Abstract
Stromal interaction molecule 1 (STIM1) is critical for store-operated Ca2+ entry (SOCE) and T cell activation. T helper 1 (TH1) cells, which express T-bet (encoded by TBX21), mediate immunity to intracellular pathogens. Although SOCE is known to regulate other TH lineages, its role in Th1 differentiation remains unclear. Here, we report a patient with an intronic loss-of-function mutation in STIM1, which abolishes SOCE and causes immunodeficiency. We demonstrate that SOCE promotes nuclear factor of activated T cells (NFAT) binding to conserved noncoding sequence (CNS)-12 in the TBX21 enhancer and enables NFAT to synergize with STAT1 to mediate TBX21 expression. While SOCE-deficient CD4+ T cells have reduced expression of TBX21 in the absence of interleukin-12 (IL-12), their expression of IL-12 receptors β1 and β2 is increased, sensitizing them to IL-12 signaling and allowing IL-12 to rescue T-bet expression. Our study reveals that the STIM1-SOCE–NFAT signaling axis is essential for the differentiation of Th1 cells depending on the cytokine milieu.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA-seq and ATAC-seq data generated for this study have been deposited in the GEO database under accession numbers GSE179625 and GSE253187. Publicly available datasets that were reanalyzed for this study are: GSE64409, GSE98726, GSE67443, GSE183883, GSE207265, GSE144586, GSE204946, GSE96724, GSE92531, GSE67451, GSE93014, GSE157597 and GSE161096. Source dataare provided with this paper.
References
Szabo, S. J. et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000).
Lazarevic, V. & Glimcher, L. H. T-bet in disease. Nat. Immunol. 12, 597–606 (2011).
Yang, R. et al. Human T-bet governs innate and innate-like adaptive IFN-gamma immunity against mycobacteria. Cell 183, 1826–1847.e1831 (2020).
Pritchard, G. H., Kedl, R. M. & Hunter, C. A. The evolving role of T-bet in resistance to infection. Nat. Rev. Immunol. 19, 398–410 (2019).
Lighvani, A. A. et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc. Natl Acad. Sci. USA 98, 15137–15142 (2001).
Afkarian, M. et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4(+) T cells. Nat. Immunol. 3, 549–557 (2002).
Schulz, E. G., Mariani, L., Radbruch, A. & Hofer, T. Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-gamma and interleukin-12. Immunity 30, 673–683 (2009).
Szabo, S. J., Dighe, A. S., Gubler, U. & Murphy, K. M. Regulation of the interleukin (IL)-12R beta 2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J. Exp. Med. 185, 817–824 (1997).
Sekiya, T. & Yoshimura, A. In vitro Th differentiation protocol. Methods Mol. Biol. 1344, 183–191 (2016).
van de Berg, P. J. et al. Human cytomegalovirus induces systemic immune activation characterized by a type 1 cytokine signature. J. Infect. Dis. 202, 690–699 (2010).
Yu, H. R. et al. IL-12-independent Th1 polarization in human mononuclear cells infected with varicella-zoster virus. Eur. J. Immunol. 35, 3664–3672 (2005).
Krueger, P. D. et al. Two sequential activation modules control the differentiation of protective T helper-1 (Th1) cells. Immunity 54, 687–701.e684 (2021).
Nembrini, C., Abel, B., Kopf, M. & Marsland, B. J. Strong TCR signaling, TLR ligands, and cytokine redundancies ensure robust development of type 1 effector T cells. J. Immunol. 176, 7180–7188 (2006).
Lee, H. H. et al. Delayed maturation of an IL-12-producing dendritic cell subset explains the early Th2 bias in neonatal immunity. J. Exp. Med. 205, 2269–2280 (2008).
Feske, S. Calcium signalling in lymphocyte activation and disease. Nat. Rev. Immunol. 7, 690–702 (2007).
Hogan, P. G., Lewis, R. S. & Rao, A. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu. Rev. Immunol. 28, 491–533 (2010).
Vaeth, M., Kahlfuss, S. & Feske, S. CRAC channels and calcium signaling in T cell-mediated immunity. Trends Immunol. 41, 878–901 (2020).
Placek, K. et al. Integration of distinct intracellular signaling pathways at distal regulatory elements directs T-bet expression in human CD4(+) T cells. J. Immunol. 183, 7743–7751 (2009).
