Abstract
Streptococcus pneumoniae (pneumococcus) is the most common respiratory pathogen worldwide. Nasopharyngeal carriage with S. pneumoniae is the major source of lower respiratory tract infection and horizontal spread among children. Investigating nasopharyngeal S. pneumoniae is crucial for clinicians to control pneumococcus disease. Here, we retrospectively analyzed clinical information of 5,960 hospitalized children, focusing on pneumonia children less than five years with positive nasopharyngeal pneumococcal cultures. Nasopharyngeal aspirates (NPAs) were collected between June 2009 and December 2016, which were outside the pneumococcal conjugate vaccine(PCV) period. NPAs were subjected to common bacterial culture and antibiotic susceptibility tests, and serotypes were identified by both multiplex PCR and DNA sequencing. Results clearly revealed that clinical manifestations of the children whose NPAs were S. pneumoniae culture positive were serious, especially in those less than twelve months old. Fifteen different serotypes of nasopharyngeal S. pneumoniae were detected, the most common ones being 19F (35.2%), 6A/B (23.8%), 19A (11.4%), 15B/C (9.3%) and 23F (7.8%). Eight serotypes, accounting for 85.5% of the isolates, corresponded to the PCV13 serotypes. Approximately one-third of all S. pneumoniae strains were susceptible to penicillin. Overall, we consider nasopharyngeal S. pneumoniae culture is beneficial in assessing the situations of pneumonia children. Moreover, PCV13 could be useful in preventing pneumococcal disease in Chongqing, China.
Similar content being viewed by others
Introduction
Streptococcus pneumonia (pneumococcus) is a significant human pathogen that can cause pneumonia, otitis media, septicemia and meningitis, and constitutes an important cause of death among children under the age of five years1. Nasopharyngeal carriage with S. pneumoniae can be a reservoir of lower respiratory tract (LRT)infection, and is a major prerequisite towards the development of pneumococcal diseases2,3,4,5. The environment of the LRT is normally not sterile6,7; nasopharyngeal microbes can in fact be microaspirated in healthy individuals, and have a higher prevalence during seasons of respiratory diseases8,9. Ongoing surveillance of nasopharyngeal S. pneumoniae characteristics is, therefore, a significant source of epidemiological information. Nonetheless, data on nasopharyngeal S. pneumoniae strains in China have been limited, and here, we describe 7½ years (June 2009-December 2016) retrospective longitudinal study that characterized nasopharyngeal S. pneumoniae strains among a large cohort of pneumonia children. The study is based on S. pneumoniae bacterial culture, serotype distribution and antibiotic susceptibility. The characteristics of nasopharyngeal S. pneumoniae strains of pneumonia children are summarized here, which should be an important resource for clinicians as well as for local and national immunization programs.
Results
Prevalence and clinical correlates of nasopharyngeal S. pneumoniae
Series of clinical data were compared between nasopharyngeal S. pneumoniae culture-positive and -negative cohorts of a range of ages, grouped as less than 12 months (m), 13–36 m and 37–59 m (Table 1). Among these age groups, S. pneumoniae culture-positive rates were 13.9%, 20.4% and 18.9%, respectively. The following differences were observed, especially in children that were less than 12 m old. First, there were several factors associated with pneumococcal carriage and disease. They were more likely to have siblings (Chi-square test, p values were 0.019, 0.016 and 0.561 in the three age groups, respectively), histories of more than 5 days of prehospital antibiotic usage (Chi-square test, p = 0.002, 0.026, 0.222), repeated respiratory tract infection(RTI) (Chi-square test, p = 0.000, 0.37, 0.576), and the history of repeated wheezing (Chi-square test, p = 0.000, 0.042, 0.002). Second, the clinical manifestations were more serious in the positive groups. For example, the lengths of hospital stay were significantly longer in the positive groups (Mann-Whitney U test, p = 0.023, 0.013, 0.106). The morbidities of persistent or chronic pneumonia were more prevalent in the positive groups (Chi-square test, p = 0.000, 0.136, 0.277). Symptoms, such as fever (Chi-square test, p = 0.001, 0.992, 0.632) and wheeze (Chi-square test, p = 0.031, 0.004, 0.530), were also more severe in the positive groups. Third, inflammatory responses, both in the blood and the lungs, were more pronounced in the S. pneumoniae-positive groups, such as the counts of leukocyte (Mann-Whitney U test, p = 0.019, 0.005, 0.936), neutrophil (Mann-Whitney U test, p = 0.001, 0.756, 0.212)and thrombocyte(Mann-Whitney U test, p = 0.031, 0.254, 0.922). Another inflammatory marker, C-reactive protein (CRP), was also higher in positive groups (Chi-square test, p = 0.006, 0.148, 0.931).
