Abstract
Staphylococcus equorum strain KM1031 is resistant to chloramphenicol, erythromycin and lincomycin. To shed light on the genetic factors underlying these antibiotic resistances, we determined the global gene expression profile of S. equorum KM1031 using RNA sequencing. During chloramphenicol, erythromycin and lincomycin treatment, 8.3% (183/2,336), 16.0% (354/2,336), and 2.9% (63/2,336) of S. equorum KM1031 genes exhibited significant differences in expression, respectively. These three antibiotics upregulated genes related to efflux and downregulated genes related to transporters. Antibiotic treatment also upregulated osmoprotectant-related genes involved in salt tolerance. To identify specific genes functionally related to antibiotic resistance, we compared the genome of strain KM1031 with those of three S. equorum strains that are sensitive to these three antibiotics. We identified three genes of particular interest: an antibiotic biosynthesis monooxygenase gene (abm, AWC34_RS01805) related to chloramphenicol resistance, an antibiotic ABC transporter ATP-binding protein gene (msr, AWC34_RS11115) related to erythromycin resistance, and a lincosamide nucleotydyltransferase gene (lnuA, AWC34_RS13300) related to lincomycin resistance. These genes were upregulated in response to the corresponding antibiotic; in particular, msr was upregulated more than fourfold by erythromycin treatment. Finally, the results of RNA sequencing were validated by quantitative real-time PCR. This transcriptomic analysis provides genetic evidence regarding antibiotic stress responses of S. equorum strain KM1031.
Similar content being viewed by others
Introduction
Four coagulase-negative staphylococci (Staphylococcus carnosus, S. equorum, S. succinus, and S. xylosus) are frequently detected in naturally fermented meat products and cheese1,2,3. These species are recognized as benign bacteria4. S. equorum is commonly used in starter cultures for meat and cheese fermentation5,6, and has been reported to contribute to the aromas of fermented foods through production of low-molecular-weight aromatic compounds such as esters, amino acids, aldehydes, and free fatty acids7,8.
Staphylococcus equorum has also been identified as the dominant species in jeotgal, a high-salt-fermented seafood produced in Korea9. S. equorum strain KS1039 was selected as a starter candidate among many jeotgal-derived S. equorum strains after a series of safety assessments10. These safety assessments showed that most S. equorum isolates from jeotgal were susceptible to 15 types of antibiotic and were nonhemolytic10. Sequencing of the complete genome of S. equorum strain KS1039 demonstrated the absence of virulence genes found in the well-known pathogen S. aureus11. In addition, genomic insights into strain KS1039 suggested its usefulness as a starter culture for aroma enhancement, bacteriocin production, foreign plasmid restriction, and nutrient optimization12.
In our previous study, we undertook a comparative genomic analysis of six phenotypically different S. equorum strains from Korean high-salt seafoods, including the multi-drug resistant strain KM1031, to assess the safety of S. equorum. Our results suggested that antibiotic resistance was linked to acquired antibiotic resistance genes13. However, information from genome sequencing is insufficient to capture the dynamics of gene expression under antibiotic stress. Strain KM1031 exhibited resistance to chloramphenicol, erythromycin, lincomycin and penicillin G based on disk diffusion analysis13. Herein, to better understand the responses of S. equorum strain KM1031 to these antibiotics, we conducted transcriptomic analysis of this strain following administration of each antibiotic. The results revealed expression of genes generally associated with antibiotic treatment as well as changes in the expression of specific genes depending on the antibiotic. This comparative transcriptomic study provides new insights into the antibiotic resistance mechanisms of S. equorum.
Materials and methods
Bacterial strain and culture conditions
Staphylococcus equorum strain KM1031 was previously isolated from the fermented seafood myeolchi-jeotgal and showed resistance to chloramphenicol, erythromycin, and lincomycin10. The complete genome sequence of strain KM1031 has been determined (GenBank accession nos. CP013980–CP013983)13. In this study, strain KM1031 was cultured in tryptic soy broth (TSB; BD, NJ, USA) at 30 °C for 24 h.
Extraction and purification of RNA from S. equorum KM1031
An overnight culture of S. equorum KM1031 grown in TSB was used to inoculate fresh TSB medium to a final concentration of 1% (w/v), followed by incubation at 30 °C. When the optical density at 600 nm (OD600) reached 0.5, the culture was divided in three and chloramphenicol (15 μg/mL), erythromycin (5 μg/mL), and lincomycin (30 μg/mL) were added to each tube. Thereafter, the cells were further incubated at 30 °C for 2 h. Controls were prepared using the same conditions without antibiotics. Cells were collected by centrifugation and total RNA was extracted using TRIzol™ reagent (Invitrogen, Carlsbad, CA, USA) after treatment with lysostaphin (40 μg/mL) at 37 °C for 20 min according to the manufacturer’s instructions.