Kiani, A. et al. Regulation of interferon-gamma gene expression by nuclear factor of activated T cells. Blood 98, 1480–1488 (2001).
Ma, J., McCarl, C. A., Khalil, S., Luthy, K. & Feske, S. T-cell-specific deletion of STIM1 and STIM2 protects mice from EAE by impairing the effector functions of Th1 and Th17 cells. Eur. J. Immunol. 40, 3028–3042 (2010).
Dietz, L. et al. NFAT1 deficit and NFAT2 deficit attenuate EAE via different mechanisms. Eur. J. Immunol. 45, 1377–1389 (2015).
Vaeth, M. et al. Store-operated Ca2+ entry controls clonal expansion of T cells through metabolic reprogramming. Immunity 47, 664–679 (2017).
Gamadia, L. E. et al. Primary immune responses to human CMV: a critical role for IFN-gamma-producing CD4(+) T cells in protection against CMV disease. Blood 101, 2686–2692 (2003).
Weinberg, A. et al. Varicella-zoster virus-specific immune responses to herpes zoster in elderly participants in a trial of a clinically effective zoster vaccine. J. Infect. Dis. 200, 1068–1077 (2009).
Malley, R. et al. CD4+ T cells mediate antibody-independent acquired immunity to pneumococcal colonization. Proc. Natl Acad. Sci. USA 102, 4848–4853 (2005).
Feske, S., Giltnane, J., Dolmetsch, R., Staudt, L. M. & Rao, A. Gene regulation mediated by calcium signals in T lymphocytes. Nat. Immunol. 2, 316–324 (2001).
Hermann-Kleiter, N. & Baier, G. NFAT pulls the strings during CD4+ T helper cell effector functions. Blood 115, 2989–2997 (2010).
Vaeth, M. et al. Store-operated Ca(2+) entry in follicular T cells controls humoral immune responses and autoimmunity. Immunity 44, 1350–1364 (2016).
Wang, Y. H. et al. Distinct roles of ORAI1 in T cell-mediated allergic airway inflammation and immunity to influenza A virus infection. Sci. Adv. 8, eabn6552 (2022).
Vaeth, M. et al. ORAI2 modulates store-operated calcium entry and T cell-mediated immunity. Nat. Commun. 8, 14714 (2017).
Kim, K. D. et al. Calcium signaling via Orai1 is essential for induction of the nuclear orphan receptor pathway to drive Th17 differentiation. J. Immunol. 192, 110–122 (2014).
Zhu, J. F. et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity 37, 660–673 (2012).
Oh-Hora, M. et al. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 9, 432–443 (2008).
Martinez, G. J. et al. The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity 42, 265–278 (2015).
Klein-Hessling, S. et al. NFATc1 controls the cytotoxicity of CD8(+) T cells. Nat. Commun. 8, 511 (2017).
Fang, D. et al. Differential regulation of transcription factor T-bet induction during NK cell development and T helper-1 cell differentiation. Immunity 55, 639–655.e637 (2022).
Raisner, R. et al. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 24, 1722–1729 (2018).
Yukawa, M. et al. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J. Exp. Med. 217, e20182009 (2020).
van Rietschoten, J. G. I. et al. Silencer activity of NFATc2 in the interleukin-12 receptor beta 2 proximal promoter in human T helper cells. J. Biol. Chem. 276, 34509–34516 (2001).
Baksh, S. et al. NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol. Cell 10, 1071–1081 (2002).
Wei, G. et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35, 299–311 (2011).
Cui, K. et al. Restraint of IFN-gamma expression through a distal silencer CNS-28 for tissue homeostasis. Immunity 56, 944–958.e946 (2023).
Schijns, V. E. C. J. et al. Mice lacking IL-12 develop polarized Th1 cells during viral infection. J. Immunol. 160, 3958–3964 (1998).
Oxenius, A., Karrer, U., Zinkernagel, R. M. & Hengartner, H. IL-12 is not required for induction of type 1 cytokine responses in viral infections. J. Immunol. 162, 965–973 (1999).
Fieschi, C. & Casanova, J. L. The role of interleukin-12 in human infectious diseases: only a faint signature. Eur. J. Immunol. 33, 1461–1464 (2003).
