Abstract
Much remains unknown regarding T follicular helper 17 (TFH17) cells commonly found in autoimmune patients. We previously showed that (and here ask why) egress of gut segmented filamentous bacteria (SFB)-induced TFH cells from Peyer’s patches (PP) to systemic sites promotes arthritis. We found splenic TFH17 cells are gut derived. Functional analyses using fate-mapping mice revealed a c-Maf-dependent and SFB-induced TH17-to-TFH cell reprogramming that dominantly occurs in PPs. Unlike conventional TFH cells, TH17-derived TFH cells are highly migratory and atypically concentrated in the dark zone of germinal centers (GCs). Compared to conventional TFH cells, TH17-derived TFH cells express higher levels of TFH-associated functional molecules and more robustly conjugate with B cells. Gain- and loss-of-function studies demonstrated their dominance in promoting GC B cells and arthritis. Notably, murine gut TH17-derived TFH signatures exist in rheumatoid arthritis patients. Thus, gut T cell plasticity generates atypical, potent TFH cells promoting systemic autoimmunity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Single-cell sequencing data supporting the results of this study have been deposited in the GEO under accession numbers GSE259259, GSE259301, GSE279838 and GSE290034. Publicly available datasets reanalyzed in the study can be accessed under this accession link: https://ngdc.cncb.ac.cn/gsa-human/browse/HRA000155. The ontology gene sets (MSigDB version v2023.2) used in the GSEA are available at https://www.gsea-msigdb.org/gsea/index.jsp. Source data are provided with this paper.
Code availability
Scripts used to analyze the RA data were generated by R 4.1 or 4.3.2 and Seurat 4.2.0 or 5.0.3, and have been deposited with the link https://github.com/Wu-lab-script/Wu_Lab.
References
Bach, J. F. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 347, 911–920 (2002).
Victora, G. D. & Nussenzweig, M. C. Germinal centers. Annu Rev. Immunol. 40, 413–442 (2022).
Crotty, S. T follicular helper cell biology: a decade of discovery and diseases. Immunity 50, 1132–1148 (2019).
Ueno, H., Banchereau, J. & Vinuesa, C. G. Pathophysiology of T follicular helper cells in humans and mice. Nat. Immunol. 16, 142–152 (2015).
Seth, A. & Craft, J. Spatial and functional heterogeneity of follicular helper T cells in autoimmunity. Curr. Opin. Immunol. 61, 1–9 (2019).
Victora, G. D. et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605 (2010).
Lee, S. E., Rudd, B. D. & Smith, N. L. Fate-mapping mice: new tools and technology for immune discovery. Trends Immunol. 43, 195–209 (2022).
Jacobsen, J. T. et al. Expression of Foxp3 by T follicular helper cells in end-stage germinal centers. Science 373, eabe5146 (2021).
Al Khabouri, S. & Gerlach, C. T cell fate mapping and lineage tracing technologies probing clonal aspects underlying the generation of CD8 T cell subsets. Scand. J. Immunol. 92, e12983 (2020).
Fang, D. et al. Transient T-bet expression functionally specifies a distinct T follicular helper subset. J. Exp. Med. 215, 2705–2714 (2018).
Hirota, K. et al. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 14, 372–379 (2013).
Ruff, W. E., Greiling, T. M. & Kriegel, M. A. Host-microbiota interactions in immune-mediated diseases. Nat. Rev. Microbiol. 18, 521–538 (2020).
Wu, H. J. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Teng, F. et al. Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer’s patch T follicular helper cells. Immunity 44, 875–888 (2016).
Huang, H., Benoist, C. & Mathis, D. Rituximab specifically depletes short-lived autoreactive plasma cells in a mouse model of inflammatory arthritis. Proc. Natl Acad. Sci. USA 107, 4658–4663 (2010).
Lee, J. Y. et al. The transcription factor KLF2 restrains CD4+ T follicular helper cell differentiation. Immunity 42, 252–264 (2015).
Mora, J. R. et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature 424, 88–93 (2003).
Iwata, M. et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity 21, 527–538 (2004).
Ansaldo, E. et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184 (2019).
Andersson, A. et al. Estrogen regulates T helper 17 phenotype and localization in experimental autoimmune arthritis. Arthritis Res Ther. 17, 32 (2015).