Distribution of nasopharyngeal S. pneumoniae serotypes
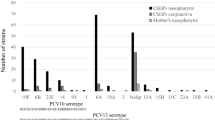
Using multiplex PCR and DNA sequencing, fifteen different serotypes were identified among 193 nasopharyngeal S. pneumoniae culture-positive pneumonia children whose demographic characteristics and clinical data were summarized in Table 2. The most common serotypes were 19 F(35.2%, 95% CI: 28.5–42.4), 6 A/B (23.8%, 95% CI: 18–30.5), 19 A(11.4%, 95% CI: 7.3–16.8), 15B/C(9.3%, 95% CI: 5.6–14.3), 23 F(7.8%, 95% CI: 4.4–12.5) and 14(5.2%, 95% CI: 2.5–9.3)(Fig. 1). The results clearly showed that 19 F and 6 A/B were the most common serotypes, accounting for more than half of all serotypes, whereas 19 A, 15B/C, 23 F and 14 were relatively less prevalent. The other serotypes were seldom detected in this study. All detected serotypes were validated by sequencing. The table of serotypes distribution (Supplementary Table 1) and the detected sequences are provided in the online Supplementary Information files. Currently, two PCVs are dominant: PCV10 and PCV13. In this study, six serotypes of PCV10 representing 73.6% (142/193, 95% CI: 67.4–79.8%) and eight serotypes of PCV13 representing 85.5% (165/193, 95% CI: 80.5–90.5%) were observed, respectively.
Distribution of nasopharyngeal S. pneumoniae serotypes among pneumonia children in Chongqing. Using multiplex PCR and DNA sequencing, fifteen different serotypes were identified among 193 nasopharyngeal S. pneumoniae culture positive pneumonia children. The most common serotypes were 19F(68/193, 35.2%), 6A/B(46/193, 23.8%), 19A(22/193, 11.4%), 15B/C(18/193, 9.3%), 23F(15/193, 7.8%) and 14(10/193, 5.2%), other serotypes were seldom detected. The table of serotypes distribution and the detected sequences have been provided in the online Supplementary Information file. Eight serotypes of PCV13 representing 85.5% were observed.
Antibiotic susceptibility of different S. pneumoniae serotypes
The antibiotic susceptibility tests were performed with nine classes of agents by the Kirby-Bauer disc diffusion method (Table 3). The outcomes were divided into susceptible, intermediate and resistant to specific antibiotics. The different antibiotics susceptibility of S. pneumoniae strains were summarized, which we generalize here. First, 63 (32.6%, 95% CI: 26.1–39.8) and 66 (34.2%, 95% CI: 27.5–41.4) of the 193 S. pneumoniae strains analyzed in the study were respectively susceptible and resistant to penicillin. Second, nearly all of the detected S. pneumoniae strains were susceptible to vancomycin(100%, 95% CI:98.1–100), linezolid(100%, 95% CI:98.1–100), levofloxacin(100%, 95% CI:98.1–100) and chloramphenicol(92.2%, 95% CI:87.5–95.6). Third, most of the S. pneumoniae strains were resistant to clindamycin (81.3%, 95% CI:75.1–86.6), tetracycline (88.6%, 95% CI:83.3–92.7), sulfamethoxazole(89.6%, 95% CI:84.5–93.6) and erythromycin(96.9%, 95% CI:93.4–98.8). 94.3% (182/193, 95% CI:90–97.1) of all strains in the NPAs were MDR strains in this study, and about 66.3% (128/193, 95% CI: 59.6–73%) of all strains were resistant to erythromycin, sulfamethoxazole, tetracycline and clindamycin simultaneously. No PDR S. pneumoniae strain has been detected so far. Lastly, antibiotic susceptibilities of different S. pneumoniae serotypes were summarized in Table 4. The penicillin resistance of 19F and 19A serotypes were similar, which might suggest that the 19 serogroup S. pneumoniae overall is more resistant to penicillin than other serogroups. Tables of antibiotic susceptibility presented as percentages of the total (%) and 95% CI were provided in the online Supplementary Information files (Supplementary Table 2 and Table 3)
Discussion
About 800,000 children die each year due to pneumococcal disease2. As potential pathogen, S. pneumoniae can colonize the nasopharynx at low density without causing symptoms in healthy children, and are less likely to be detected by culture methods10,11. This study documented that positive S. pneumoniae culture of nasopharyngeal aspirates is an important reference for clinicians. S. pneumoniae culture-positive children had several specific characteristics, such as more than one siblings, history of repeated wheezing or respiratory tract infection (RTI) and more common antibiotic usage. Several previous studies had shown a clear association between siblings and the isolation of nasopharyngeal S. pneumoniae12,13. This is likely because