Upham, J. W. et al. Development of interleukin-12-producing capacity throughout childhood. Infect. Immun. 70, 6583–6588 (2002).
Arulanandam, B. P., Van Cleave, V. H. & Metzger, D. W. IL-12 is a potent neonatal vaccine adjuvant. Eur. J. Immunol. 29, 256–264 (1999).
Metzger, D. W. Interleukin-12 as an adjuvant for induction of protective antibody responses. Cytokine 52, 102–107 (2010).
Iwata, S. et al. The transcription factor T-bet limits amplification of type I IFN transcriptome and circuitry in T helper 1 cells. Immunity 46, 983–991.e984 (2017).
Kaufmann, U. et al. Selective ORAI1 inhibition ameliorates autoimmune central nervous system inflammation by suppressing effector but not regulatory T cell function. J. Immunol. 196, 573–585 (2016).
Tsvilovskyy, V. et al. Deletion of Orai2 augments endogenous CRAC currents and degranulation in mast cells leading to enhanced anaphylaxis. Cell Calcium 71, 24–33 (2018).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Desmet, F. O. et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 (2009).
Yeo, G. & Burge, C. B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11, 377–394 (2004).
Jaganathan, K. et al. Predicting splicing from primary sequence with deep learning. Cell 176, 535–548.e524 (2019).
Erdogmus, S. et al. Cavbeta1 regulates T cell expansion and apoptosis independently of voltage-gated Ca(2+) channel function. Nat. Commun. 13, 2033 (2022).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Qin, Q. et al. LISA: inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol. 21, 32 (2020).
Wickham, H. et al. Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 (2019).
Meadows, S. K. et al. Epitope tagging ChIP-seq of DNA binding proteins using CETCh-seq. Methods Mol. Biol. 2117, 3–34 (2020).
Picard, C. et al. Brief report: STIM1 mutation associated with a syndrome of immunodeficiency and autoimmunity. New Engl. J. Med. 360, 1971–1980 (2009).
Acknowledgements
We acknowledge technical support from the Genome Technology Center (grant no. RRID: SCR_017929), and Cytometry and Cell Sorting Laboratory (grant no. RRID: SCR_019179) at NYU Langone Health. We thank the genomics and bioinformatics cores at WCMQ for support with the WGS and initial analyses. Both cores at supported by the Biomedical Research Program (BMRP) program funded by Qatar Foundation. We thank J. Wang (NYUSOM) for providing the 3A9 cells used in this study. This study was funded by National Institutes of Health (NIH) grant nos. AI097302 and AI130143 to S.F., a grant from the Qatar Foundation to K.M. under the BMRP at Weill Cornell Medicine Qatar. Additional funding was provided by postdoctoral fellowships from the German Research Foundation (DFG) to S.K. (grant no. KA 4514/1-1) and an NIH F30 training grant no. AI164803 (to A.Y.T.).
Author information
Authors and Affiliations
Contributions
L.Z., Y.-H.W., S.K., J.Y., L.N. and M.J. conducted experiments. L.Z., M.J., S.K., A.Y.T., M.M., D.R., D.P. and K.H., analyzed data and interpreted results. K.M. supervised WGS and interpreted results. L.Z., Y.-H.W., S.K. and S.F., designed experiments. T.E.K. and M.C. provided patient data and blood samples. L.Z. and S.F. wrote the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.F. is a scientific cofounder and consultant of Calcimedica, and an inventor on a patent Regulators of NFAT (WO/2007/081804) related to this paper. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: L. A. Dempsey, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Clinical findings in patient with STIM1 c.497+776A>G mutation.
a, Anhidrosis of palm after exertion. b, Persistent dental hypoplasia of newly formed adult teeth of the PT at 6 years of age. c, Dilation of pupils in both eyes.