Total RNA from each sample was subjected to an rRNA-removal process based on the subtractive hybridization/bead capture system of the Ribo-Zero kit (Epicentre Biotechnologies, Madison, WI, USA). Purified RNA samples were used for mRNA-Seq library construction using the Illumina TruSeq RNA Sample Preparation kit v2 (Illumina, San Diego, CA, USA). RNA-Seq was performed using two Illumina HiSeq runs to generate single-end reads around 100 bp in length. All RNA-Seq data analyzed in this study, including whole transcriptome profiles (Supplementary Tables S1 and S2), were deposited in the Sequence Read Archive (SRR10807062–SRR10807065). Using the CLRNASeq program (ChunLab, Seoul, South Korea), sequencing reads were mapped to the S. equorum KM1031 genome and normalized. The normalization methods used in the RNA-Seq analysis included Reads Per Kilobase of transcript per Million mapped reads (RPKM), Relative Log Expression (RLE), and Trimmed Mean of M-value (TMM) (Table S3). Because the coefficient of variation values for the RLE and TMM methods were lower than that for RPKM and because TMM was previously reported to be the most effective normalization method14, TMM was used for normalization of gene expression levels. The p-value for TMM was calculated using edgeR and the fold-change value was calculated as [TMMantibiotic/TMMcontrol]. For subsequent experiments, differentially expressed genes (DEGs) with an absolute log2 [fold change] > 2 were filtered and visualized using the CLRNASeq program. Clusters of orthologous groups (COG) analysis15 was used for functional grouping of all strain KM1031 genes. The proportion of DEGs in each functional group was calculated.
Quantitative real-time PCR (qRT-PCR)
The expression levels of specific genes were validated using qRT-PCR. qRT-PCR was performed using a C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA) with IQ™ SYBR®Green Supermix (Bio-Rad). Thermal cycling consisted of 3 min at 95 °C, followed by 40 cycles of 10 s at 95 °C and 30 s at 60 °C. The primers used for the detection of target genes are listed in Supplementary Table S4. Expression levels of all genes were quantified in duplicate using three independent experiments. These analyses were performed on the same batches of RNA as those used for transcriptomic experiments. The 16S rRNA gene was used as the reference gene for normalization. Results were normalized using the comparative cycle threshold method16.
Comparative genomics of S. equorum strains
For comparative genomic analysis within S. equorum strains, genome sequence data for strains KM1031 [chloramphenicol, erythromycin and lincomycin resistant (CRERLR); GenBank accession: CP013980–CP013983], C2014 [chloramphenicol, erythromycin and lincomycin sensitive (CSESLS); GenBank accession: CP013714–CP013719], and KS1039 (CSESLS; GenBank accession: CP013114.1) were obtained from the NCBI database (http://ncbi.nlm.nih.gov/genomes).
Cloning of the abm and msr genes and assessment of their roles in antibiotic resistance
To assess whether antibiotic resistance was dependent on the abm and msr genes, both full-length genes from S. equorum strain KM1031 were cloned and recombinantly expressed in Escherichia coli. PCR amplifications from strain KM1031 using specific primer sets (Supplementary Table S5) were performed using a T-3000 thermocycler (Biometra, Göttingen, Germany). The PCR mixture was prepared according to the manual for Inclone Taq DNA polymerase (Inclone Biotech, Daejeon, South Korea). Samples were preheated for 5 min at 95 °C and then amplified using 30 cycles of 1 min at 95 °C, 30 s at 55 °C, and 1 min at 72 °C. The PCR products for abm and msr were digested with XhoI and EcoRI, respectively, and then inserted into pYJ335 and pCL55 digested with the same enzymes. The resulting plasmids were designated pYJ335-abm and pCL55-msr, respectively.
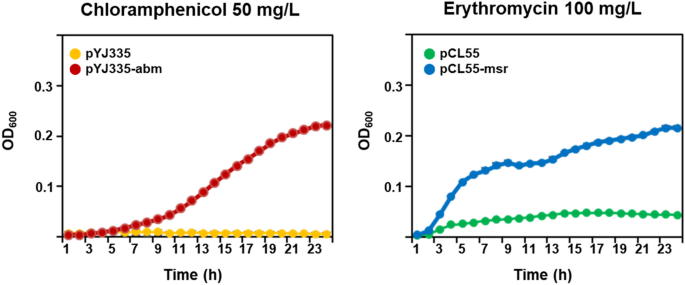
To assess the roles of abm and msr in chloramphenicol and erythromycin resistance, respectively, E. coli DH5α (pYJ335-abm) and E. coli DH5α (pCL55-msr) cultures in Luria–Bertani medium (LB; BD) were normalized to an OD600 of 0.5 and diluted tenfold. A 10 μL aliquot was subcultured into LB medium containing 50 mg/L chloramphenicol or 100 mg/L erythromycin. The antibiotic resistance of E. coli transformants was determined by examining their growth. E. coli cells transformed with empty plasmids were used as negative controls.