close contact can transmit nasopharyngeal S. pneumoniae between siblings in the same family. Children with repeated wheezing were more likely to have positive nasopharyngeal S. pneumoniae detection, which is in agreement with other references14,15. Nasopharyngeal S. pneumoniae species appeared to contribute to respiratory symptoms16, and therefore, avoidance of exposure to S. pneumoniae pathogen or PCV inoculation should be beneficial for repeated RTI or wheezing in children17. As noted, the clinical manifestations of culture-positive children were obviously more serious than those of the negative ones, especially in younger children. The positive group not only had a longer recovery time, but also displayed higher levels of inflammatory markers than the negative group, which was consistent with previous reports15,18. These results supported S. pneumoniae carriage as a prerequisite for pneumococcal infection or diseases5,19. Nasopharyngeal colonization of S. pneumoniae resulted in increased numbers of mucosal and systemic inflammatory cells and higher concentrations of proinflammatory cytokines, which may impact on disease severity20,21,22. High bacterial load in the nasopharynx and local inflammatory reactions were indeed shown to be important in bacterial invasion of the LRT23. The above mentioned reasons may cause transmission of the nasopharyngeal S. pneumoniae into the LRT and aggravate the conditions of the children. Moreover, the younger children exhibited more serious clinical manifestations, probably due to their immature and weaker immunity. Taken together, these findings lead us to conclude that the children who tested positive for nasopharyngeal S. pneumoniae culture had certain risk factors and serious clinical manifestations, especially in children less than 12 m of age.
Various factors may promote or facilitate nasopharyngeal S. pneumoniae invasion of the LRT. Polysaccharide capsules, for example, may play a crucial role in the process24,25, which is not only significant for S. pneumoniae classification, but is also a cardinal determinant of vaccine target. Currently, according to the biochemical structures of the polysaccharide capsule and immunological distinction, S. pneumoniae can be divided into 48 serogroups and 97 serotypes26. The different serotypes have diverse characteristics, such as activation of complement, invasive ability and influence on biofilm formation27,28. It is believed that serotype epidemiology is quite variable both geographically and temporally. Prior to 2000, a large number of epidemiological studies reported that 19F, 6B, 23F and 14 serotypes accounted for the most common pneumococcal serotypes detected in the nasopharynx or in invasive diseases in the United States and several other countries29. Following the widespread use of PCVs, the incidence of pneumococcal diseases dramatically declined, bringing significant benefit to the developing countries30,31,32. PCVs not only protected the vaccinated individuals against disease but also reduced the carriage of vaccine serotypes that could induce herd effects across whole populations33,34. As shown in our study, 19F and 6A/B were the most common serotypes detected in nasopharynx in Chongqing while 19A, 15B/C, 23F, 14 and 22F were also detected, consistent with studies in other Chinese cities35,36,37,38. However, in these studies, S. pneumoniae strains were isolated from patients with invasive pneumococcal disease. These serotypes were mainly the PCV serotypes, likely because the PCVs had not yet been introduced in the national compulsory immunization program in China. Compared with the serotypes of nasopharyngeal S. pneumoniae strains detected in other studies39, there were some serotypes that were seldom detected or not detected in this study, such as serotypes 1, 3 and 5. Geographical division may well be a reason for this difference. Current United States guidelines on vaccine use recommend that children aged 2 to 59 m receive PCV13 as routine care40. Moreover, PCV13 covers serotypes of significantly higher invasive propensity, such as 1, 3, 5, 7F, and 19A39,41, of which 19A has exhibited high prevalence in China, as we have also shown. Furthermore, PCV13 covered the major proportion of serotypes in this study. We thus suggest that PCV13 could indeed be an effective strategy for prevention of invasive pneumococcal disease in Chongqing, and even nationally in China.
