4. Free Radic. Biol. Med. 65, 419–427 (2013).
Weerapana, E. et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010).
Wang, C., Weerapana, E., Blewett, M. M. & Cravatt, B. F. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85 (2014).
Backus, K. M. et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016).
Qin, W. et al. S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate. Nat. Chem. Biol. 15, 983–991 (2019).
Abegg, D. et al. Proteome-wide profiling of targets of cysteine reactive small molecules by using ethynyl benziodoxolone reagents. Angew. Chem. Int. Ed. Engl. 54, 10852–10857 (2015).
Szychowski, J. et al. Cleavable biotin probes for labeling of biomolecules via azide–alkyne cycloaddition. J. Am. Chem. Soc. 132, 18351–18360 (2010).
Yang, F., Gao, J., Che, J., Jia, G. & Wang, C. A dimethyl-labeling-based strategy for site-specifically quantitative chemical proteomics. Anal. Chem. 90, 9576–9582 (2018).
Kuljanin, M. et al. Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries. Nat. Biotechnol. 39, 630–641 (2021).
Yang, F., Jia, G., Guo, J., Liu, Y. & Wang, C. Quantitative chemoproteomic profiling with data-independent acquisition-based mass spectrometry. J. Am. Chem. Soc. 144, 901–911 (2022).
Spillantini, M. G. & Goedert, M. Tau protein pathology in neurodegenerative diseases. Trends Neurosci. 21, 428–433 (1998).
Iqbal, K., Liu, F. & Gong, C. X. Tau and neurodegenerative disease: the story so far. Nat. Rev. Neurol. 12, 15–27 (2016).
Wakamatsu, K. et al. The oxidative pathway to dopamine-protein conjugates and their pro-oxidant activities: implications for the neurodegeneration of Parkinson’s disease. Int. J. Mol. Sci. 20, 2575 (2019).
Koike, H. et al. Thimet oligopeptidase cleaves the full-length Alzheimer amyloid precursor protein at a β-secretase cleavage site in COS cells. J. Biochem. 126, 235–242 (1999).
Wang, Y. & Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 5–21 (2016).
Barre, P. & Eliezer, D. Structural transitions in Tau K18 on micelle binding suggest a hierarchy in the efficacy of individual microtubule-binding repeats in filament nucleation. Protein Sci. 22, 1037–1048 (2013).
Kadavath, H. et al. Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl Acad. Sci. USA 112, 7501–7506 (2015).
Moreira, G. G. et al. Dynamic interactions and Ca2+-binding modulate the holdase-type chaperone activity of S100B preventing Tau aggregation and seeding. Nat. Commun. 12, 6292 (2021).
Shelanski, M. L., Gaskin, F. & Cantor, C. R. Microtubule assembly in the absence of added nucleotides. Proc. Natl Acad. Sci. USA 70, 765–768 (1973).
Lee, J. C. & Timasheff, S. N. In vitro reconstitution of calf brain microtubules: effects of solution variables. Biochemistry 16, 1754–1764 (1977).
Tseng, H. C., Lu, Q., Henderson, E. & Graves, D. J. Phosphorylated Tau can promote tubulin assembly. Proc. Natl Acad. Sci. USA 96, 9503–9508 (1999).
Johnson, E. C. B. et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 26, 769–780 (2020).
Budnik, B., Levy, E., Harmange, G. & Slavov, N. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 19, 161–172 (2018).
Lepack, A. E. et al. Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 368, 197–201 (2020).
Zhao, H. et al. α-synuclein dopaminylation presented in plasma of both healthy subjects and Parkinson’s disease patients. Proteomics Clin. Appl. 14, e1900117 (2020).
Ballatore, C., Lee, V. M. & Trojanowski, J. Q. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci. 8, 663–672 (2007).
Arakhamia, T. et al. Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell 180, 633–644 (2020).
Fitzpatrick, A. W. P. et al. Cryo-EM structures of Tau filaments from Alzheimer’s disease. Nature 547, 185–190 (2017).
Falcon, B. et al. Structures of filaments from Pick’s disease reveal a novel Tau protein fold. Nature 561, 137–140 (2018).
Falcon, B. et al. Novel Tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423 (2019).
Falcon, B. et al. Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol. 136, 699–708 (2018).
Arakhamia, T. et al. Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell 184, 6207–6210 (2021).
Shi, Y. et al. Structure-based classification of tauopathies. Nature 598, 359–363 (2021).