Extended Data Fig. 2 Altered gene expression in CD4+ T cells of STIM1 deficient patient.
a, Principal component analysis (PCA) of differentially expressed genes (DEGs) detected by RNA-Seq. CD4+ T cells from the patient (PT) with STIM1 c.497+776 A > G mutation and a healthy donor (HD) were left unstimulated or stimulated with anti-CD3/28 for 6 hours. b, Venn diagram of up- and down-regulated DEGs after stimulation of CD4+ T cells from HD and the PT. An adjusted P value < 0.05 and absolute log2 fold change (LFC) > 0.5 were used as cut-off. c, Gene set enrichment analysis (GSEA) of DEGs in CD4+ T cells from the PT and a HD. Normalized enrichment score (NES) and adjusted P values are as indicated for the following datasets: NFAT TF pathway (M60: PID), P < 0.01; alpha/beta T cell differentiation (GO: 0046632), P < 0.05; Hallmark IFN-gamma response (M5913), P < 0.001. d, LFC in expression levels of DEGs related to the NFAT TF pathway (M60: PID). Data are from one experiment with 2 samples per cohort and stimulation condition. e, mRNA levels of CD44, NR4A1/Nur77 and CD38 in CD4+ T cells from the PT, his father (Fa), mother (Mo) and a HD stimulated with anti-CD3/28 for 6 hours and analyzed by RNA-seq.
Extended Data Fig. 3 Attenuated IFN-γ and T-bet response in CD4+ T cells of STIM1 deficient patient.
a, mRNA levels of IFNG, IL2 and TNF in CD4+ T cells from the patient (PT), his father (Fa), mother (Mo) and a HD stimulated with anti-CD3/28 for 6 hours and analyzed by RNA-seq. b, Metascape analysis of all downregulated differentially expressed genes (DEGs) in HD, Fa, Mo vs. the PT. c, IPA upstream regulator analysis of DEGs in CD4+ T cells from the PT vs. all three controls (HD, Fa and Mo) ranked by P value and separated by upstream signaling molecules and transcription factors. Colors indicate the activation Z score. d, Epigenetic Landscape In Silico deletion Analysis (LISA) of transcriptional regulators of downregulated DEGs in T cells from the PT vs. all three controls (HD, Fa and Mo) ranked by P value. Note that LISA uses different ChIP-Seq datasets to identify transcriptional regulators; T-bet is identified as ‘T’ using GSE81881 (human Th1 cells) indicated here as T-bet*, and as Tbx21 using GSE33802 (mouse Th1 cells) indicated as Tbx21#. Fold changes were shrunk using the apeglm method. DEGs were considered significant if shrinkage of the log2 fold change (LFC) absolute value was > 0.5, and the adjusted P value < 0.05.
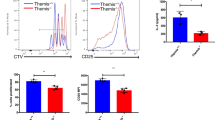
Extended Data Fig. 4 SOCE is required for T-bet expression in human and mouse CD4+ T cells.
a, Venn Diagram (left) of differential gene expression of potential T-bet targets in anti-CD3/28 stimulated CD4+ T cells (6 hours) from the STIM1 deficient PT and a HD (left). T-bet targets were identified by (i) T-bet binding using ChIP-Seq, (ii) positive regulation by T-bet using RNA-Seq, and (iii) reduction of H3K4me1 histone methylation in T-bet-deficient cells. Z scores of mRNA expression of T-bet target gene that fulfill all three selection criteria (right). b, GSEA analysis (left) and heatmap of DEGs (right) related to Th1 specific genes. c, Cytokine expression in CD4+ T cells from a HD and the PT restimulated with 20 nM PMA and 1 μM ionomycin for 6 hours. Bar graphs showing relative median fluorescence intensities (rel. MFI) in the patients T cells normalized to HD. d, Intracellular Ca2+ levels in HD T cells treated with DMSO or 1 μM BTP2. Cells were loaded with 2 μM Fura-2 and stimulated with 1 μM Thapsigargin (TG) in Ca2+ free Ringer solution, followed by perfusion with 2 mM extracellular Ca2+. e, T-bet protein levels in anti-CD3/28 stimulated CD4+ T cells from a HD with DMSO or 1 μM BTP2 (24 hours). Bar graphs showing relative delta MFI (rel. ΔMFI) of T-bet levels in stimulated HD T cells normalized to cells treated with DMSO. f, Intracellular Ca2+ levels in murine WT CD4+ T cells treated with DMSO or 1 μM BTP-2. g, T-bet protein levels in anti-CD3/28 stimulated CD4+ T cells from WT mice with DMSO or 1 μM BTP-2. Data are the means ± SEM for c, e and g. Data are from at least 3 independent experiments. Statistical analysis is performed by two-sided Student’s t test. * P < 0.05.