Results
Comprehensive transcriptome analysis of S. equorum strain KM1031 under antibiotic stress
In a previous study, S. equorum strain KM1031 showed resistance to chloramphenicol, erythromycin, lincomycin, and penicillin G based on disk diffusion analysis 10,13. However, in the MIC results, it was determined that strain KM1031 was sensitive to the penicillin G because it did not grow at 1 mg/L of penicillin G. To understand the bacterial response and adaptations during three antibiotic stress excluding penicillin G, RNA was isolated from S. equorum strain KM1031 following antibiotic stress for RNA-Seq analysis. RNA-Seq data were acquired, mapped, and normalized as described in the “Methods” section (Supplementary Tables S1 and S2). A total of 2,336 strain KM1031 genes were categorized using COG analysis. Antibiotic treatment affected the expression of several genes in strain KM1031 (Supplementary Figs. S1 and S2). After mRNA abundance was compared between control and antibiotic-exposed cells, genes showing a log2 (fold-change) greater than 2 or less than − 2 were considered to be DEGs (Supplementary Tables S6 and S7; Supplementary Fig. S1). In strain KM1031 cells exposed to chloramphenicol, erythromycin, and lincomycin stress, 8.3% (183/2,336), 16.0% (354/2,336), and 2.9% (63/2,336) of genes exhibited significant differences in their expression, respectively (Fig. 1B).
Classification of differentially expressed genes (DEGs) based on predicted functions. (A) DEG analysis from RNA-Seq data comparing untreated Staphylococcus equorum strain KM1031 with strain KM1031 treated with antibiotics. The x-axis shows log-scaled Trimmed Mean of M-value (TMM) data for strain KM1031, and the y-axis shows log-scaled TMM values for cells treated with chloramphenicol, erythromycin, and lincomycin, respectively. Total gene expression in the two conditions was filtered to identify significantly down- or upregulated genes with the criteria P-value ≤ 0.05 and fold-change ≥ 2. (B) Genes upregulated or downregulated by twofold or more following treatment of the bacterium with antibiotics were grouped into functional categories based on the Clusters of Orthologous Groups database.
Following chloramphenicol treatment, 75 genes were significantly upregulated and 108 genes were significantly downregulated (Supplementary Tables S6 and S7; and Fig. 1). Significant upregulation was observed for genes associated with translation, ribosomal structure, and biogenesis (22.7%; 17/75) as well as genes associated with amino acid transport and metabolism (20.0%; 15/75). By contrast, genes associated with “function unknown” (24.1%; 26/108) and transcription (19.4%; 21/108) were downregulated.
Following erythromycin treatment, 214 genes were significantly upregulated and 140 genes were significantly downregulated. Significant upregulation was observed for genes associated with translation, ribosomal structure, and biogenesis (28.5%; 61/214) as well as genes associated with “function unknown” (11.2%; 24/214). By contrast, genes associated with “function unknown” (29.3%; 41/140) and transcription (14.3%; 20/140) were downregulated.
Following lincomycin treatment, 43 genes were significantly upregulated and 20 genes were significantly downregulated. Significant upregulation was observed for genes associated with “function unknown” (25.6%; 11/43) as well as genes associated with translation, ribosomal structure, and biogenesis (14.0%; 6/43). By contrast, genes associated with “function unknown” (45.0%; 9/20) and transcription (20.0%; 9/20) were downregulated.