The rising occurrence of antibiotic resistance enables S. pneumoniae to be an alarming threat to children’s health. In fact, three major risk factors (antibiotic use, younger age and attending day-care facility) have been identified for nasopharyngeal-resistant S. pneumoniae42. Most importantly, association between carriage or infection with resistant S. pneumoniae and antibiotic use is now widely accepted. Many clinical studies have indeed linked the usage of antibiotics to community-wide antibiotic resistance43,44. It is now confirmed that antibiotic selection pressure enhances antibiotic resistance, and is linked to a reduction of susceptible bacterial strains, shift of the competitive balance, and dissemination of the existing resistant clone(s). The situation is particularly grave in China, where antibiotic usage is popular, as bacterial pathogens occur more frequently in developing countries. Reports have shown that a longer duration of carriage leads to higher incidence of resistance due to the greater risk of antibiotic exposure45. In the S. pneumoniae positive groups, over 30% of children received antibiotics for longer than 5 days before hospitalization, and thus, it is possible that the antibiotics contributed to the observed antibiotic resistance. As shown in our study, almost all S. pneumoniae strains were resistant to clindamycin, sulfamethoxazole, tetracycline and erythromycin, and the most common pattern was co-resistance to former four drugs. The results were consistent with previous reports46,47. Antibiotic-resistant S. pneumoniae could be detected if the antibiotic treatment is conducted within 4 weeks preceding the susceptibility test12,48. But these antibiotics have seldom been used for treatment in Chongqing for a long time. It was also reported that antibiotic resistance was due to the spread of strains belonging to a limited number of clones49. It was speculated that S. pneumoniae clones were stably resistant to former four antibiotics in Chongqing, which may be widely spread by over-use of antibiotics. And the detailed mechanisms of antibiotics resistance await further research. Conversely, S. pneumoniae strains have remained susceptible to other antibiotics, such as vancomycin, linezolid, levofloxacin and chloramphenicol, which lend hope to the treatment of resistant S. pneumoniae. Specifically, the former two antibiotics are better choices, while levofloxacin and chloramphenicol are cautiously used in children in the pediatric clinic. It was found that approximately one-third of all S. pneumoniae was susceptible to penicillin, which is also consistent with other studies43. Taken together, efforts to promote judicious antibiotic use in children appear to be the most appropriate measures to control the spread of antibiotic-resistant clones.
Lastly, we would like to point out potential limitations of our study. First, this study was conducted in a relatively isolated hospital population, and the mild pneumonia children that did not require hospitalization were, therefore, excluded. Second, this was a retrospective, single-center study, and thus, larger and continuous multicenter prospective studies are needed, which should provide crucial data to assess the national immunization program and the effects of vaccines and antibiotics on S. pneumoniae strains. Thirdly, serotyping NPAs were only possible for about 50% of all samples, as NPA samples with a DNA concentration lower than 20 ng/ul did not detected serotypes further. Finally, we have described the characteristics of the nasopharyngeal carriage S. pneumoniae, which should be valuable in monitoring S. pneumoniae epidemiology. Nonetheless, it may be more appropriate to collect S. pneumoniae from the lower respiratory tract to comprehensively monitor invasive S. pneumoniae characteristics in the future.
Methods
Ethics statement
The study was approved by the Ethics Committee of the Children’s Hospital of Chongqing Medical University (Permit number 2015–77). It was conducted in compliance with principles of the declaration of Helsinki. Informed consent was obtained from each parent or guardian on behalf of the children participants, prior to enrollment.
Research subjects and sample collection
During the period from June 2009 to December 2016, a total of 9923 children were hospitalized at the Department of Respiration in Children’s Hospital of Chongqing Medical University. A total of 5960 cases in this period were randomly selected and analyzed (minimum: 50 cases/month, 600 cases/year). In the primary diagnosis, children with no pneumonia were excluded. Pneumonia was diagnosed according to WHO clinical criteria50, lung auscultation with moist rales or evidence of patchy alveolar opacities on chest radiographs. Cases with immune dysfunction/immunodeficiency or heart disease were excluded sequentially. Cases that were positive for other nasopharyngeal bacteria by culture or which S. pneumoniae was co-detected with other bacteria were also excluded, so that the focus was mainly on nasopharyngeal S. pneumoniae strains. Overall, 2583 cases were eligible, consisting of positive nasopharyngeal S. pneumoniae culture in 417 cases and no bacteria in 2166 cases. The conditions of children less than 5 years old were of utmost concern. They were further divided into three age groups: 0–12 m, 13–36 m, and 37–59 m. The demographic and clinical information of the children were collected after admission. NPAs and venous blood were collected within 24 h by trained clinical personnel in accordance with standard protocols. Venous bloods were used for detection and quantification of inflammation markers, such as the leukocytes, neutrophil, thrombocyte and CRP. The normal ranges of these markers are listed in Table 1. Clinical criteria for diagnosis of severe pneumonia was defined by WHO on the basis of cough, tachypnea, difficult breathing, and general danger signs (central cyanosis, inability to breastfeed or drink, severe chest indrawing, head nodding, reduced level of consciousness and convulsions)51. Persistent or chronic pneumonia were defined as the course of pneumonia for 1–3 months or more than3 months, respectively. Chest radiographs were reviewed by specialists. NPA samples with a DNA concentration greater than 20 ng/ul were considered eligible, so only 193 NPA samples (193/390, 49.5%) from children under 5 years of age were further tested for S. pneumoniae serotypes. The screening, eligibility and enrollment of children with pneumonia are summarized in Fig. 2.
