Abstract
Terrestrial geothermal springs, reminiscent of early Earth conditions, host diverse and abundant populations of Archaea. In this study, we reconstructed 2,949 metagenome-assembled genomes (MAGs) from 152 metagenomes collected over six years from 48 geothermal springs in Tengchong, China. Among these MAGs, 1,431 (49%) were classified as high-quality, while 1,518 (51%) were considered as medium-quality. Phylogenomic analysis revealed that these MAGs spanned 12 phyla, 27 classes, 67 orders, 147 families, 265 genera, and 475 species. Notably, 575 (19%) MAGs represented new taxa at various taxonomic levels, and 2,075 (70%) lacked nomenclature and effective descriptions. The most abundant phyla of archaeal genomes were Thermoproteota, Thermoplasmatota, and Micrarchaeota. The DRTY, ZMQ, and ZZQ geothermal springs were predominated by Archaea, particularly by Thermoproteia and Thermoplasmata. These draft genomes provide new data for studying species diversity and function within terrestrial geothermal spring archaeal communities, thus contributing to the conservation and utilization of thermophilic and hyperthermophilic microbial resources.
Similar content being viewed by others
Background & Summary
Environments with temperatures below 55 °C are ubiquitous on Earth, while those exceeding 55–60 °C are relatively rare and mostly associated with geothermal habitats1. Terrestrial geothermal springs represent typical extreme environments, reminiscent of early Earth conditions, and are considered ideal sites for exploring the origin and evolution of life2,3. The temperature of these geothermal springs often exceeds the growth limits of eukaryotic organisms (approaching 60 °C)4, fostering diverse ancient and metabolically unique life forms, particularly housing unique and abundant resources of thermophilic and hyperthermophilic Archaea.
Archaea play an essential role in biogeochemical cycles5, facilitating unique metabolic processes like methanogenesis, anaerobic alkanotrophy, and chemolithotrophic ammonia oxidation6,7,8. Beyond their ecological importance, extremophilic Archaea are a valuable resource for novel enzymes used in biotechnology9,10, such as L-asparaginase11. Thermophilic Archaea show promise in medicine by converting carbohydrate-rich substances into hydrogen, which can be used in therapeutic applications9,12. Their ability to survive in extreme temperatures also makes them useful in preventing the growth of heat-sensitive pathogens. Additionally, Archaea produce metabolites that influence host-microbe interactions, with the gut and oral cavity showing the highest diversity13. These microorganisms are closely linked to human diseases, including cancer, particularly methanogenic Archaea, whose metabolites may affect the tumor microenvironment and contribute to carcinogenesis. However, their limited ability to be cultured has impeded a more comprehensive understanding of their functions14.
As the sole magmatic hydrothermal source of terrestrial springs in inland China, the Tengchong geothermal springs, situated within the Mediterranean-Himalayan Geothermal Belt, exhibit diverse physicochemical characteristics including numerous springs, wide ranges of temperature and pH15. The advent of high-throughput sequencing has bypassed the necessity for microbial cultivation and enabled the discovery of numerous previously unknown taxonomic groups. Extensive research has been conducted on the microbial diversity of the Tengchong geothermal springs using the 16S rRNA gene amplicon sequencing16,17,18. However, amplicon analysis, targeting one or a few genes, often exhibits amplification bias and fails to provide comprehensive functional profiling of microorganisms when assessing community diversity. In contrast, metagenomics offers extensive gene information about microbes through high-throughput sequencing, enabling the identification of various uncultured microbes and their ecological traits. With advancements in metagenomic technology, several specific archaeal lineages have already been discovered from Tengchong geothermal spring samples without cultivation19,20,21,22,23. In this study, we further revealed the hidden microbial diversity of Tengchong geothermal springs by retrieving and assembling metagenomic sequences.
By conducting metagenomic sequencing on 152 sediment samples from 48 geothermal pools and streams located in Tengchong County, Yunnan Province, China, collected over six years from 2016 to 2021, we successfully reconstructed 2,949 metagenome-assembled genomes (MAGs) of Archaea (Fig. 1a; Supplementary Table 1). Among them, 1,431 (49%) were considered high-quality (completeness >90% with contamination <5%)24, while the remaining 1,518 (51%) MAGs were classified as medium-quality with an average completeness of 76% (Fig. 1b; Supplementary Table 2). Most MAGs, comprising 2,672 (91%), possessed fewer than 200 scaffolds, averaging 60 for high-quality and 123 for medium-quality MAGs (Fig. 1c). Additionally, the majority of MAGs, accounting for 2,256 (77%), exhibited an N50 length exceeding 10,000 bp, with the longest reaching 2.18 Mbp (Supplementary Table 2), indicating exceptional assembly quality. A significant positive correlation between genome size and N50 length was observed, suggesting that larger genome assemblies contained less fragments (Fig. 1d). Overall, these findings underscore the good quality of genome assembly in this study.