Mullershausen, F. et al. Persistent signaling induced by FTY720-phosphate is mediated by internalized S1P1 receptors. Nat. Chem. Biol. 5, 428–434 (2009).
Baeyens, A., Fang, V., Chen, C. & Schwab, S. R. Exit strategies: S1P signaling and T cell migration. Trends Immunol. 36, 778–787 (2015).
Yang, Y. et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156 (2014).
Bradley, C. P. et al. Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis Th17 cells expressing dual TCRs. Cell Host Microbe 22, 697–704 (2017).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
LaBranche, T. P. et al. Characterization of the KRN cell transfer model of rheumatoid arthritis (KRN-CTM), a chronic yet synchronized version of the K/BxN mouse. Am. J. Pathol. 177, 1388–1396 (2010).
Hall, J. A. et al. Transcription factor RORα enforces stability of the Th17 cell effector program by binding to a Rorc cis-regulatory element. Immunity 55, 2027–2043 (2022).
Ivanov, I. I. et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Vacchio, M. S. et al. A Thpok-directed transcriptional circuitry promotes Bcl6 and Maf expression to orchestrate T follicular helper differentiation. Immunity 51, 465–478 (2019).
Kroenke, M. A. et al. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol. 188, 3734–3744 (2012).
Reinhardt, R. L., Liang, H. E. & Locksley, R. M. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10, 385–393 (2009).
Yeh, C. H., Finney, J., Okada, T., Kurosaki, T. & Kelsoe, G. Primary germinal center-resident T follicular helper cells are a physiologically distinct subset of CXCR5hiPD-1hi T follicular helper cells. Immunity 55, 272–289.e277 (2022).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Crotty, S. Follicular helper CD4 T cells (TFH). Annu Rev. Immunol. 29, 621–663 (2011).
Zotos, D. et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J. Exp. Med. 207, 365–378 (2010).
Linterman, M. A. et al. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J. Exp. Med. 207, 353–363 (2010).
Misiak, J., Tarte, K. & Amé-Thomas, P. in T Follicular Helper Cells: Methods and Protocols (eds Espéli, M. & Linterman, M.) 163–173 (Springer, 2015).
Bauquet, A. T. et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10, 167–175 (2009).
Wu, X. et al. Single-cell sequencing of immune cells from anticitrullinated peptide antibody positive and negative rheumatoid arthritis. Nat. Commun. 12, 4977 (2021).
Yang, X. O. et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 28, 29–39 (2008).
Gaffen, S. L., Jain, R., Garg, A. V. & Cua, D. J. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol. 14, 585–600 (2014).
Lavelle, E. C. & Ward, R. W. Mucosal vaccines: fortifying the frontiers. Nat. Rev. Immunol. 22, 236–250 (2022).
Allen, C. D. et al. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 5, 943–952 (2004).
Cinamon, G. et al. Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nat. Immunol. 5, 713–720 (2004).
Vinuesa, C. G., Linterman, M. A., Yu, D. & MacLennan, I. C. Follicular helper T cells. Annu. Rev. Immunol. 34, 335–368 (2016).
Silva-Cayetano, A. et al. Spatial dysregulation of T follicular helper cells impairs vaccine responses in aging. Nat. Immunol. 24, 1124–1137 (2023).
Goronzy, J. J. & Weyand, C. M. Immune aging and autoimmunity. Cell. Mol. Life Sci. 69, 1615–1623 (2012).
Volkov, M., van Schie, K. A. & van der Woude, D. Autoantibodies and B cells: the ABC of rheumatoid arthritis pathophysiology. Immunol. Rev. 294, 148–163 (2020).
Vergroesen, R. D. et al. B-cell receptor sequencing of anti-citrullinated protein antibody (ACPA) IgG-expressing B cells indicates a selective advantage for the introduction of N-glycosylation sites during somatic hypermutation. Ann. Rheum. Dis. 77, 956–958 (2018).
Chang, Y. et al. TGF-β specifies TFH versus TH17 cell fates in murine CD4+ T cells through c-Maf. Sci. Immunol. 9, eadd4818 (2024).
Gabrysova, L. et al. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4+ T cells. Nat. Immunol. 19, 497–507 (2018).