Flow chart of screening, eligibility and enrollment of children with pneumonia for comparison of nasopharyngeal S. pneumoniae culture. During the period from June 2009 and December 2016, a total of 9923 children with respiratory tract infection were hospitalized at the Department of Respiration in Children’s Hospital of Medical University of Chongqing, of which 5960 cases were randomly selected(minimum: 50 cases/month, 600 cases/ year). The numbers of NPA samples each year were also listed. 1343 cases with no pneumonia primary diagnosis, 24 cases with immune dysfunction/deficiency, 485 cases with heart disease and 1525 cases of bacterial common culture with detection or co-detection other bacteria were excluded sequentially. Lastly, 2583 cases were eligible, among which 417 cases were S. pneumoniae culture positive. The conditions of children less than 5 years old were of utmost concern, among which S. pneumoniae culture-positive rate was 16.6% (390/2355). They were further divided into three age groups: 0–12 m, 13–36 m and 37–59 m. NPAs with S. pneumoniae culture positive were not all detected serotypes. DNA concentrations of NPA samples more than 20 ng/μl were qualified. Finally, 193 NPAs were detected for S. pneumoniae serotypes further. The numbers of different age group children were shown. The primary diagnose not pneumonia contained: Upper respiratory airway infection: 124, Bronchitis:191, Bronchiolitis:514, Asthma:362, Other:152. Immune dysfunction/deficiency contained: primary immune deficiency or secondary immune deficiency/dysfunction:24. Heart disease contained: Atrial septal defect: 246, Interventricular septal defect:69, Patent ductus arteriosus:43, others:127. Detection or co-detection with other bacteria contained: Haemophilusparainfluenzae:359, Cardamorasia:174, Haemophilusinfluenzae:156, Escherichia coli:149, Staphylococcus aureus:129, Klebsiella pneumoniae:158, Others:191, co- detections:209.
NPA preparation
NPAs were collected into two tubes: one was immediately used for common bacteria culture and antibiotic susceptibility test by standard microbiological methods in the clinical bacteriology laboratory; the other one was sent to the respiratory laboratory for future analysis. The specimens were kept at 4 °C for a maximum of 4 h, and preserved at −80 °C until further use. DNA in the NPAs were extracted using a QIAamp DNA Mini Kit (Qiagen, Germany), following the manufacturer’s instructions. The concentrations of extracted DNA were then determined, and those exceeding 20 ng/ul were considered qualified. The DNA was preserved at −80 °C for subsequent tests.
Bacterial culture and antibiotic susceptibility test
NPA specimens were inoculated on blood plates and chocolate plates within 2 hours of collection, and the plates were cultured at 35 °C for 24–48 hours in a 5–10% CO2 environment. S. pneumoniae was identified by colony morphology, gram staining, catalase test, optochin test, and biliary lysis test. Antibiotic sensitivity tests were performed using the Kirby-Bauer disc diffusion method to determine the sensitivity of all strains to vancomycin, linezolid, levofloxacin, chloramphenicol, penicillin, clindamycin, sulfamethoxazole, tetracycline, and erythromycin. Antibiotic susceptibility was determined according to the Clinical and Laboratory Standards Association (CLSI) guidelines of the year. S. pneumoniae ATCC49619 was included as the control strain. Multi-drug resistance (MDR) S. pneumoniae was defined as resistant to more than 3 classes of antibiotics, while pan-drug resistance (PDR) was defined as resistant to all antibiotics, including glycopeptides and linezolid.