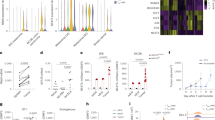
Extended Data Fig. 5 Regulation of TBX21 expression by CNS-12.
a, Analysis of chromatin accessibility of Tbx21 locus in murine CD4+ T cell by DNase I hypersensitive sites sequencing (DNase-seq). Cells were stimulated with 20 ng/ml phorbol myristate acetate (PMA) and 2 μM calcium ionophore (CaI) for 4 hours (GSE67451). b, Analysis of chromatin accessibility of Tbx21 locus in murine CD8+ T cells by ATAC-Seq. Cells were stimulated with 10 ng/ml PMA and 0.5 μM ionomycin (Iono) or left unstimulated (resting) for 2 hours with or without 2 μM cyclosporin A (CsA) (GSE93014). c, Analysis of chromatin accessibility, RNA polymerase II (pol II) and H3K27ac binding in TBX21 gene locus in human CD4+ and CD8+ T cells. ATAC-seq data of human naive CD4+ T cells with or without anti-CD3/CD28 for 1 day (Top, GSE161096). ATAC-seq data of human naïve CD8+ T cells with or without anti-CD3/CD28 for 2 days (Middle, GSE212699). ChIP-Seq analysis of Pol II and H3K27ac binding to the TBX21 gene locus in human T cells activated with anti-CD3/CD28 Dynabeads for 24 hours (Bottom, GSE183883). d, Analysis of NFATc2 binding to CNS-12 of Tbx21 in CD4+ T cells from WT and Stim1/2CD4 mice by ChIP-qPCR. T cells were stimulated with anti-CD3/CD28 and cultured for 5 days, then restimulated with 20 nM PMA and 1 μM ionomycin for 1 hour followed by ChIP-qPCR. e, ChIP-Seq analysis of p300, H3K27ac and JUNB binding to the Tbx21 gene locus in mouse CD4+ T cells. Murine CD4+ T cell stimulated with anti-CD3/CD28 antibodies for 5 days (Top, GSE207265) or with 20 ng/ml phorbol myristate acetate (PMA) and 2 μM calcium ionophore (CaI) for 4 hours (Bottom, GSE67443). The statistical significance of differences between ChIP-seq peaks of test and control groups was calculated using MACS2 v2.1.1., and for ATAC-seq peaks using the DESEQ2 package. ***P < 0.001; **P < 0.01; *P < 0.05.
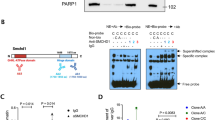
Extended Data Fig. 6 Effects of NFAT and STAT1 on Tbx21 transcription.
a, Two-step model of T-bet expression. b-c, Representative flow cytometry plots (left) and relative median fluorescence intensities (Rel. MFI, right) of T-bet (b), and percentage of T-bet+ cells (c). Murine WT CD4+ T cells were pretreated with DMSO or 1 μM FK506 for 30 min and then stimulated with anti-CD3/CD28 antibodies with or without exogenous IL-12 (10 ng/ml) or IFN-γ (100 ng/ml) and/or anti-IL-4 antibodies (5 ug/ml) for 3-4 days. The bar graph in (b) shows relative MFI of T-bet normalized to T-bet levels in DMSO-treated CD4+ T cells without exogenous IL-12 or IFN-γ. d, T-bet protein levels in CD4+ T cells from a HD and the PT with STIM1 mutation stimulated with anti-CD3/CD28 for 4 days with or without exogenous IFN-γ (100 ng/ml). Shown are relative delta MFI (Rel. ΔMFI) levels of T-bet normalized to T-bet in HD T cells without IFN-γ. e-f, Tbx21 mRNA (e) and T-bet protein (f) levels in mouse WT CD4+ T cells pretreated with anti-IFN-γ antibodies or left untreated for 1 hour followed by anti-CD3/CD28 stimulation for 24 hours (RT-qPCR) or 3-4 days (flow cytometry). The bar graph in (f) shows relative MFI of T-bet normalized to T-bet levels in DMSO-treated CD4+ T cells. g, ChIP-seq analysis of H3K27ac binding to the Tbx21 locus in mouse CD4+ T cells. Cells were activated by anti-CD3/CD28 stimulation either under neutral conditions (nc, with anti-IFN-γ and anti-IL-4 antibodies) or treatment with 100 ng/ml IFN-γ for 3 days (GSE96724). h, ChIP-Seq analysis of H3K27ac binding to the Tbx21 locus in murine naïve CD4+ T cells, Th1, Th2 and Th17 cells (GSE144586). Data are the means ± SEM for b to f. Data are from at least 3 independent experiments. Statistical analysis is performed by two-sided Student’s t test. *** P < 0.001; **P < 0.01; *P < 0.05.