Brockmann, L. et al. Intestinal microbiota-specific Th17 cells possess regulatory properties and suppress effector T cells via c-MAF and IL-10. Immunity 56, 2719–2735 (2023).
Heath, W. R. et al. Expression of two T cell receptor alpha chains on the surface of normal murine T cells. Eur. J. Immunol. 25, 1617–1623 (1995).
Padovan, E. et al. Expression of two T cell receptor alpha chains: dual receptor T cells. Science 262, 422–424 (1993).
Tlustochowicz, W. et al. Efficacy and safety of subcutaneous and intravenous loading dose regimens of secukinumab in patients with active rheumatoid arthritis: results from a randomized phase II study. J. Rheumatol. 43, 495–503 (2016).
Genovese, M. C. et al. Efficacy and safety of secukinumab in patients with rheumatoid arthritis: a phase II, dose-finding, double-blind, randomised, placebo controlled study. Ann. Rheum. Dis. 72, 863–869 (2013).
Dahdal, S. et al. Residual activatability of circulating Tfh17 predicts humoral response to thymodependent antigens in patients on therapeutic immunosuppression. Front. Immunol. 9, 3178 (2018).
Gao, X. et al. T follicular helper 17 (Tfh17) cells are superior for immunological memory maintenance. eLife 12, e82217 (2023).
Xu, M. et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018).
Dixit, D. et al. S1PR1 inhibition induces proapoptotic signaling in T cells and limits humoral responses within lymph nodes. J. Clin. Invest. 134, e174984 (2024).
Badr, M. E., Zhang, Z., Tai, X. & Singer, A. CD8 T cell tolerance results from eviction of immature autoreactive cells from the thymus. Science 382, 534–541 (2023).
Dauner, J. G., Chappell, C. P., Williams, I. R. & Jacob, J. Perfusion fixation preserves enhanced yellow fluorescent protein and other cellular markers in lymphoid tissues. J. Immunol. Methods 340, 116–122 (2009).
Kim, S. T. et al. Human extrafollicular CD4+ Th cells help memory B cells produce Igs. J. Immunol. 201, 1359–1372 (2018).
Ise, W. et al. T follicular helper cell-germinal center B cell interaction strength regulates entry into plasma cell or recycling germinal center cell fate. Immunity 48, 702–715 (2018).
Sage, P. T., Paterson, A. M., Lovitch, S. B. & Sharpe, A. H. The coinhibitory receptor CTLA-4 controls B cell responses by modulating T follicular helper, T follicular regulatory, and T regulatory cells. Immunity 41, 1026–1039 (2014).
Ahmed, R. et al. Human stem cell-like memory T cells are maintained in a state of dynamic flux. Cell Rep. 17, 2811–2818 (2016).
Gattinoni, L. et al. A human memory T cell subset with stem cell-like properties. Nat. Med. 17, 1290–1297 (2011).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G. D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5, e13984 (2010).
Acknowledgements
This work was supported by grants R01AI107117 and R01HL148347 to H.-J.J.W. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank S. Badr and A. Singer for providing thymi and advice for S1PR1 staining. We thank C. Birchmeier for generating and agreeing to provide and D. Littman for providing the Maffl/fl mice. We thank P. Yan and X. Chen at the Genomics core of The Ohio State University for consulting on the preparation of 10x Genomics samples and library processing.
Author information
Authors and Affiliations
Contributions
H.-J.J.W. conceived the study. T.F. designed and performed most experiments and analyzed the data. C.T. performed and designed non-coding-associated bioinformatic analysis and several mRNA and T-B cell doublet studies. K.C.S. designed, and M.P.C. and H.K. performed microscope-related studies. Y.L. generated the volcano plot, and J.L. and G.X. generated violin plots. M.P.C., M.G., L.B., C.E., P.V. and T.N.T provided assistance for mouse experiments and colony management. S.L. and R.L. provided AKK commensals. W-L.L. contributed to the design and analysis of S1PR1 experiments. J.L., B.H. and W.J. helped recruit patients with RA and provided clinical guidance for inclusion/exclusion criteria. M.W., X.Z. and G.C.T. provided the access to the public scRNA-seq data of patients with RA39. H.-J.J.W., T.F. and C.T. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Nadine Cerf-Bensussan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: L. A. Dempsey, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
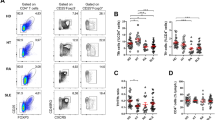
Extended Data Fig. 1 Gut commensal SFB but not Akkermansia muciniphila (AKK) induce TFH17 cells.