Effects of antibiotics on efflux proteins and transporters
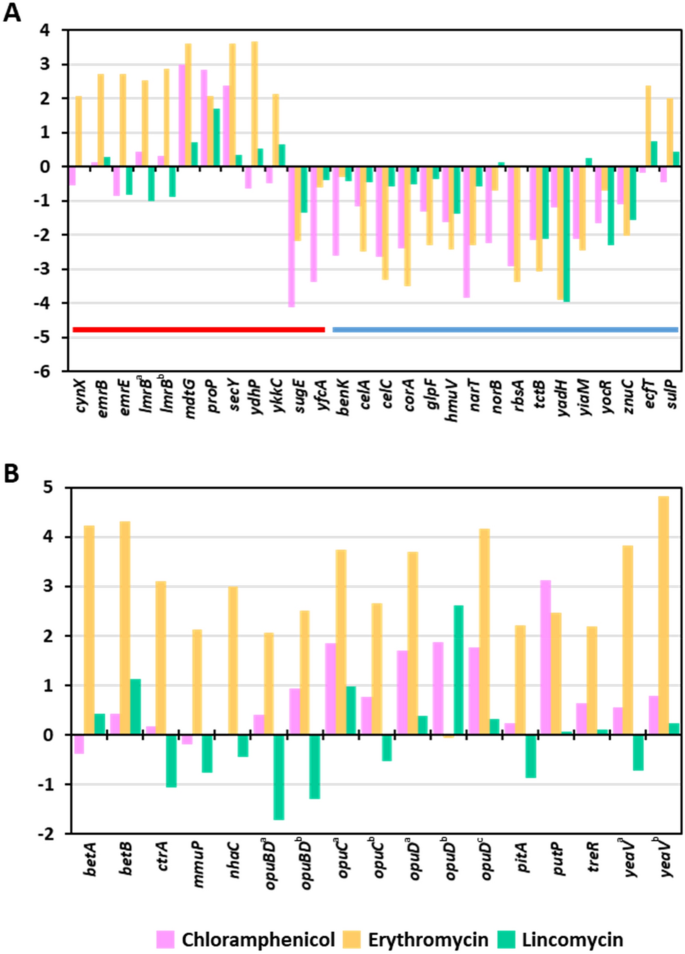
Transporter and efflux proteins are required for antibiotics to enter or be expelled from bacteria17,18. Thus, we hypothesized that antibiotics would alter the expression of efflux- and transporter-related genes in S. equorum. DEGs were screened using the keywords “efflux” and “transporter.” Among DEGs following treatment with chloramphenicol, erythromycin and lincomycin, 7.1% (13/183), 6.8% (24/354), and 4.8% (3/63), respectively, were related to transporters and efflux (Fig. 2A; Supplementary Table S8). Chloramphenicol and erythromycin treatment (especially the former) upregulated efflux-related genes and downregulated transporter-related genes. Similar results were observed for lincomycin, although expression changes were less dramatic compared with the other two antibiotics.
Effects of antibiotics on expression of genes related to salt tolerance
Accumulation or release of compatible solutes such as glycine betaine, proline betaine, and carnitine confers salt tolerance by facilitating the response of cells to osmotic pressure19. Interestingly, osmoprotectant-related genes, such as those involved in the synthesis of trehalose, glycine betaine, choline, and proline, were upregulated following chloramphenicol and erythromycin treatment (Fig. 2B; Supplementary Table S8), while lincomycin only slightly affected the expression of a few genes related to salt tolerance. Zhu and Dai20 reported that overexpression of efflux pumps required for salt tolerance led to decreased antibiotic susceptibility. Our results suggest that strain KM1031 express salt tolerance-related genes to counter the effects of antibiotics, especially chloramphenicol and erythromycin.
Responsive genes to three antibiotics based on transcriptomic and comparative genomic analyses
We hypothesized that some genes in S. equorum strain KM031 might be specifically and functionally (i.e., mechanistically) related to chloramphenicol and erythromycin resistance. To identify such genes, we undertook comparative genomic analysis of strains KM1031 (CRERLR), C2014 (CSESLS), and KS1039 (CSESLS).
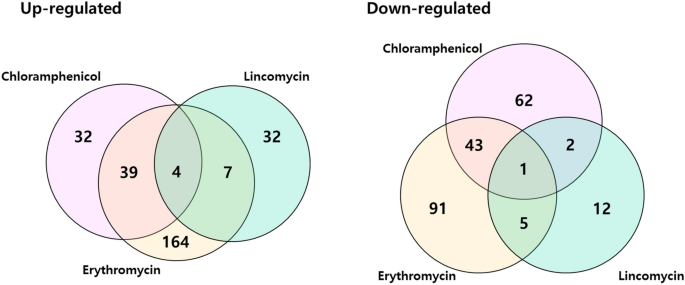
We plotted Venn diagrams of genes that were significantly differentially expressed in S. equorum strain KM031 in response to chloramphenicol, erythromycin and lincomycin (Fig. 3). Four genes (AWC34_RS06585, AWC34_RS08650, AWC34_RS10220, and AWC34_RS12080) were upregulated by all three antibiotics, while one (AWC34_RS11270) was downregulated by all three antibiotics (Supplementary Tables S6 and S7). These genes were detected in one or more of the complete genome sequences of the antibiotic-sensitive S. equorum strains C2014 and KS1039 based on comparative genomic analysis.
Venn diagram of differentially expressed genes (DEGs) of S. equorum strain KM1031 following treatment with chloramphenicol, erythromycin and lincomycin. Overlapping regions represent genes that were differentially expressed in strain KM1031 (compared with untreated cells) on treatment with two or three of the antibiotics. The numbers outside overlapping regions indicate the numbers of significantly differentially expressed genes that were affected by each antibiotic individually.