Zhang, W. et al. Novel Tau filament fold in corticobasal degeneration. Nature 580, 283–287 (2020).
Lövestam, S. et al. Assembly of recombinant Tau into filaments identical to those of Alzheimer’s disease and chronic traumatic encephalopathy. eLife 11, e76494 (2022).
Maza, J. et al. Enzymatic modification of N-terminal proline residues using phenol derivatives. J. Am. Chem. Soc. 141, 3885–3892 (2019).
Savitski, M. M. et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346, 1255784 (2014).
Barghorn, S., Biernat, J. & Mandelkow, E. Purification of recombinant Tau protein and preparation of Alzheimer-paired helical filaments in vitro. Methods Mol. Biol. 299, 35–51 (2005).
Wang, L. et al. pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21, 2985–2991 (2007).
Lee, W., Tonelli, M. & Markley, J. L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015).
Johnson, B. A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 (2004).
Wang, Q. et al. Quantitative chemoproteomics reveals dopamine’s protective modification of Tau. Zenodo https://doi.org/10.5281/zenodo.14511272 (2024).
Acknowledgements
We thank the Computing Platform of the Center for Life Science for supporting the LC–MS/MS data analysis. We thank B. Ma for help with animal experiments and sample collection. C.W. acknowledges support from the National Natural Science Foundation of China (21925701) and the National Key Research and Development Projects (2022YFA1304700). This work was also supported by the National Natural Science Foundation of China (92153301 and 22321005 to C.W.; 92353302 to S.Z. and D.L.; 22425704, 82188101 and 31872716 to C.L.; 22307001 to W.X.), the Beijing National Laboratory for Molecular Sciences (BNLMS-CXTD-202401 to C.W.), the Science and Technology Commission of Shanghai Municipality (22JC1410400 to C.L.), Chinese Academy of Sciences (CAS) Projects for Young Scientists in Basic Research (YSBR-009 and YSBR-095 to C.L.), the Shanghai Basic Research Pioneer Project (to C.L.), the Shanghai Pilot Program for Basic Research (JCYJ-SHFY-2022-005 to C.L.), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB1060000 to S.Z. and C.L.). We thank the staff members of the NMR System (https://cstr.cn/31129.02.NFPS.NMRSystem) at the National Facility for Protein Science in Shanghai (https://cstr.cn/31129.02.NFPS) for providing technical support and assistance in data collection and analysis. We thank Y. Li’s Lab at Peking University for providing samples. We thank Z. Tan from the C.W. lab and members from F. Gai’s Lab at Peking University for providing technical support and assistance in SPPS experiments. We thank X. Fu from the C.W. lab for providing assistance in structure analysis.
Author information
Authors and Affiliations
Contributions
C.W., C.L., Y.C. and W.X. conceptualized the experiments and supervised the work. Q.W. performed the DIA-ABPP experiment, reactivity characterization, DAyne synthesis, biochemical characterization of dopamination of hTau and MS structural characterization of dopamination of hTau. Z.L. constructed the Tau mutant plasmids and performed the expression and purification of Tau proteins, ThT kinetic assay, TP assay, TEM characterization, NMR structural characterization of dopamination of hTau and sample preparation for the cell seeding assay. S.Z. took part in the NMR data analysis. W.Z. and Z.H. performed the cell seeding assay. Y.W. validated the endogenous dopamination on Tau, including coelution and carrier validation, supervised by W.X. Y.L. performed the computational analysis. F.Y. gave constructive suggestions for the experimental operations and data analysis of DIA-ABPP. Q.W., Z.L, Y.W., S.Z., W.Z., D.L., Z.H., W.X., C.L. and C.W. wrote and revised the manuscript with input from all authors. C.W., C.L. and W.X. acquired funding for the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Reactivity of dopamine (DA) with N-acetyl-cysteine (NAC).
a, Reaction scheme of DA with NAC. b, Base peak of the DA-NAC adducts ([M + H]+ = 315) as analyzed by LC-MS.