(a) Cells from the popliteal lymph nodes (pLN) of both SFB− and SFB + K/BxN mice were stimulated with PMA/ionomycin for 4 h. Representative plots and quantitative data indicating the number of TFH17 cells in pLN are shown (n = 4 mice/ SFB − , and 11 mice/ SFB + , data combined from 2 independent assays). (b-c) Splenocytes from both SFB− and SFB + K/BxN mice were stimulated with PMA/ionomycin for 4 h. Representative plots of percentage and quantitative data indicating the numbers of (b) TFH1 and (c) TFH2 cells are shown (n = 5 mice/group, data from one representative experiment). (d) Splenocytes and PP cells were harvested and subjected to TH17 analysis on day 10 after AKK colonization. Representative plots and quantitative data of the percentage and number of TH17 cells are shown (n = 13 mice/ AKK − , and 18 mice/group, combined from 4 experiments). (e) Splenocytes and PP cells from both AKK− and AKK + K/BxN mice were harvested and subjected to TFH17 analysis on day 10 after AKK colonization. Representative plots and quantitative data of the percentage and number of TFH17 cells are shown (n = 13 mice/ AKK − , and 18 mice/group, combined from 4 independent assays). (f) Quantitative data show the number of splenic α4β7+ IL-17− TFH and α4β7+ TFH17 in SFB− and SFB + K/BxN mice (n = 10 mice/ SFB − , and 13 mice/ SFB + , data combined from 3 independent assays). (g) Representative plots of KikR+ and KikG+ cells in TFH17 and IL-17− TFH cells from the spleens of sham surgery KikGR.K/BxN mice are shown. As these mice never went through photoconversion surgery, all the cells should be KikG+. The few KikR+ cells established the background noise in this procedure. In the Extended Data Fig. 1, data are presented as mean values ± s.e.m., and two-tailed unpaired t-test were used in a-f.
Extended Data Fig. 2 The expression of S1PR1 mRNA in Tfh cells, and the controls of S1PR1 protein staining.
(a) Gating strategy of non-TH17/TFH, Rorγt+ T, Rorγt− TFH and Rorγt+ TFH cells in K/BxN mice. PP cells from SFB + K/BxN mouse were stained with Abs against CD4, TCRβ, Foxp3, Rorγt, CXCR5 and PD-1. (b) IL-17− TFH (eGFP−TdTom−CXCR5+PD-1+) and TFH17 (eGFP+TdTom+CXCR5+PD-1+) cells were sorted from PPs of Current-Ex TH17 fate-mapping mice expressing both TH17-fate reporter TdTom and IL-17 “current” reporter eGFP. S1pr1 transcripts were determined by qRT-PCR. Data were first normalized with the HPRT housekeeping gene, and then further normalized to the average expression of S1pr1 in IL-17− TFH cells (value set as 1) in each experiment. (n = 6 mice/ IL-17− TFH, and 5 mice/TFH17, data combined from 3 independent experiments). (c) Data show the specificity of anti-S1PR1 staining. (I) PP cells from SFB + K/BxN mice were treated in vitro with or without FTY720P for 1 hr at 37 °C and then stained for CD4, TCRβ, and S1PR1. Representative contour plots from the indicated FTY720P concentration are shown. (II) Thymocytes from WT and S1PR1 cKO (CD4CreS1PR1fl/fl) mice were stained with the Abs against CD4, CD8, TCRβ, and S1PR1. Representative plots show the percentage of S1PR1 on CD4 single-positive T cells from WT and S1PR1 cKO thymocytes.
Extended Data Fig. 3 Characterization of TFH cells with or without TH17-fate reporter in both autoimmune and non-autoimmune mice.