Overview of the 2,949 archaeal MAGs. (a) Workflow of archaeal MAG reconstruction. (b) Genomic completeness and contamination of high-quality (red dots) and medium-quality (blue dots) archaeal MAGs. (c) Number of scaffolds in high-quality (red dots) and medium-quality (blue dots) archaeal MAGs. (d) The Pearson correlation between genomic size and N50 length among archaeal MAGs.
According to the Genome Taxonomy Database (GTDB)25, these archaeal MAGs were classified into 12 phyla, 27 classes, 67 orders, 147 families, 265 genera, and 475 species (Fig. 2a; Supplementary Table 2). The hotspots of novelty were observed in phyla Altiarchaeota, B1Sed10-29, Micrarchaeota, Nanoarchaeota, and Thermoproteota. A substantial number of MAGs (n = 2,104) were attributed to the phylum Thermoproteota, particularly the orders Sulfolobales, Caldarchaeales, and B26-1, followed by phyla Thermoplasmatota and Micrarchaeota (Fig. 2a,b). Out of the 2,374 MAGs classifiable in GTDB, 2,075 (70%) lacked nomenclature and effective descriptions, with 9 belonging to the archaeal phylum B1Sed10-29 (Fig. 2c). Notably, 575 (19%) MAGs lacked a previously identified species representative in GTDB, suggesting potential novel taxa, comprising 14 orders, 36 families, 240 genera, and 285 species. Additionally, the genome size and GC content of Aenigmatarchaeota, Altiarchaeota, Iainarchaeota, B1Sed10-29, Micrarchaeota, and Nanoarchaeota (all known as the DPANN superphylum) were significantly smaller than those of other phyla (Fig. 2d,e). The largest genome size of MAGs was detected in Asgardarchaeota, which was considered as the archaeal ancestor of eukaryotes26. In contrast, Hadarchaeota displayed the highest GC content.
Phylogeny of archaeal MAGs. (a) Phylogenetic tree displaying all strain-level archaeal MAGs (n = 603) at the order level. Lineages within the same phylum are displayed in the same color. Pie charts indicate potential novelty at the family- (blue), genus- (purple), and species-level (red) within the corresponding order. The colors, ranging from light to dark, indicate the proportion of MAGs that are newly identified, existing but without nomenclature, and existing with nomenclature, respectively. Green columns represent the number of archaeal MAGs within each order. (b) Distribution of all 2,949 archaeal MAGs at the phylum level. (c) Potential taxonomic novelty of archaeal MAGs at various taxonomic levels. Unnamed MAGs indicate those present in GTDB without nomenclature; New MAGs indicate those newly discovered in this study. (d) Genome size of MAGs across different archaeal phyla. (e) GC content of MAGs across different archaeal phyla. The letters indicate the grouping of results from the least significant difference (LSD) test for significance analysis of data across phyla.
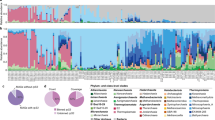
The abundance of archaeal MAGs varied across different samples. On average, Archaea accounted for 34.7% of the community, with a maximum relative abundance of 95.2%. Notably, 54 samples exhibited dominance by Archaea, constituting over 50% of the community (Fig. 3a). Specifically, the DRTY, ZMQ, and ZZQ pools were predominantly populated by Archaea rather than Bacteria. Moreover, Thermoplasmata and Thermoproteia were prevalent in the DRTY pools, while Thermoproteia dominated the ZMQ and ZZQ pools (Fig. 3b). In the SRZB springs, Archaea were also abundant, primarily dominated by Thermoproteia. In contrast, other springs except the JZ-1 pool harbored less prominent of Archaea, with dominance observed in Methanomethylicia, Thermoproteia, Nitrososphaeria_A, Nitrososphaeria, Bathyarchaeia, and Nanoarchaeia.
Archaeal composition across 152 Tengchong geothermal spring samples. (a) Relative abundance of Archaea within microbial communities. (b) Heatmap of the relative abundance of each archaeal class within archaeal communities. Samples are clustered by geothermal spring pools, and detailed information for each sample is provided in Supplementary Table 1.