Interestingly, an antibiotic ABC transporter ATP-binding protein-encoding gene (msr, AWC34_RS11115) was identified among genes specifically upregulated in response to erythromycin. Msr is annotated, among other things, as an erythromycin resistance ATP-binding protein. This gene was suggested to be responsible for erythromycin resistance in a previous genomic study of S. equorum strain KM103113. Reynolds et al. reported that Msr gives rise to erythromycin resistance via an active transport process21. Collectively, these findings strongly suggest that the antibiotic ABC transporter ATP-binding protein-coding gene (AWC34_RS11115) confers erythromycin resistance in strain KM1031. Chloramphenicol- and lincomycin-specific response genes were not identified among DEGs. Therefore, we conclude that most commonly up- and downregulated genes under antibiotic pressure are associated with general environmental responses, and not responses to chloramphenicol, erythromycin, and/or lincomycin specifically.
We hypothesized that antibiotic exposure might increase the expression of genes that are specifically related to antibiotic resistance (i.e., that encode proteins that are involved in the molecular-level resistance of the bacteria to the drug). Thus, we took the set of genes that were upregulated in S. equorum strain KM031 (not DEGs) in response to any of the antibiotics and subtracted genes detected in the two CSESLS strains. This left 65 strain KM1031-specific genes (Table 1). The msr (AWC34_RS11115) gene was among them. In a previous study, we suggested that an antibiotic biosynthesis monooxygenase-encoding gene (abm AWC34_RS01805) and a lincosamide nucleotydyltransferase-encoding gene (lnuA, AWC34_RS13300) might confer resistance to chloramphenicol and lincomycin, respectively13. These genes were also among the KM1031-specific genes and abm and lnuA were slightly upregulated by exposure to chloramphenicol and lincomycin, respectively (Table 1). The lincomycin-resistance phenotype of lnuA in strain KM1031 has already been reported22. These results imply that abm may confer resistance to chloramphenicol, although abm was not significantly upregulated by chloramphenicol treatment.
To investigate the effect of the abm and msr genes on chloramphenicol and erythromycin resistance, the genes AWC34_RS01805 and AWC34_RS11115 were PCR amplified and then cloned into the pYJ335 and pCL55 vectors, respectively. The resulting plasmids were designated pYJ335-abm for the gene AWC34_RS01805 and pCL55-msr for the gene AWC34_RS11115. E. coli transformants harboring pYJ335-abm and pCL55-msr grew under chloramphenicol and erythromycin pressure, respectively (Fig. 4). Collectively, these results suggested that chloramphenicol and erythromycin treatment modified the expression of the abm and msr genes in S. equorum strain KM031, and that the gene products encoded by these genes contributed to the phenotypic resistance of E. coli cells to these antibiotics.
Effects of antibiotics on two-component systems
Although we identified specific genes that may be responsible for the observed antibiotic resistance of S. equorum strain KM031, the transcriptional regulators of specific antibiotic resistance gene expression remained unclear. Two-component systems (TCSs) are the most common systems for bacterial signal transduction in response to environmental signals such as antibiotics and salts. TCS signaling is involved in bacterial resistance to antibiotics. Several TCSs have been detected in S. equorum genomes13. However, most TCS genes were not markedly up- or down-regulated in our experiments, except the WalKR TCS (Table 2). Although walKR genes were not significantly differentially expressed, expression of these genes increased following treatment with chloramphenicol, erythromycin, and lincomycin. The WalKR TCS regulates genes responsible for cell wall metabolism and homeostasis, as well as genes involved in stress responses, virulence, and biofilm formation23,24,25,26. Although the WalR consensus binding site (5′-TGTWAH N5 TGTWAH-3′)27 was not identified upstream of abm, msr or lnuA, we hypothesize that the WalKR TCS might be related to expression of these three antibiotic-specific responsive genes.
Validation of RNA-Seq data by qRT-PCR
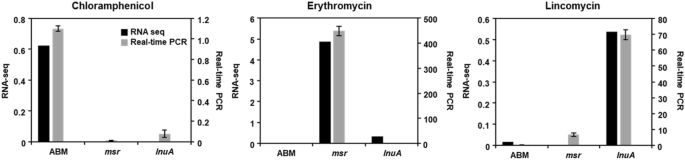
qRT-PCR was used to validate the S. equorum strain KM031 transcriptional profiles obtained by RNA-Seq analysis. As shown in Fig. 5, the expression patterns for each gene (abm, msr, and lnuA) were similar by qRT-PCR and RNA-Seq. Expression of the abm, msr and lnuA genes increased following exposure to chloramphenicol, erythromycin, and lincomycin, respectively. In addition, expression of walKR genes was increased following exposure the three antibiotics, although not significantly (Supplementary Fig. S3). Thus, our RNA-Seq data were confirmed by qRT-PCR.
Validation of RNA sequencing data by quantitative real-time PCR (qRT-PCR). Genes related to resistance to chloramphenicol, erythromycin, and lincomycin were selected for validation under different antibiotic pressures. Data are expressed as log2 fold-changes in gene expression between control and antibiotic-treated samples. In qRT-PCR, 16S rRNA gene expression was used for normalization of target gene expression.