(a) The schematic diagram depicts the TH17 fate-mapping model. Diagram shown is the breeding scheme for K/BxB6g7 TH17 fate-mapping mice. Later, after fully crossing KRN.IL-17Cre to Rosa-STOPfl/fl-ZsGreen (or TdTom) mice, we also generated the K/BxN TH17 fate-mapping mice (please see Supplementary Table 1). Note: CD4+ Tconv stands for Foxp3− conventional CD4+ T cells. (b) PP cells from both SFB− and SFB + K/BxB6g7 TH17 fate-mapping mice. Representative plots and quantitative data of the percentage and number of ZsGreen+ T are shown (n = 5 mice/SFB − , and 8 mice/SFB + , data combined from 4 independent assays). (c) PP cells from both SFB− and SFB + K/BxB6g7 TH17 fate-mapping mice were stimulated with PMA/ionomycin for 4 h. Representative plots and quantitative data indicating the percentage and number of TH17 cells are shown (n = 6 mice/SFB − , and 10 mice/SFB + , data combined from 4 independent assays). (d) Representative plots and quantitative data of the percentage and number of ZsGreen− TFH and ZsGreen+ TFH cells from SFB− and SFB+ non-autoimmune B6g7 TH17 fate-mapping mice are shown (PP: n = 13 mice/SFB − , and 17 mice/SFB + , data combined from 8 independent assays; Spleen: n = 13 mice/SFB − , and 14 mice/SFB + , data combined from 7 independent assays). In the Extended Data Fig. 3, data are presented as mean values ± s.e.m. Two-tailed unpaired t-test in b-d.
Extended Data Fig. 4 Characterization of WT and TH17ΔMaf K/BxB6g7 TH17 fate-mapping mice.
(a) SFB fail to induce Bcl-6 in PP TH17 and TFH cells from both SFB− and SFB + K/BxN mice. Representative histogram plots and quantitative gMFI of Bcl-6 expression in TH17 (Rorγt+ T) and TFH cells are shown (n = 9 mice/SFB − , and 15 mice/SFB + , data combined from 3 independent assays). (b) Confirmation of c-Maf deletion in TH17 lineage cells of TH17∆Maf mice. Representative plots and quantitative gMFI of c-Maf in ZsGreen– and ZsGreen+ Tconv are shown (n = 5 mice/WT, and 6 mice/TH17∆Maf, data combined from 3 independent assays). (c) TH17 cells are comparable between WT and TH17ΔMaf mice. Representative plots and quantitative data of PP and splenic TH17 cells are shown (n = 7 mice/WT, and 8 mice/TH17∆Maf, data combined from 3 independent assays). (d) ZsGreen+ T cells (containing both PD-1−/loCXCR5−/lo non-TFH cells and the PD-1+CXCR5+ TFH cells) are reduced in the PP but not spleen of TH17ΔMaf mice. Representative plots and quantitative data of PP and splenic ZsGreen+ T are shown (n = 7 mice/group, data combined from 3 independent assays). (e) The percentage of non-TFH cells in ZsGreen+ T increased in PP (but not spleen) with little change in their number in both tissues. Thus, this increase % is due to the reduction of the other major component of ZsGreen+ T cells, the ZsGreen+ TFH cells, in PPs (shown in Fig. 3c). This data also suggested that the reduction of ZsGreen+ T cells in the PP of TH17∆Maf mice shown in Extended Data Fig. 4d is majorly due to the reduction of the TFH and not non-TFH component. Representative plots and quantitative data show the percentages and number of PP and splenic non-TFH cells gated from ZsGreen+ T cells (n = 7 mice/WT, and 8 mice/TH17∆Maf, data combined from 3 independent assays). In the Extended Data Fig. 4, data are presented as mean values ± s.e.m. Two-way ANOVA with Sidak’s multiple comparisons test in a, b, and two-tailed unpaired t-test in c-e.
Extended Data Fig. 5 Characterization of murine conventional TFH and TFH17Der cells, and gating strategy of the TFH-B cell doublet.