This study provides a detailed overview of archaeal diversity in terrestrial geothermal springs, with the number of reconstructed MAGs exceeding those reported in similar studies from other terrestrial thermal environments27,28,29, positioning the Tengchong geothermal springs as a significant biodiversity hotspot. The exploration of archaeal diversity suggests valuable future research directions. The dominance of Thermoproteota and the rich species diversity within the DPANN superphylum indicate unique adaptations to extreme conditions. The small genome sizes of DPANN Archaea may reflect a strategy of genomic streamlining in resource-limited environments. The study also opens promising prospects for industrial enzyme production and biomedical applications, particularly through novel archaeal lineages, whose unique metabolic pathways could lead to innovative therapeutics and biotechnological tools. However, the study has limitations, such as its focus on Tengchong’s magmatic geothermal springs, which may not be representative of global geothermal springs. Furthermore, functional predictions need validation through multi-omics approaches, like transcriptomics and metabolomics, or experimental methods to confirm metabolic capabilities.
To the best of our knowledge, this study stands out as one of the few to recover such a large number of archaeal genomes from terrestrial geothermal ecosystems. The collection of archaeal genomes from geothermal springs holds significant potential to deepen our comprehension of species diversity, community structure, and functional dynamics within these microbial ecosystems30. This, in turn, can substantially aid in the conservation and exploitation of thermophilic and hyperthermophilic microbial resources, open new perspectives for exploring the clinical and industrial relevance of Archaea in both natural and human-associated environments, and advance our understanding of the origin of eukaryotes and even the fundamental processes underlying life itself.
Methods
Sample collection, DNA extraction, and metagenomic sequencing
Sediment samples were collected using sterile spatulas and spoons, then stored in liquid nitrogen before transportation to the laboratory. Total genomic DNA was extracted from approximately 20 g of sediment using the PowerSoil DNA Isolation kit (MoBio), following the manufacturer’s protocol. Paired-end sequencing was performed on Illumina Hiseq 4000 instruments at Guangdong Magigene Biotechnology Co., Ltd. (Guangdong, China), with each sample yielding approximately 30 Gbp (2 × 150 bp) of raw sequence data.
Quality control and assembly
Raw reads were preprocessed using custom Perl scripts, as previously described31 (https://github.com/hzhengsh/qualityControl). Briefly, three steps were employed to obtain clean reads of high quality: 1. Removal of duplicated reads resulting from PCR amplification; 2. Discarding reads containing excessive “N”s; 3. Trimming bases with low quality at both ends. Quality reads from each sample were then de novo assembled separately using SPAdes (v.3.9.0)32 with the following parameters: -meta -k 21,33,55,77,99,127.
Genome binning and refinement
For each sample, high-quality reads were mapped onto assembled scaffolds longer than 2,500 bp using BBMap (v.38.92; http://sourceforge.net/projects/bbmap/) with the following parameters: k = 15, minid = 0.97, build = 1. The resulting file was utilized to calculate sequence depth and perform genome binning using three tools: MaxBin2 (v.2.2.7)33, CONCOCT (v.1.1.0)34, and MetaBAT (v.2.12.1)35. The optimal bins were selected using DASTool (v.1.1.3)36. Clean reads for each selected bin were then recruited using BBMap (with the same parameters as mentioned previously) and were reassembled using SPAdes (v.3.15.2) with the parameters: -careful -k 21,33,55,77,99,127. Scaffolds with depths falling below [1st quartile – 1.5× (3rd quartile - 1st quartile)] or above [3rd quartile +1.5× (3rd quartile - 1st quartile)] were considered outliers and removed. The quality of genomes, including completeness, contamination, and heterogeneity was assessed using CheckM (v.1.1.3)37. A total of 2,949 Archaea bins were obtained with completeness >50% and contamination <10%.
Taxonomic assignment and phylogenetic analysis of MAGs
The taxonomy of all 2,949 archaeal MAGs was determined using the classify_wf workflow of GTDB-Tk (v.2.1.0)38 with GTDB release 207 and default parameters. Phylogeny was inferred and used to validate the taxonomic assignments. The program dRep (v.3.2.2)39 was utilized to dereplicate MAGs at 99% ANI at the strain level, resulting in 603 representative MAGs. A phylogenetic tree comprising these 603 representative MAGs from the present study and reference genomes from GTDB was constructed through multiple sequence alignments of 53 concatenated conserved archaeal marker genes retrieved from GTDB-Tk. Poorly aligned regions were trimmed using trimAl (v.1.4.rev22)40 with the parameter: -automated1. Phylogenetic inference was conducted using IQ-TREE (v.1.6.12)41 with 1,000 ultrafast bootstrapping iterations. The best-fit model of LG + F + R10 was determined using ModelFinder42, supported by Bayesian information criterion (BIC). Finally, the phylogenetic tree was visualized using iTOL (v.6)43.