Multiplex PCR and sequencing
S. pneumoniae capsular serotypes were determined both by multiplex PCR and DNA sequencing. Twenty eight oligonucleotide primers, described previously52, were divided into 7 groups and used to detect S. pneumoniae serotypes (Supplementary Table 4), which included not only all common serotypes detected in China but also the PCV 13 serotypes. Multiplex PCR were performed in 25 μl volumes, each reaction mixture containing the following: 1 × PCR buffer (20 mM Tris-HCl, pH 8.0, 100mMKCl, 1 mM dithiothreitol, 0.1 mM EDTA, 0.5% Tween 20, 0.5% NonidetP-40), 6.25 μM of each deoxy nucleoside triphosphate, 62.5 μM of MgCl2, and 1.25 U of Taq DNA polymerase. All samples were analyzed using a commercial detection kit (TaKaRaEx Taq, RR01AM, Dalian, China and Applied Biosystems, Japan). The PCR parameters were: 95 °C for 5 min, followed by 35 amplification cycles of 95 °C for 45 s, 57 °C for 45 s, 72 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR products were analyzed by electrophoresis in 2% NuSieve agarose gels. Specific target primers were then used for further amplification, and the products were sent for sequencing to the Beijing Genomics Institute (BGI).
Statistical analysis
Continuous variables that do not satisfied the normal distribution were expressed as median with inter-quartile range(IQR);Categorical variables were reported as numbers (n), percentages of the total (%) and 95% confidence intervals (95% CI). Comparison between two groups, the Mann-Whitney U test and Chi-square test were used. Fisher’s exact tests were appropriately performed. Comparison among more than three groups, p values were adjusted by Bonferroni correction for multiple comparisons. All tests were two-sided considered statistically significant. SPSS (version 21.0) was used for all analyses.
References
O’Brien, K. L. et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 374, 893–902 (2009).
Weiser, J. N. The pneumococcus: why a commensal misbehaves. J Mol Med. 88, 97–102 (2010).
Siegel, S. J. & Weiser, J. N. Mechanisms of bacterial colonization of the respiratory tract. Annu Rev Microbiol. 69, 425–444 (2015).
Kadioglu, A., Weiser, J. N., Paton, J. C. & Andrew, P. W. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 6, 288–301 (2008).
Bogaert, D., De Groot, R. & Hermans, P. W. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 4, 144–154 (2004).
Aho, V. T. E. et al. The microbiome of the human lower airways: a next generation sequencing perspective. World Allergy Organ J. 8, 23 (2015).
Segal, L. N., Rom, W. N. & Weiden, M. D. Lung microbiome for clinicians. New discoveries about bugs in healthy and diseased lungs. Ann Am Thorac Soc. 11, 108–116 (2014).
Sullivan, A., Hunt, E., MacSharry, J. & Murphy, D. M. The microbiome and the pathophysiology of asthma. Respir Res. 17, 163 (2016).
Venkataraman, A. et al. Application of a neutral community model to assess structuring of the human lung microbiome. MBio. 6, e02284–14 (2015).
Chien, Y. W. et al. Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J. 32, 72–77 (2013).
Da Gloria Carvalho, M. et al. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes. J Clin Microbiol. 48, 1611–1618 (2010).
Koliou, M. G. et al. Risk factors for carriage of Streptococcus pneumoniae in children. BMC Pediatrics. 18, 144 (2018).
Toivonen, L. et al. Burden of recurrent respiratory tract infections in children. J Pediatr Inf Dis. 35, 362–369 (2016).
Esposito, S. et al. Streptococcus pneumoniae colonisation in children and adolescents with asthma: impact of the heptavalent pneumococcal conjugate vaccine and evaluation of potential effect of thirteen-valent pneumococcal conjugate vaccine. BMC Infect Dis. 16, 12 (2016).
Sohail, I., Ghosh, S., Mukundan, S., Zelewski, S. & Khan, M. N. Role of Inflammatory Risk Factors in the Pathogenesis of Streptococcus pneumoniae. Front immunol. 9, 2275 (2018).
Alsuwaidi, A. R. et al. Nasopharyngeal isolates and their clinical impact on young children with asthma: a pilot study. J Asthma Allergy. 11, 233–243 (2018).
Cardozo, D. M. et al. Prevalence and risk factors for nasopharyngeal carriage of Streptococcus pneumoniae among adolescents. J Med Microbiol. 57, 185–189 (2008).
Petraitiene, S. et al. The influence of Streptococcus pneumoniae nasopharyngeal colonization on the clinical outcome of the respiratory tract infections in preschool children. BMC Infect Dis. 15, 403 (2015).