Extended Data Fig. 7 Regulation of Il12rb1 and Il12rb2 genes.
a-b, ChIP-Seq analysis of HDAC1, 4 and 7 binding to Il12rb1 (a) and Il12rb2 (b) loci in mouse Th17 cells (GSE92531). Boxes highlight potential gene regulatory regions. c, Analysis of HDAC1 binding to the promoter of Il12rb1 by ChIP-qPCR using mouse CD4+ T cells. Cells from WT mice were stimulated with anti-CD3/CD28 for 5 days, incubated with DMSO or 1 μM FK506 for 30 min and restimulated with 20 nM PMA and 1 μM ionomycin for 1 hour. d, Il12rb1 and Il12rb2 mRNA levels in CD4+ T cells from WT or Stim1/2CD4Cre mice that were cultured with DMSO or 100 nM TSA for 24 hours in the presence of 10 ng/ml IL-12. e, Analysis of chromatin accessibility, T-bet binding and GATA3 binding to the Il12rb2 locus. Top two rows show ATAC-Seq data from CD4+ T cells of WT OTII (WT) and Orai1/2CD4 (Orai1fl/fl Orai2−/− Cd4Cre) OT-II mice that had been injected into TCRα−/− host mice followed by infection with the PR8-OVA strain of influenza A virus (IAV) for 8 days. The middle two rows show ATAC-Seq data of CD8+ T cells from WT mice that were left untreated or pretreated with 2 μM cyclosporin A (CsA) for 15 mins and stimulated with 10 ng/ml PMA and 0.5 μM ionomycin (iono) or left unstimulated (resting) for 2 hours (GSE93014). Bottom rows show ChIP-Seq data from naïve mouse CD4+ T cells and Th1 cells (GSE204946). Boxes highlight potential gene regulatory regions. The table in e (right) summarizes information about chromatin accessibility and TF binding to the Il12rb2 locus. Data in c-d are the means ± SEM; statistical analysis was performed by two-sided Student’s t test. ***P < 0.001; **P < 0.01; *P < 0.05.
Extended Data Fig. 8 Model of SOCE- and NFAT-dependent T-bet expression and Th1 differentiation.
In CD4+ T cells, TCR engagement activates STIM1 to induces SOCE through ORAI1 Ca2+ channels. Increased intracellular Ca2+ activates NFAT to promote the production of IFN-γ, which binds to the IFN-γ receptor (IFNGR) and activates STAT1. Moreover, NFAT directly binds to CNS-12 of TBX21. STAT1 and NFAT synergize to induce T-bet expression and thus, Th1 differentiation. NFAT also binds to IL12RB1 and IL12RB2 genes and inhibits their expression, likely by acting as a partner of HDAC proteins (IL12RB1) and other negative regulators (IL12RB2). Lack of SOCE following TCR stimulation impairs T-bet expression when IL-12 is not present, but sensitizes T cells to IL-12 signaling by enhancing the expression of IL-12Rβ1 and IL-12Rβ2, thus promoting TBX21 expression and Th1 differentiation when IL-12 is available. Created with BioRender.com.
Supplementary information
Supplementary Information
Supplementary Note, Figs. 1–5 and Tables 1–6.
Supplementary Data Table 1
qPCR primers.
Supplementary Data 2
Source data for supplementary figures.
Source data
Source Data Figs. 1–7 and Extended Data Figs. 2–7
Statistical source data in one file.
Source Data Figs. 1 and 2
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhong, L., Wang, YH., Kahlfuss, S. et al. STIM1-mediated NFAT signaling synergizes with STAT1 to control T-bet expression and TH1 differentiation. Nat Immunol 26, 484–496 (2025). https://doi.org/10.1038/s41590-025-02089-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-025-02089-8