Data Records
The 2,949 MAGs of Archaea described in this study have been deposited in NCBI under the BioProject PRJNA544494 (ref. 44), with BioSample IDs ranging from SAMN18253264 to SAMN18253267, SAMN18253269, SAMN18253270, SAMN18838809, SAMN19656016 to SAMN19656018, SAMN28867992 to SAMN28867995, SAMN28867997 to SAMN28867999, SAMN31028420 to SAMN31028426, SAMN31028428 to SAMN31028439, SAMN31028763, SAMN34195732, SAMN36035244 to SAMN36035357. The accession numbers of genomes range from JAVYKE000000000 to JAWCTO000000000, with detailed accessions for each MAG recorded in Supplementary Table 2.
Technical Validation
The raw reads underwent preprocessing for quality control following the procedure outlined in previous studies31. To guarantee the quality of the MAGs, we chose assembled scaffolds longer than 2,500 bp for metagenomic binning, as suggested in previous studies19,23. The MAGs were rigorously evaluated based on the following criteria: (1) completeness >50% and contamination <10%; (2) absence of potential chimerism in the genome sequences (see Supplementary Table 2 for details); and (3) exclusion of potentially misassigned contigs from the genome sequences.
Code availability
Custom scripts utilized in this study are available at https://github.com/hzhengsh/qualityControl. The parameters and versions of all bioinformatics tools employed for the analysis are detailed in the Methods section.
References
Brock, T. D. Introduction: an overview of the thermophiles. Thermophiles: general, molecular, and applied microbiology 1–16 (1986).
Nisbet, E. G. & Sleep, N. H. The habitat and nature of early life. Nature 409, 1083–1091 (2001).
Damer, B. & Deamer, D. The Hot Spring Hypothesis for an Origin of Life. Astrobiology 20, 429–452 (2020).
López-López, O., Cerdán, M. E. & González-Siso, M. I. Hot Spring Metagenomics. Life 3, 308–320 (2013).
Baker, B. J. et al. Diversity, ecology and evolution of Archaea. Nat Microbiol 5, 887–900 (2020).
Evans, P. N. et al. An evolving view of methane metabolism in the Archaea. Nat Rev Microbiol 17, 219–232 (2019).
Santoro, A. E., Buchwald, C., McIlvin, M. R. & Casciotti, K. L. Isotopic Signature of N2O Produced by Marine Ammonia-Oxidizing Archaea. Science 333, 1282–1285 (2011).
Borrel, G. et al. Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat Microbiol 4, 603–613 (2019).
Straub, C. T. et al. Biotechnology of extremely thermophilic archaea. FEMS Microbiology Reviews 42, 543 (2018).
Elleuche, S., Schröder, C., Sahm, K. & Antranikian, G. Extremozymes — biocatalysts with unique properties from extremophilic microorganisms. Current Opinion in Biotechnology 29, 116–123 (2014).
Dumina, M. et al. A Novel L-Asparaginase from Hyperthermophilic Archaeon Thermococcus sibiricus: Heterologous Expression and Characterization for Biotechnology Application. International Journal of Molecular Sciences 22, 9894 (2021).
Uppar, A. P. et al. Applications of Extremophiles in Therapeutics. in Trends in Biotechnology of Polyextremophiles (eds. Shah, M. P. & Dey, S.) 231–242, https://doi.org/10.1007/978-3-031-55032-4_10 (Springer Nature Switzerland, Cham, 2024).
Cai, M., Kandalai, S., Tang, X. & Zheng, Q. Contributions of Human-Associated Archaeal Metabolites to Tumor Microenvironment and Carcinogenesis. Microbiology Spectrum 10, e02367–21 (2022).
Lewis, W. H., Tahon, G., Geesink, P., Sousa, D. Z. & Ettema, T. J. G. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol 19, 225–240 (2021).