Birgit, S. et al. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines. 11, 841–855 (2012).
Wilson, R. et al. Protection against Streptococcus pneumoniae lung infection after nasopharyngeal colonization requires both humoral and cellular immune responses. Mucosal Immunol. 8, 627–639 (2015).
Yu, D. et al. Impact of bacterial colonization on the severity, and accompanying airway inflammation, of virus-induced wheezing in children. Clin Microbiol Infect. 16, 1399–1404 (2010).
Richards, L., Ferreira, D. M., Miyaji, E. N., Andrew, P. W. & Kadioglu, A. The immunising effect of pneumococcal nasopharyngeal colonisation; protection against future colonisation and fatal invasive disease. Immunobiology. 215, 251–263 (2010).
Short, K. R., Reading, P. C., Wang, N., Diavatopoulos, D. A. & Wijburg, O. L. Increased nasopharyngeal bacterial titers and local inflammation facilitate transmission of Streptococcus pneumoniae. MBio. 3, e00255–12 (2012).
Manso, A. S. et al. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun. 5, 5055 (2014).
Li, J. et al. Epigenetic switch driven by DNA inversions dictates phase variation in Streptococcus pneumoniae. Plos Pathog. 12, e1005762 (2016).
Geno, K. A. et al. Pneumococcal capsules and their types: Past, Present, and Future. Clin Microbiol Rev. 28, 871–899 (2015).
Domenech, M., Araujo-Bazan, L., Garcia, E. & Moscoso, M. In vitro biofilm formation by Streptococcus pneumoniae as a predictor of post-vaccination emerging serotypes colonizing the human nasopharynx. Environ Microbiol. 16, 1193–1201 (2014).
Kadioglu, A. et al. Upper and lower respiratory tract infection by Streptococcus pneumoniae is affected by pneumolysin deficiency and differences in capsule type. Infect Immun 70, 2886–2890 (2002).
Johnson, H. L. et al. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the pneumococcal global serotype project. Plos Med. 7, e1000348 (2010).
Kaplan, S. L. et al. Decrease of invasive pneumococcal infections in children among 8 children’s hospitals in the United States after the introduction of the 7-valent pneumococcal conjugate vaccine. Pediatrics. 113, 443–449 (2004).
Harboe, Z. B. et al. Early effectiveness of heptavalent conjugate pneumococcal vaccination on invasive pneumococcal disease after the introduction in the Danish childhood immunization programme. Vaccine. 28, 2642–2647 (2010).
Bettinger, J. A. et al. The effect of routine vaccination on invasive pneumococcal infections in Canadian children, Immunization Monitoring Program, Active 2000–2007. Vaccine. 28, 2130–2136 (2010).
Collins, A. M. et al. First human challenge testing of a pneumococcal vaccine. Double-blind randomized controlled trial. Am J Respir Crit Care Med. 192, 853–858 (2015).
Fitzwater, S. P., Chandran, A., Santosham, M. & Johnson, H. L. The worldwide impact of the seven-valent pneumococcal conjugate vaccine. Pediatr Infect Dis J. 31, 501–508 (2012).
Pan, F. et al. Serotype distribution, antimicrobial susceptibility, and molecular epidemiology of Streptococcus pneumoniae isolated from children in Shanghai, China. Plos One. 10, e0142892 (2015).
Zhao, C. et al. Phenotypic and genotypic characteristic of invasive pneumococcal isolates from both children and adult patients from a multicenter surveillance in China 2005–2011. PloS One. 8, e82361 (2013).
Li, Q. H. et al. Spread of multidrug-resistant clonal complex 271 of serotype 19F Streptococcus pneumoniae in Beijing, China: characterization of serotype 19F. Epidemiol Infect. 141, 2492–2496 (2013).
Kang, L. H. et al. Molecular epidemiology of pneumococcal isolates from children in China. Saudi Med J. 37, 403–413 (2016).
Greenberg, D. et al. Nasopharyngeal pneumococcal carriage during childhood community-acquired alveolar pneumonia:Relationship between specific serotypes and co-infecting viruses. J Infect Dis. 215, 1111–1116 (2017).
Nuorti, J. P. & Whitney, C. G. Centers for Disease Conrol and Prevention(CDC). Prevention of pneumococcal disease among infantsand children — Use of 13-valent pneumococcal conjugate vaccine and 23-valent pneumococcal polysaccharide vaccine-Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep. 59, 1–18 (2010).