Discussion
We sought to identify the genes that confer resistance to the antibiotics chloramphenicol, erythromycin and lincomycin in S. equorum strain KM031. Using transcriptomic analysis, we confirmed that abm, msr, and lnuA are associated with resistance to chloramphenicol, erythromycin, and lincomycin, respectively.
Chloramphenicol binds to residues A2451 and A2452 of the 23S rRNA and inhibits peptidyl transferase activity by hampering the binding of transfer RNA to the A site of the ribosome28. The most common mechanism of resistance to chloramphenicol in bacteria is enzymatic inactivation of chloramphenicol by acetylation, mainly via acetyltransferases, or, in some cases, by chloramphenicol phosphotransferases. Antibiotic biosynthesis monooxygenase oxidizes phenolic compounds and acts as a superoxide scavenger as a defense mechanism in the host29. It is also involved in dehalogenation of various aromatic and aliphatic compounds30. Chloramphenicol is a halogen and phenyl-containing antibiotic. Therefore, we suggest that antibiotic biosynthesis monooxygenase (abm, AWC34_RS01805) might contribute to the loss of chloramphenicol activity in S. equorum strain KM031 through phenol-oxidation and/or dehalogenation. This hypothesis requires further study.
Erythromycin inhibits protein synthesis and subsequent structural and functional processes by binding to the 23S rRNA31. Erythromycin interferes with aminoacyl translocation, preventing the transfer of the tRNA bound at the A site of the rRNA complex to the P site of the rRNA complex, and consequently prevents movement along mRNA. The most common mechanisms of resistance to erythromycin in bacteria are target modification through methylation of 23S rRNA catalyzed by the product of the erm gene and efflux of erythromycin31,32. Some reports suggest that msr encodes an antibiotic ABC transporter ATP-binding protein and confers resistance to erythromycin through energy-dependent efflux of erythromycin21,33. However, Sharkey et al.34 suggested that Msr confers erythromycin resistance through ribosomal protection. Strain KM1031 possesses, strain-specifically, an msr (AWC34_RS11115) gene. The product of the msr gene contains three conserved motifs: Walker A (GXXGXGKST), Walker B (hhhhDEPTNXLD, where h is a hydrophobic residue), and the signature motif (LSGGE)35,36. Transmembrane prediction software (TMHMM Serve v.2.20) did not predict a transmembrane ___domain in the MsrA of S. equorum strain KM1031. Therefore, we suggest that S. equorum strain KM1031Msr might confer erythromycin resistance through ribosomal protection, rather than by acting as an efflux pump.
Lincomycin belongs to the lincosamides, which interact with the A and P sites of the 50S ribosome37. Although the binding sites of the lincosamides differ from those of chloramphenicol, lincosamides also inhibit protein synthesis by inhibition of peptidyl transferases38. The most common mechanisms of resistance to lincomycin in bacteria are enzymatic inactivation by acetylation, mainly via acetyltransferases, and efflux39. LnuA modifies lincomycin by AMP addition onto the hydroxyl groups of the methylthiolincosamide via nucleotidyltransferase reaction40. Strain KM1031 possesses a lincosamide nucleotidyltransferase-encoding gene (lnuA; AWC34_RS13300), and thus we assume that LnuA confers resistance to lincomycin via lincomycin modification.
Ahmad et al. suggested that antibiotic resistance genes may be regulated by TCSs41. However, we found that TCSs were slightly upregulated, although not significantly, by antibiotic treatment in this study. Thus, if TCSs are involved in the expression of antibiotic resistance genes in strain KM1031, their activity may be regulated by phosphorylation, rather than by changes in their expression levels following antibiotic exposure. The expression of the walKR TCS was shown to increase slightly in S. equorum strain KM1031 under antibiotic pressure both by RNA-Seq and RT-PCR analyses (Table 2; and Supplementary Fig. S3). Although we did not identify the DNA binding motif of WalR upstream of the three putative antibiotic resistance genes reported in this study, we suggest that the walKR TCS may be involved in expression of antibiotic resistance genes.
Apart from TCSs, global regulators may be involved in the expression of antibiotic-specific response genes. Using the keyword “regulator,” 138 regulator genes were detected in the genome of S. equorum KM1031 (Supplementary Table S9); 10 and two of these genes were significantly upregulated by erythromycin and lincomycin, respectively. Prior results suggested that such regulators are involved in the expression of antibiotic resistance genes, but the underlying mechanism remains unclear42,43. Because regulators bind to their targets as dimers, we checked for direct repeat sequences upstream of abm, msr, and lnuA using Tandem Repeats Finder44, but no repeats or palindromes were identified. Additional studies will be required to understand the regulation of antibiotic resistance-related gene expression in S. equorum KM1031.