Guo, Q. & Wang, Y. Geochemistry of hot springs in the Tengchong hydrothermal areas, Southwestern China. Journal of Volcanology and Geothermal Research 215–216, 61–73 (2012).
Hedlund, B. P. et al. A review of the microbiology of the Rehai geothermal field in Tengchong, Yunnan Province, China. Geoscience Frontiers 3, 273–288 (2012).
Weiguo, H. et al. A Comprehensive Census of Microbial Diversity in Hot Springs of Tengchong, Yunnan Province China Using 16S rRNA Gene Pyrosequencing. PLoS ONE 8, 1–15 (2013).
Li, H. et al. The impact of temperature on microbial diversity and AOA activity in the Tengchong Geothermal Field, China. Sci Rep 5, 17056 (2015).
Hua, Z.-S. et al. Genomic inference of the metabolism and evolution of the archaeal phylum Aigarchaeota. Nat Commun 9, 1–11 (2018).
Hua, Z.-S. et al. Insights into the ecological roles and evolution of methyl-coenzyme M reductase-containing hot spring Archaea. Nat Commun 10, 4574 (2019).
De Anda, V. et al. Brockarchaeota, a novel archaeal phylum with unique and versatile carbon cycling pathways. Nat Commun 12, 2404 (2021).
Qu, Y.-N. et al. Panguiarchaeum symbiosum, a potential hyperthermophilic symbiont in the TACK superphylum. Cell Reports 42 (2023).
Qi, Y.-L. et al. Comparative Genomics Reveals Thermal Adaptation and a High Metabolic Diversity in “Candidatus Bathyarchaeia”. mSystems 6, https://doi.org/10.1128/msystems.00252-21 (2021).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35, 725–731 (2017).
Rinke, C. et al. A standardized archaeal taxonomy for the Genome Taxonomy Database. Nat Microbiol 6, 946–959 (2021).
Liu, Y. et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature 593, 553–557 (2021).
Sriaporn, C., Campbell, K. A., Van Kranendonk, M. J. & Handley, K. M. Bacterial and archaeal community distributions and cosmopolitanism across physicochemically diverse hot springs. ISME Communications 3, 80 (2023).
Guo, L. et al. Temperature governs the distribution of hot spring microbial community in three hydrothermal fields, Eastern Tibetan Plateau Geothermal Belt, Western China. Science of The Total Environment 720, 137574 (2020).
Inskeep, W. P. et al. Metagenomes from High-Temperature Chemotrophic Systems Reveal Geochemical Controls on Microbial Community Structure and Function. PLOS ONE 5, e9773 (2010).
Qi, Y.-L. et al. Analysis of nearly 3000 archaeal genomes from terrestrial geothermal springs sheds light on interconnected biogeochemical processes. Nat Commun 15, 4066 (2024).
Hua, Z.-S. et al. Ecological roles of dominant and rare prokaryotes in acid mine drainage revealed by metagenomics and metatranscriptomics. The ISME Journal 9, 1280–1294 (2015).
Bankevich, A. et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of Computational Biology 19, 455–477 (2012).
Wu, Y.-W., Simmons, B. A. & Singer, S. W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016).
Alneberg, J. et al. Binning metagenomic contigs by coverage and composition. Nat Methods 11, 1144–1146 (2014).
Kang, D. D., Froula, J., Egan, R. & Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165 (2015).
Sieber, C. M. K. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 3, 836–843 (2018).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2020).
Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. The ISME Journal 11, 2864–2868 (2017).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol Biol Evol 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., von Haeseler, A. & Jermiin, L. S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat Methods 14, 587–589 (2017).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49, W293–W296 (2021).
NCBI GenBank https://identifiers.org/ncbi/bioproject: PRJNA544494 (2021).
Acknowledgements
We thank the entire staffs from Yunnan Tengchong Volcano and Spa Tourist Attraction Development Corporation for strong support. This work was financially supported by National Natural Science Foundation of China (42207145, Y.L.Q.; 32170014, Z.S.H.; 92251302, W.J.L.).
Author information
Authors and Affiliations
Contributions
Y.L.Q. and Z.S.H. designed the project and conducted the sampling. Y.L.Q. and H.T.Z. generated and processed the data. Y.L.Q., M.L., W.J.L. and Z.S.H. wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qi, YL., Zhang, HT., Li, M. et al. Recovery of nearly 3,000 archaeal genomes from 152 terrestrial geothermal spring metagenomes. Sci Data 12, 151 (2025). https://doi.org/10.1038/s41597-025-04493-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-04493-z