(a) GSEA of scRNA-seq data of ZsGreen+ compared to ZsGreen− TFH cells. The significant 5 pathway clusters with gene sets (nodes, p < 0.01) upregulated in ZsGreen+ TFH cells were visualized with Cytoscape and EnrichmentMap. The graph represents the annotated results for clustered gene set comparisons. Sets of genes are organized in a network, where each set is a node and edges connect similar sets. Automated networks layout groups of related sets of genes into a network of pathway clusters. (b) GSEA of scRNA-seq data of B cells conjugated with ZsGreen+ TFH cells compared to B cells conjugated with ZsGreen− TFH cells. The significant 5 pathway clusters with 3 nodes (gene sets, p < 0.01) upregulated in B cell conjugated with ZsGreen+ TFH cells were visualized with Cytoscape and EnrichmentMap. (c) Heatmap of clustered genes in the leading-edge subsets of gene sets related to cell adhesion (Extended Data Fig. 5b). Red and blue colors indicate positively and negatively regulated transcripts, respectively, upon comparing B cells conjugated with ZsGreen+ TFH cells to the B cells conjugated with ZsGreen− TFH cells. Darker the color, the greater the fold difference. (d) (I) Gating scheme for Fig. 4f is shown, illustrating the gating strategy of the indicated T cell type for T-B conjugate analysis, without excluding doublets. Additionally, the gating strategy in (II) that excludes doublets is shown as a comparison. Doublets were excluded by FSC-A and FSC-H gating in (II) for single-cell analysis. Note, the extra PD-1loCXCR5+ cells in gate (I), but not gate (II), indicate the B cells that are bound to T cells and are typically excluded using the single-cell analysis. In the Extended Data Fig. 5, ANOVA test is used in scRNA-seq analysis in a-c.
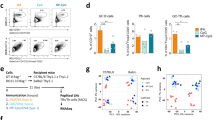
Extended Data Fig. 6 The evaluation of T-B cell doublet’s stability.
(a) Splenocytes from CTV labeled (CTV + ) TdTom and unlabeled (CTV − ) ZsGreen K/BxN TH17 fate-mapping mice were mixed at 1:1 ratio in vitro (mixed group). For the control group, Splenocytes from ZsGreen K/BxN TH17 fate-mapping mice were used without mixing with CTV+ cells (unmixed group). Representative plots and quantitative data indicating (I) CD4+ T cells gated from singlet gate were further gated for CD4 vs. CTV. This control group showing a ~ 1:1 ratio of T cells from CTV+ and CTV − TH17 fate-mapping mice. (II) ZsGreen+ TFH cells were gated to include doublets (purposefully not exclude the doublets) were further gated for CD4 vs. B220. The T-B conjugates were then gated for CD4 vs. CTV (n = 2/unmixed group; n = 16/mixed group, from two experiments). The CTV + T-B conjugates of ZsGreen+ TFH cells indicates that the B cells were from CTV+ TdTom mouse. (b) CD4+ T cells were enriched from the spleens of TH17 fate-mapping mice using positive selection with CD4 microbeads (Miltenyi Biotec). The enriched CD4+ T cells (which included single cells and T-B doublets) were stained with antibodies against CD4, TCRβ, CD19, GITR, CXCR5, and PD-1. Doublets with B cells bound to non-TH17/TFH, TH17, ZsGreen− TFH, or ZsGreen+ TFH cells were sorted. Non-TH17/TFH and B cell conjugates were then combined with TH17/B cell conjugates as the non-TFH doublet group. The conjugates were examined under a microscope, and two representative images of conjugates from the (I) non-TFH group, (II) ZsGreen− TFH group, and (III) ZsGreen+ TFH group are shown. (c) Compiled data on the percentage of surviving conjugates from Extended Data Fig. 6b, I-III using 16 randomly selected fields in each group with 4 samples (each sample/symbol represented one mouse except one sample was a combination of two mice) is shown. To calculate the surviving doublet %, we counted each cell number in each field and divided that number by two; this is the “would-be” doublet number. We then divided the observed real doublet number by the would-be doublet number x100 to obtain the surviving doublet % in each field. In Extended Data Fig. 6, data are presented as mean values ± s.e.m. One-way ANOVA with Tukey’s multiple comparisons test in c.
Extended Data Fig. 8 LZ or DZ analysis of ZsGreen− TFH and ZsGreen+ TFH cells.