Generally, the molecular mechanisms through which bacteria become drug resistant involve antibiotic efflux, antibiotic inactivation, or alteration of the antibiotic target site in the bacterium. We suggest that alteration of chloramphenicol and lincomycin activity by the products of the abm and lnuA genes, respectively, contribute to the resistance of S. equorum strain KM1031 to these antibiotics, and that msr confers the erythromycin resistance of strain KM1031 by ribosomal protection. Changes in expression of genes related to efflux, transport, and salt tolerance may nonspecifically contribute to resistance to all three antibiotics. However, further studies are required to define the specific mechanisms of resistance, including gene regulation and the mechanism of acquisition of relevant genes. This information will help reduce the antibiotic resistance of food bacteria such as starters involved in food fermentation.
Data availability
RNA-Seq data analyzed in this study were deposited in the Sequence Read Archive (SRR10807062–SRR10807065). The data presented in this study are available in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
References
Coton, E. et al. Biodiversity of coagulase-negative Staphylococci in French cheeses, dry fermented sausages, processing environments and clinical samples. Int. J. Food Microbiol. 137(2–3), 221–229 (2010).
Casaburi, A., Blaiotta, G., Mauriello, G., Pepe, O. & Villani, F. Technological activities of Staphylococcus carnosus and Staphylococcus simulans strains isolated from fermented sausages. Meat Sci. 71(4), 643–650 (2005).
Blaiotta, G., Casaburi, A. & Villani, F. Identification and differentiation of Staphylococcus carnosus and Staphylococcus simulans by species-specific PCR assays of sodA genes. Syst. Appl. Microbiol. 28(6), 519–526 (2005).
Heo, S., Lee, J.-H. & Jeong, D.-W. Food-derived coagulase-negative Staphylococcus as starter cultures for fermented foods. Food Sci. Biotechnol. 29(8), 1023–1035 (2020).
Place, R. B., Hiestand, D., Gallmann, H. R. & Teuber, M. Staphylococcus equorum subsp. linens, subsp. nov., a starter culture component for surface ripened semi-hard cheeses. Syst. Appl. Microbiol. 26(1), 30–37 (2003).
Schleifer, K. H., Kilpperbalz, R. & Devriese, L. A. Staphylococcus arlettae sp. nov., Staphylococcus equorum sp. nov. and Staphylococcus kloosii sp. nov.: Three new coagulase-negative, novobiocin-resistant species from animals. Syst. Appl. Microbiol. 5(4), 501–509 (1984).
Deetae, P., Bonnarme, P., Spinnler, H. E. & Helinck, S. Production of volatile aroma compounds by bacterial strains isolated from different surface-ripened French cheeses. Appl. Microbiol. Biotechnol. 76(5), 1161–1171 (2007).
Fulladosa, E. et al. Volatile profile and microbiological characterization of hollow defect in dry-cured ham. Meat Sci. 86(3), 801–807 (2010).
Guan, L., Cho, K. H. & Lee, J. H. Analysis of the cultivable bacterial community in jeotgal, a Korean salted and fermented seafood, and identification of its dominant bacteria. Food Microbiol. 28(1), 101–113 (2011).
Jeong, D.-W., Han, S. & Lee, J.-H. Safety and technological characterization of Staphylococcus equorum isolates from jeotgal, a Korean high-salt-fermented seafood, for starter development. Int. J. Food Microbiol. 188, 108–115 (2014).
Jeong, D.-W., Na, H., Ryu, S. & Lee, J.-H. Complete genome sequence of Staphylococcus equorum KS1039 isolated from Saeu-jeotgal, Korean high-salt-fermented seafood. J Biotechnol. 219, 88–89 (2016).
Lee, J.-H., Heo, S. & Jeong, D.-W. Genomic insights into Staphylococcus equorum KS1039 as a potential starter culture for the fermentation of high-salt foods. BMC Genom. 19(1), 136 (2018).
Jeong, D.-W., Heo, S., Ryu, S., Blom, J. & Lee, J.-H. Genomic insights into the virulence and salt tolerance of Staphylococcus equorum. Sci. Rep. 7(1), 5383 (2017).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11(3), R25 (2010).
Tatusov, R. L., Koonin, E. V. & Lipman, D. J. A genomic perspective on protein families. Science 278(5338), 631–637 (1997).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29(9), e45 (2001).
Rahman, T., Yarnall, B. & Doyle, D. A. Efflux drug transporters at the forefront of antimicrobial resistance. Eur. Biophys. J. 46(7), 647–653 (2017).
Greene, N. P., Kaplan, E., Crow, A. & Koronakis, V. Antibiotic resistance mediated by the MacB ABC transporter family: A structural and functional perspective. Front. Microbiol. 9, 950 (2018).