Extended Data Fig. 2 Profiling of DA-hypersensitive cysteines through DIA-ABPP.
a, Three independent biological replicate DIA-ABPP experiments showed good correlations between data sets within the same DA concentration or across different DA concentrations. b, Venn diagram showing the number of quantified cysteines from three independent biological replicates of DIA-ABPP experiments at all nine DA concentrations. c, IC50 values of dopaminated cysteines from targets that have been known with DA modifications. #N/A denotes that the IC50 values could not be confidently calculated during the curve fitting step in the corresponding replicate. The asterisks denote cysteine sites modified by probe. d, IC50 curves of the top 4 hyper-sensitive dopaminated cysteines that were quantified from the DIA-ABPP experiments. Curves are drawn with Graphpad Prism 7. Values represent means ± SD from three independent biological replicate experiments. e, Locations of DA-hypersensitive cysteines in the corresponding proteins shown in d. Cysteine side chains are shown in stick (magenta and yellow) and backbones are shown in cartoon (cyan) with PyMOL. Protein names and PDB ids are shown below. f, g, Pie charts illustrating the classifications of structural (f) and functional (g) annotations of the DA-hypersensitive cysteines in UniProt.
Extended Data Fig. 3 Synthesis and structural characterization of DAyne.
a, Chemical synthesis of DAyne. b, 1H-NMR spectrum. 1H NMR (400 MHz, CDCl3) δ ppm 6.81 (d, J = 8.1 Hz, 1H), 6.75 (d, J = 2.1 Hz, 1H), 6.59 (dd, J = 8.0, 2.0 Hz, 1H), 3.53 (t, J = 7.0 Hz, 2H), 2.71 (dt, J = 31.3, 7.0 Hz, 2H), 2.54–2.45 (m, 2H), 2.37 (t, J = 7.3 Hz, 2H), 2.00 (t, J = 2.6 Hz, 1H). c, FTMS spectrum. Calcd. for C13H16NO3+ [M + H]+ m/z, 234.112470; found, 234.112844.
Extended Data Fig. 4 Domain organizations of Tau.
a, Domain schematic of human 3 R and 4 R Tau, of which both contain projection ___domain and a microtubule binding ___domain (top), 3 R and 4 R hTau are characterized by three (R1-R3-R4) or four (R1-R2-R3-R4) microtubule-binding repeats. Protein sequence alignment of the four microtubule-binding repeats from different species are shown at the bottom based on the UniProt. For Human (P10636-1), Mouse (P10637-1), Rhesus macaque (P57786-1), Rat (P19332-1), Goat (O02828-1), Bovine (P29172-1), canonical sequences are shown, besides, Human brain Tau (P10636-8), Mouse brain Tau (P10637-2) and Rat brain Tau (P19332-5) sequences are shown, because these isoforms are widely-researched, and in our study, we used human / mouse brain Tau sequence for illustration. The alignment indicating that the two cysteines (Cys291 and Cys322 in hTau) are highly conserved. NTD, N-terminal ___domain, PRD, proline rich ___domain, CTD, C-terminal ___domain. b, Domain schematic of K18 with the two cysteines labeled. The primary sequence of K18 is shown at the bottom.
Extended Data Fig. 5 Biochemical validation of dopamination in Tau40.
a, Scheme of validating dopamination of hTau by DAyne using in-gel fluorescence imaging. b, Single or double cysteine mutants abolished DAyne’s labeling of Tau40. c, DAyne’s labeling of Tau40 can be competed by native DA. For b and c, fluorescence and Coomassie brilliant blue signals were collected in parallel from the same gel. Similar results were obtained in three independent biological replicates.
Extended Data Fig. 6 Dopamination of hTau is cysteine-dependent as revealed by MS and NMR.
a, b, MS2 spectra of hTau peptides with dopaminated Cys322 in the form of DAQ/LDAC (a) and DHI (b). For a, similar results were observed in two independent biological replicates. For b, similar results were observed in three independent biological replicates. Fragment ions from the DAQ/LDAC and DHI-modified peptide were shown. c, Overlay of the 2D 1H-15N HSQC spectra of 50 μM 15N-labeled K18 C291A (left) and K18 C322A (right) in the presence of 50 μM DA ± 2 nM mTYR with the incubation time of 1 (pink), 3 (orange) and 5 h (blue), respectively. d, Residue-specific intensity ratios (I/I0) from c are shown. The ___domain organization of K18 is shown on top. The two cysteines containing segments at R2 (290KCGS293) and R3 (321KCGS324) of K18 are labeled.