(a) Another example of PPs cryostat sections from SFB + K/BxN TH17 fate-mapping mice with the same staining as indicated in Fig. 6a is shown. Arrows indicate the ZsGreen+ TFH cells. (b-c) CD4+ T cells and ZsGreen+ T cells numbers in each GC LZ or DZ were counted from the experiment in Fig. 6a. Cell number are shown (data combined from 28 PP GCs of 13 mice, 4 independent assays). (d) Non-TH17/ TFH, ZsGreen− TFH and ZsGreen+ TFH cells were sorted from PPs of SFB + K/BxN mice. Cxcr4 transcripts were determined by qRT-PCR. Data were first normalized with the HPRT housekeeping gene, and then further normalized to one of the non-TH17/ TFH samples (value set as 1); (n = 14 mice per group. In the ZsGreen+ TFH group, some samples were pooled due to small cell numbers, data combined from 2 independent assays. Each dot represents one mouse, except in the ZsGreen+ TFH group). (e) Gating strategy for non-TH17/TFH, Rorγt+ T, ZsGreen− TFH and ZsGreen+ TFH cells in K/BxN TH17 fate-mapping mice is shown. (f) Representative plots and quantitative gMFI of Bcl-6 and CD40L in splenic non-TH17/TFH, TH17, ZsGreen− TFH and ZsGreen+ TFH cells from SFB + K/BxN fate-mapping mice are shown (n = 10 mice/group, data are representative of 2 independent experiments). In Extended Data Fig. 8, data are presented as mean values ± s.e.m. Two-tailed paired t-test in b, c. One-way ANOVA with Tukey’s multiple comparisons test in d, f.
Extended Data Fig. 9 Confirmation of CXCR4 ablation in TFH17Der cells of CXCR4 cKO mice.
(a) IL-17Cre-driven CXCR4 deletion specifically impaired CXCR4 expression in TdTom+ TFH cells, but not TH17 and TdTom− TFH cells. CD4+ T cells from WT and CXCR4 cKO mice were transferred into SFB+ TCRα−/−.BxN mice recipients for two weeks before tissue harvest. Representative plots and quantitative CXCR4 gMFI data in TH17 (CD4+TCRβ+Foxp3−TdTom+Rorγt+CXCR5−/loPD-1−/lo), TdTom− TFH (CD4+TCRβ+Foxp3−TdTom−CXCR5+PD-1+), and TdTom+ TFH (CD4+TCRβ+Foxp3−TdTom+CXCR5+PD-1+) from WT and CXCR4 cKO groups are shown (n = 9 mice/WT, and 6 mice/cKO, data combined from 2 independent assays). (b) IL-17Cre-driven CXCR4 deletion doesn’t impact the number of TdTom+ TFH cells. Representative plots and quantitative data indicating the percentages and numbers of the splenic and PP TdTom− TFH and TdTom+ TFH from WT and CXCR4 cKO groups are shown (n = 9 mice/WT, and 6 mice/cKO, data combined from 2 independent assays). In Extended Data Fig. 9, data are presented as mean values ± s.e.m. Two-way ANOVA with Sidak’s multiple comparisons test in a, and two-tailed unpaired t-test in b.
Extended Data Fig. 10 B cell-only control group of in vitro TFH and B co-culture experiments.
Representative data show the percentage of GC-like B cells and IgG1 expression on day 7 from B cell only group without the addition of TFH cells.
Supplementary information
Supplementary Information
Supplementary Table 1.
Supplementary Table 2
List of gene clusters upregulated in ZsGreen+ Tfh cells compared to ZsGreen⎼ Tfh cells.
Supplementary Table 3
List of gene clusters upregulated in B cells conjugated with ZsGreen+ Tfh cells compared to B cells conjugated with ZsGreen⎼ Tfh cells.
Supplementary Table 4
List of genes upregulated in ZsGreen+ Tfh cells compared to ZsGreen⎼ Tfh cells in PPs and spleen.
Supplementary Table 5
List of gene clusters upregulated in CXCR4+ ZsGreen+ Tfh cells compared to CXCR4⎼ ZsGreen+ Tfh cells.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4.
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fan, T., Tai, C., Sleiman, K.C. et al. Aberrant T follicular helper cells generated by TH17 cell plasticity in the gut promote extraintestinal autoimmunity. Nat Immunol 26, 790–804 (2025). https://doi.org/10.1038/s41590-025-02125-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-025-02125-7