Wood, J. M. et al. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A 130(3), 437–460 (2001).
Zhu, M. & Dai, X. High salt cross-protects Escherichia coli from antibiotic treatment through increasing efflux pump expression. mSphere 3(2), e0009518 (2018).
Reynolds, E., Ross, J. I. & Cove, J. H. Msr(A) and related macrolide/streptogramin resistance determinants: Incomplete transporters?. Int. J. Antimicrob. Agents 22(3), 228–236 (2003).
Lee, J.-H. & Jeong, D.-W. Characterization of mobile Staphylococcus equorum plasmids isolated from fermented seafood that confer lincomycin resistance. PLoS ONE 10(10), e0140190 (2015).
Howell, A. et al. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49(6), 1639–1655 (2003).
Dubrac, S., Boneca, I. G., Poupel, O. & Msadek, T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189(22), 8257–8269 (2007).
Howden, B. P. et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog. 7(11), e1002359 (2011).
Delaune, A. et al. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect. Immun. 80(10), 3438–3453 (2012).
Ayala, E. et al. A biochemical characterization of the DNA binding activity of the response regulator VicR from Streptococcus mutans. PLoS ONE 9(9), e108027 (2014).
Gu, Z., Harrod, R., Rogers, E. J. & Lovett, P. S. Anti-peptidyl transferase leader peptides of attenuation-regulated chloramphenicol-resistance genes. Proc. Natl. Acad. Sci. U. S. A. 91(12), 5612–5616 (1994).
Lemieux, M. J. et al. The crystal structure of Rv0793, a hypothetical monooxygenase from M. tuberculosis. J. Struct. Funct. Genom. 6(4), 245–257 (2005).
Arora, P., Srivastava, A. & Singh, V. P. Application of monooxygenases in dehalogenation, desulphurization, denitrification and hydroxylation of aromatic compounds. J. Bioremed. Biodegrad. 1(3), 112 (2010).
Vazquez-Laslop, N. & Mankin, A. S. How macrolide antibiotics work. Trends Biochem. Sci. 43(9), 668–684 (2018).
Lovmar, M. et al. Erythromycin resistance by L4/L22 mutations and resistance masking by drug efflux pump deficiency. EMBO J. 28(6), 736–744 (2009).
Ross, J. I. et al. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 4(7), 1207–1214 (1990).
Sharkey, L. K., Edwards, T. A. & O’Neill, A. J. ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio 7(2), e01975 (2016).
Mendez, C. & Salas, J. A. ABC transporters in antibiotic-producing actinomycetes. FEMS Microbiol. Lett. 158(1), 1–8 (1998).
Burnie, J. P. et al. Identification of an immunodominant ABC transporter in methicillin-resistant Staphylococcus aureus infections. Infect. Immun. 68(6), 3200–3209 (2000).
Matzov, D. et al. Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus. Nucleic Acids Res. 45(17), 10284–10292 (2017).
Cocito, C., Di Giambattista, M., Nyssen, E. & Vannuffel, P. Inhibition of protein synthesis by streptogramins and related antibiotics. J. Antimicrob. Chemother. 39(Suppl A), 7–13 (1997).
Schwarz, S. et al. Lincosamides, streptogramins, phenicols, and pleuromutilins: Mode of action and mechanisms of resistance. Cold Spring Harb. Perspect. Med. 6(11), a027037 (2016).
Liu, Y., Li, R., Xiao, X. & Wang, Z. Molecules that inhibit bacterial resistance enzymes. Molecules 24(1), 43 (2018).
Ahmad, A., Majaz, S. & Nouroz, F. Two-component systems regulate ABC transporters in antimicrobial peptide production, immunity and resistance. Microbiology 166(1), 4–20 (2020).
Cuthbertson, L. & Nodwell, J. R. The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77(3), 440–475 (2013).
Depardieu, F., Podglajen, I., Leclercq, R., Collatz, E. & Courvalin, P. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 20(1), 79–114 (2007).
Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27(2), 573–580 (1999).
Funding
This work was supported by the National Research Foundation of Korea (NRF) [NRF-2019R1A2C1003639]. We thank Edanz (www.edanz.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
D.W.J. designed and supervised the study. S.H., T.K., H.E.N, G.L., and J.H.L. collected and analyzed the data. S.H., J.H.L., and D.W.J. drafted the manuscript. S.H. and D.W.J finalized the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Heo, S., Kim, T., Na, HE. et al. Transcriptomic analysis of Staphylococcus equorum KM1031 from the high-salt fermented seafood jeotgal under chloramphenicol, erythromycin and lincomycin stresses. Sci Rep 12, 15541 (2022). https://doi.org/10.1038/s41598-022-19897-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-19897-9
This article is cited by
-
Unraveling the role of M2 TAMs in ovarian cancer dynamics: a systematic review
Journal of Translational Medicine (2025)