Abstract
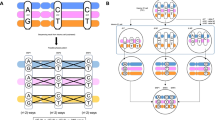
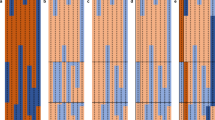
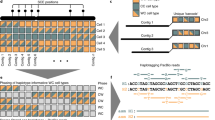
Haplotype phasing represents a pivotal procedure in genome analysis, entailing the identification of specific genetic variant combinations on each chromosome. Achieving chromosome-level genome phasing constitutes a considerable challenge, particularly in organisms with large and complex genomes. To address this challenge, we have developed a robust, gamete cell-based phasing pipeline, including wet-laboratory processes for plant sperm cell isolation, short-read sequencing and a bioinformatics workflow to generate chromosome-level phasing. The bioinformatics workflow is applicable for both plant and other sperm cells, for example, those of mammals. Our pipeline ensures high-quality single-nucleotide polymorphism (SNP) calling for each sperm cell and the subsequent construction of a high-density genetic map. The genetic map facilitates accurate chromosome-level genome phasing, enables crossover event detection and could be used to correct potential assembly errors. Our bioinformatics pipeline runs on a Linux system and most of its steps can be executed in parallel, expediting the analysis process. The entire workflow can be performed over the course of 1 d. We provide a practical example from our previous research using this protocol and provide the whole bioinformatics pipeline as a Docker image to ensure its easy adaptability to other studies.
Key points
-
This protocol describes a method for phasing plant genomes, using gamete cells isolated from pollen to enable chromosome-level phasing and crossover detection.
-
The protocol enables phasing without the need for Hi-C data or sequencing large plant populations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The test data described in this protocol are available at https://figshare.com/articles/dataset/Example_data_for_PollenSeq/25272493. Contig-level test genome is available at https://figshare.com/articles/dataset/test_contig-level_genome_for_PollenSeq/26089288. Source data for Fig. 2 can be downloaded from https://figshare.com/articles/dataset/source_data_for_Fig2_a-b/26088184, https://figshare.com/articles/dataset/Source_data_for_Fig2_c/26088187 and https://figshare.com/articles/dataset/Source_data_for_Fig_2d/26088193.
Code availability
Scripts shown here are available from github at https://github.com/zwycooky/PollenSeq.
References
Garg, S. Computational methods for chromosome-scale haplotype reconstruction. Genome Biol. 22, 101 (2021).
Leitwein, M., Duranton, M., Rougemont, Q., Gagnaire, P.-A. & Bernatchez, L. Using haplotype information for conservation genomics. Trends Ecol. Evo. 35, 245–258 (2020).
Browning, B. L. & Browning, S. R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84, 210–223 (2009).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Bhat, J. A., Yu, D., Bohra, A., Ganie, S. A. & Varshney, R. K. Features and applications of haplotypes in crop breeding. Commun. Biol. 4, 1266 (2021).
Vonholdt, B. M. et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464, 898–902 (2010).
Zhao, J., Sauvage, C., Bitton, F. & Causse, M. Multiple haplotype-based analyses provide genetic and evolutionary insights into tomato fruit weight and composition. Horticul. Res. 9, uhab009 (2022).
Blanca, J. et al. Haplotype analyses reveal novel insights into tomato history and domestication driven by long-distance migrations and latitudinal adaptations. Hortic. Res. 9, uhac030 (2022).
Yang, J. et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 3, 696–703 (2017).
Yan, M. et al. Haplotype-based phylogenetic analysis and population genomics uncover the origin and domestication of sweetpotato. Mol. Plant 17, 277–296 (2024).
Todesco, M. et al. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature 584, 602–607 (2020).
Zhang, F. et al. The landscape of gene–CDS–haplotype diversity in rice: Properties, population organization, footprints of domestication and breeding, and implications for genetic improvement. Mol., Plant 14, 787–804 (2021).
Naj, A. C. Genotype imputation in genome‐wide association studies. Curr. Protoc. Hum. Genet. 102, e84 (2019).
Liu, F., Jiang, Y., Zhao, Y., Schulthess, A. W. & Reif, J. C. Haplotype-based genome-wide association increases the predictability of leaf rust (Puccinia triticina) resistance in wheat. J. Exp. Bot. 71, 6958–6968 (2020).
Wu, X. et al. Prioritized candidate causal haplotype blocks in plant genome-wide association studies. PLoS Genet. 18, e1010437 (2022).
Dong, X. et al. Dynamic and antagonistic allele-specific epigenetic modifications controlling the expression of imprinted genes in maize endosperm. Mol. Plant 10, 442–455 (2017).
Shao, L. et al. Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc. Natl Acad. Sci. USA 116, 5653–5658 (2019).
Guk, J. Y., Jang, M. J., Choi, J. W., Lee, Y. M. & Kim, S. De novo phasing resolves haplotype sequences in complex plant genomes. Plant Biotechnol. J. 20, 1031–1041 (2022).
Mansfeld, B. N. et al. A haplotype resolved chromosome‐scale assembly of North American wild apple Malus fusca and comparative genomics of the fire blight Mfu10 locus. Plant J. 116, 989–1002 (2023).
Han, X. et al. Two haplotype-resolved, gap-free genome assemblies for Actinidia latifolia and Actinidia chinensis shed light on the regulatory mechanisms of vitamin C and sucrose metabolism in kiwifruit. Mol. Plant 16, 452–470 (2023).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21, 245 (2020).