Extended Data Fig. 7 Effects of DA in regulating hTau’s pathological aggregation ability and tubulin polymerization.
a, ThT kinetics of 25 μM K18 in the presence of DA with different concentrations (0, 1, 10 and 100 μM), all samples were added with 83.3 nM mTYR and 10 μM heparin. The error bars denote mean ± SD (n = 3 independent replicates). b, Comparison of the inhibitory effect of DA at different concentrations on K18 aggregation at the end-point from the ThT assay from a. The experiment was performed in three independent replicates, significance analysis was performed through One-way ANOVA with Tukey multiple comparisons test, P value of 0 μM versus 1 was 0.4751, 2.6 × 10−5 and 4.5 × 10−6 respectively, n.s. no significant; *P < 0.05; **P < 0.01; ***P < 0.001. c, TEM images of the K18 samples incubated 0 μM or 100 μM DA from the end-point of the ThT assay in a. Scale bar, 200 nm. d, ThT kinetics of 25 μM Tau40 cysteine mutants (C291A/C322A, C291A and C322A) in the presence of DA with different concentrations (0, 1, 10 and 100 μM), all samples were added with 83.3 nM mTYR and 10 μM heparin. The error bars denote mean ± SD (n = 3 independent replicates). e, f, Dopamination of Tau40 inhibits cell seeding of pathological Tau. e, Representative images of immunocytochemistry staining with K9JA (pan-Tau) and PHF-1 (phospho-Tau at Ser396 and Ser404) antibodies on Tau40-expressing HEK293T cells. f, Quantification of the seeding-induced pathological tau as shown in e. Data are presented as mean ± SEM with three independent biological repeats for each group. Two-tailed unpaired t test was performed to calculate P value; P value was 2.6 × 10−6, *P < 0.05; **P < 0.01; ***P < 0.001. Scale bar, 100 μm. g, Tubulin polymerization effect of DA on Tau40 cysteine mutants. DA-treated Tau40 mutants were prepared by pretreating proteins with 100 μM DA in the presence of mTYR (83.3 nM). Control proteins were prepared in the same condition without adding DA. Tubulins (20 μM) were incubated with these mutants to monitor the polymerization kinetics curves and TEM images of the samples from the end-point of tubulin polymerization are shown below the kinetic curves. Data are presented as mean ± SEM with three independent replicates for each group. Scale bar: 200 nm.
Extended Data Fig. 8 Synthesis and characterization of synthetic standard peptides with DAQ/LDAC modifications.
a, Workflow of synthesis of the CK19 standard peptides with DAQ/LDAC modifications on Cys311. The light or heavy synthetic peptides were incubated with 1 mM LDAC at 37 °C for 2 h, and then desalted prior to LC-MS/MS analysis. Multiple forms of dopaminations were found with peptide spectra match counts listed in the table, including DA, DAQ/LDAC and DHI. b, c, Characterization of the CK19 standard peptides with DAQ/LDAC modifications by LC-MS/MS. The MS1 (b) and MS2 (c) spectra are shown, respectively, which illustrate the isotopic signatures and fragmentation patterns of the modified light (red) and heavy (blue) peptides.
Extended Data Fig. 9 Atomic structures of the fibril cores of Tau filaments extracted from the brains of individuals with neurodegenerative diseases.
a–g, Cryo-EM structures of Tau filaments from the brains of patients with AD, PSP, CTE, GGT, CBD, PID and AGD respectively. The regions containing the two cysteines are zoomed in, and the nearby segments are colored cyan and displayed. AD: Alzheimer’s disease, PSP: progressive supranuclear palsy, CTE: Chronic traumatic encephalopathy, GGT: Globular glial tauopathy, CBD: Corticobasal degeneration, PID: Pick’s disease, AGD: Argyrophilic grain disease. Dopamination of hTau on cysteines cannot be formed in the pathological hTau fibril structures from AD (a), PSP (b), CTE (c), GGT (d), CBD (e) and AGD (g).
Supplementary information
Supplementary Information
Supplementary Dataset 1 and Tables 1–4.
Supplementary Dataset 1
List of the cysteines quantified from the proteomes of mouse brains by DIA-ABPP.
Supplementary Table 1
Primer (5' to 3') list for constructing Tau mutant plasmids.
Supplementary Table 2
Co-IP buffer A formula.
Supplementary Table 3
Inclusion list for standard peptide and co-IP samples.
Supplementary Table 4
Inclusion list for TMT labeling samples.
Source data
Source Data Fig. 1
Unprocessed gels.
Source Data Fig. 2
Unprocessed gels.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed gels.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data and unprocessed images.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Q., Liu, Z., Wang, Y. et al. Quantitative chemoproteomics reveals dopamine’s protective modification of Tau. Nat Chem Biol (2025). https://doi.org/10.1038/s41589-025-01849-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41589-025-01849-9