Del Amo, E. et al. High invasiveness of pneumococcal serotypes included in the new generation of conjugate vaccines. Clin Microbiol Infect. 20, 684–689 (2014).
Belongia, E. A. et al. A community intervention trial to promote judicious antibiotic use and reduce penicillinresistant Streptococcus pneumoniae carriage in children. Pediatrics. 108, 575–583 (2001).
Samore, M. H. et al. High rates of multiple antibiotic resistance in Streptococcus pneumoniae from healthy children living in isolated rural communities: Association with cephalosporin use and intrafamilial transmission. Pediatrics. 108, 856–865 (2001).
Skalet, A. H. et al. Antibiotic selection pressure and macrolide resistance in nasopharyngeal Streptococcus pneumoniae: a cluster-randomized clinical trial. PLos Med. 7, e1000377 (2010).
Lehtinen, S. et al. Evolution of antibiotic resistance is linked to any genetic mechanism affecting bacterial duration of carriage. Proc Natl Acad Sci USA 114, 1075–1080 (2017).
Zhanel, G. G. et al. Molecular characterisation of Canadian paediatric multidrug-resistant Streptococcus pneumoniae from 1998–2004. Int J Antimicrob Agents. 28, 465–471 (2006).
Nguyen, Q. H. et al. Decreased Streptococcus pneumoniae susceptibility to oral antibiotics among children in rural Vietnam: a community study. BMC Infect Dis. 10, 85 (2010).
Tyrstrup, M., Melander, E., Hedin, K., Beckman, A. & Molstad, S. Children with respiratory tract infections in Swedish primary care; prevalence of antibiotic resistance in common respiratory tract pathogens and relation to antibiotic consumption. BMC Infect Dis. 603 (2017).
Sjostrom, K. et al. Clonal success of piliated penicillin nonsusceptible pneumococci. Proc Natl Acad Sci USA 104, 12907–12912 (2007).
Khan, A. J., Khan, J. A., Akbar, M. & Addiss, D. G. Acute respiratory infections in children: a case management intervention in Abbottabad District, Pakistan. Bulletin of the World Health Organization. 68, 577–585 (1990).
WHO. Hospital care for children: guidelines for the management of common illnesses with limited resources. 2nd edn., (World Health Organization, Geneva, 2013).
Pai, R., Gertz, R. E. & Beall, B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol. 44, 124–131 (2006).
Acknowledgements
We would like to acknowledge the patients and their guardians involved in this study and the staff in the Department of Respiratory Medicine and the Key Laboratory of Developmental Diseases in Childhood at Chongqing Medical University. This work was supported by the China Special Grant for the Prevention and Control of Infectious Diseases (2012zx10004212) and National Key Specialty [2011] 873.
Author information
Authors and Affiliations
Contributions
E.M.L., X.H.X. and L.R. conceived and designed this study. Y.G., Y. Z. and H.L. assisted to collect the samples and clinical information. Y.Y.Y. performed the experiments, analyzed the data and wrote the manuscript. Y.D., J.L.and Z.X.L. reviewed and revised the manuscript. All authors approved the final manuscript and agree to be accountable for all of the work.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, YY., Xie, XH., Ren, L. et al. Epidemiological characteristics of nasopharyngeal Streptococcus pneumoniae strains among children with pneumonia in Chongqing, China. Sci Rep 9, 3324 (2019). https://doi.org/10.1038/s41598-019-40088-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-40088-6
This article is cited by
-
Prevalence and serotype distribution of nasopharyngeal carriage of Streptococcus pneumoniae among healthy children under 5 years of age in Hainan Province, China
Infectious Diseases of Poverty (2024)
-
Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae in China among children under 14 years of age post-implementation of the PCV13: a systematic review and meta-analysis (2017–2024)
Pneumonia (2024)
-
Genetic determinants of macrolide and tetracycline resistance in penicillin non-susceptible Streptococcus pneumoniae isolates from people living with HIV in Dar es Salaam, Tanzania
Annals of Clinical Microbiology and Antimicrobials (2023)
-
Molecular epidemiology of Streptococcus pneumoniae isolated from pediatric community-acquired pneumonia in pre-conjugate vaccine era in Western China
Annals of Clinical Microbiology and Antimicrobials (2021)
-
Urban Particles Elevated Streptococcus pneumoniae Biofilms, Colonization of the Human Middle Ear Epithelial Cells, Mouse Nasopharynx and Transit to the Middle Ear and Lungs
Scientific Reports (2020)