Browning, S. R. & Browning, B. L. Haplotype phasing: existing methods and new developments. Nat. Rev. Genet. 12, 703–714 (2011).
Schrinner, S. D. et al. Haplotype threading: accurate polyploid phasing from long reads. Genome Biol. 21, 252 (2020).
Kuleshov, V. et al. Whole-genome haplotyping using long reads and statistical methods. Nat. Biotechnol. 32, 261–266 (2014).
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Browning, B. L., Tian, X., Zhou, Y. & Browning, S. R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 108, 1880–1890 (2021).
Zhang, W. et al. A phased genome based on single sperm sequencing reveals crossover pattern and complex relatedness in tea plants. Plant J. l 105, 197–208 (2021).
Martin, M. et al. WhatsHap: weighted haplotype assembly for future-generation sequencing reads. J. Comput. Biol. 22, 498–509 (2015).
Duan, H. et al. Physical separation of haplotypes in dikaryons allows benchmarking of phasing accuracy in Nanopore and HiFi assemblies with Hi-C data. Genome Biol. 23, 84 (2022).
Kronenberg, Z. N. et al. Extended haplotype-phasing of long-read de novo genome assemblies using Hi-C. Nat. Commun. 12, 935 (2021).
Cheng, H. et al. Haplotype-resolved assembly of diploid genomes without parental data. Nat. Biotechnol. 40, 1332–1335 (2022).
Zhang, X., Zhang, S., Zhao, Q., Ming, R. & Tang, H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants 5, 833–845 (2019).
Li, X., Li, L. & Yan, J. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat. Commun. 6, 6648 (2015).
Luo, C., Li, X., Zhang, Q. & Yan, J. Single gametophyte sequencing reveals that crossover events differ between sexes in maize. Nat. Commun. 10, 785 (2019).
Shi, D. et al. Single-pollen-cell sequencing for gamete-based phased diploid genome assembly in plants. Genome Res. 29, 1889–1899 (2019).
Campoy, J. A. et al. Gamete binning: chromosome-level and haplotype-resolved genome assembly enabled by high-throughput single-cell sequencing of gamete genomes. Genome Biol. 21, 306 (2020).
Sun, H. et al. Linked-read sequencing of gametes allows efficient genome-wide analysis of meiotic recombination. Nat. Commun. 10, 4310 (2019).
Zhang, W. et al. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat. Commun. 11, 3719 (2020).
Zhou, Q. et al. Haplotype-resolved genome analyses of a heterozygous diploid potato. Nat. Genet. 52, 1018–1023 (2020).
Serra Mari, R. et al. Haplotype-resolved assembly of a tetraploid potato genome using long reads and low-depth offspring data. Genome Biol. 25, 26 (2024).
Sun, H. et al. Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nat. Genet. 54, 342–348 (2022).
Rommel Fuentes, R. et al. Meiotic recombination profiling of interspecific hybrid F1 tomato pollen by linked read sequencing. Plant J. 102, 480–492 (2020).
Dreissig, S., Fuchs, J., Himmelbach, A., Mascher, M. & Houben, A. Sequencing of single pollen nuclei reveals meiotic recombination events at megabase resolution and circumvents segregation distortion caused by postmeiotic processes. Front. Plant Sci. 8, 1620 (2017).
Li, R. et al. Inference of chromosome-length haplotypes using genomic data of three or a few more single gametes. Mol. Biol. Evol. 37, 3684–3698 (2020).
Lyu, R. et al. sgcocaller and comapr: personalised haplotype assembly and comparative crossover map analysis using single-gamete sequencing data. Nucleic Acids Res. 50, e118–e118 (2022).
Rowan, B. A., Patel, V., Weigel, D. & Schneeberger, K. Rapid and inexpensive whole-genome genotyping-by-sequencing for crossover localization and fine-scale genetic mapping. G3 5, 385–398 (2015).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (3211101118) NSFC-DFG collaborative project, the National Key R&D Program of China (2022YFF1003103), Fundamental Research Funds for the Central Universities (2662023PY011) to W.W., Deutsche Forschungsgemeinschaft (DFG)-Project number 468870408 to B.U. and A.R.F., and China Postdoctoral Innovation Program (BX20220127) to W.Z.
Author information
Authors and Affiliations
Contributions
W.W., B.U. and A.R.F. conceived and managed the project, and drafted the first version of the manuscript. W.Z. and A.T. prepared the bioinformatics section. X.J. prepared the wet-laboratory section. W.Z., W.W., B.U., A.R.F. and J.Y. revised and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewer(s) for their contribution to the peer review of this process.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Li, X., Li, L. & Yan, J. Nat. Commun. 6, 6648 (2015): https://doi.org/10.1038/ncomms7648
Luo, C. et al. Nat. Commun. 10, 786 (2019): https://doi.org/10.1038/s41467-019-08786-x
Zhang, W. et al. Plant J. 105, 197–208 (2021): https://doi.org/10.1111/tpj.15051
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Table 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, W., Tariq, A., Jia, X. et al. Plant sperm cell sequencing for genome phasing and determination of meiotic crossover points. Nat Protoc 20, 690–708 (2025). https://doi.org/10.1038/s41596-024-01063-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-024-01063-2