Abstract
In addition to focal lesions, diffusely abnormal white matter (DAWM) is seen on brain MRI of multiple sclerosis (MS) patients and may represent early or distinct disease processes. The role of MRI-observed DAWM is understudied due to a lack of automated assessment methods. Supervised deep learning (DL) methods are highly capable in this ___domain, but require large sets of labeled data. To overcome this challenge, a DL-based network (DAWM-Net) was trained using semi-supervised learning on a limited set of labeled data for segmentation of DAWM, focal lesions, and normal-appearing brain tissues on multiparametric MRI. DAWM-Net segmentation performance was compared to a previous intensity thresholding-based method on an independent test set from expert consensus (N = 25). Segmentation overlap by Dice Similarity Coefficient (DSC) and Spearman correlation of DAWM volumes were assessed. DAWM-Net showed DSC > 0.93 for normal-appearing brain tissues and DSC > 0.81 for focal lesions. For DAWM-Net, the DAWM DSC was 0.49 ± 0.12 with a moderate volume correlation (ρ = 0.52, p < 0.01). The previous method showed lower DAWM DSC of 0.26 ± 0.08 and lacked a significant volume correlation (ρ = 0.23, p = 0.27). These results demonstrate the feasibility of DL-based DAWM auto-segmentation with semi-supervised learning. This tool may facilitate future investigation of the role of DAWM in MS.
Similar content being viewed by others
Introduction
Multiple sclerosis (MS) is the most common non-traumatic demyelinating disease in young adults that affects more than 2.8 million people worldwide1. Treatment options include disease-modifying drugs that limit inflammation associated with acute tissue destruction and clinical attacks. Many of these treatments have limited to moderate efficacy. Noninvasive identification of biological correlates of disease load, activity, and progression could allow for the development of more effective and targeted therapies and improved outcome.
Magnetic resonance imaging (MRI) plays a key role both in diagnosis and management of MS2,3. The hallmark of MS is the presence of focal hyperintense lesions on T2-weighted (T2w) MRI in the central nervous system (CNS). In addition to focal lesions, diffusely abnormal white matter (DAWM) is observed on T2w, proton density-weighted (PDw), and T2w fluid-attenuated inversion recovery (FLAIR) MRI. DAWM appears as diffuse hyperintense regions with poorly defined margins and is predominantly observed in the periventricular and centrum semiovale regions4,5,6,7. The signal intensity of DAWM is intermediate to focal lesions and normal-appearing white matter (NAWM)8, often overlapping with that of normal-appearing gray matter (NAGM). DAWM is seen in at least 25% of MS patients6,9 and it is also detected in patients with only a few focal lesions10. Histological studies have suggested that DAWM may represent an early disease process with the potential to progress into focal lesions, while other studies have suggested that DAWM represents a pathologic process distinct from that of focal lesions7,11,12,13. Associations with DAWM have been reported for some histopathologic markers and clinical symptoms6,7,9,14,15,16,17,18,19,20,21.
Despite the potential role for DAWM as a marker of MS pathology suggested in those studies, DAWM is often not considered in a majority of imaging studies in MS. A major reason for this is the lack of reliable and automated methods to segment and quantify DAWM volume. The diffuse appearance of DAWM makes manual segmentation difficult and time consuming. This also introduces inter-reader variability. To our knowledge, inter-reader variability of DAWM segmentation has yet to be assessed. Maranzano et al. utilized automated intensity thresholding applied to normalized T2w images to discriminate DAWM and focal lesions (T2L) from NAWM22, but the performance of this technique has not yet been assessed against manual segmentation or supported by an independent study.
Deep learning (DL) methods, particularly fully convolutional neural networks (CNNs) such as the U-Net23, have consistently shown improved performance on a variety of tasks in MS, including the segmentation of T2 hyperintense lesions24,25,26, T1 hypointense lesions27, contrast-enhancing lesions28, and new and enlarging lesions29. These methods appear promising for automated DAWM segmentation. However, unlike focal MS lesions, there are currently no large datasets with DAWM labels available for supervised training of deep learning models.
In this work, we propose a new DL-based method, DAWM-Net, for segmentation of DAWM from multiparametric MRI using a semi-supervised training approach. The proposed approach initiates network training on a large set of imperfect, heuristic prior segmentations and then fine-tunes the network using a small number of consensus segmentations from expert raters. In this feasibility study, we evaluated the model performance on a withheld test set and compared its performance to a published DAWM segmentation technique.
Methods
Image dataset
The data used in this study included multiparametric MRI data accessed from the CombiRx Phase-III multi-site clinical trial (identifier: NCT00211887) on relapsing remitting MS (RRMS) patients30. MRI scans were acquired across different scanner models and vendors (General Electric Healthcare, Milwaukee, USA; Philips Healthcare, Best, the Netherlands; Siemens Healthineers, Erlangen, Germany) at both 1.5-T (85%) and 3-T (15%) field strengths. A total of 1006 patients were enrolled at baseline and scanned using a standard MRI protocol including 2D FLAIR and 2D dual-echo turbo spin echo images (yielding proton density weighted and T2w images of 0.94 mm × 0.94 mm × 3 mm voxel dimensions), and pre- and post-contrast T1-weighted images with geometry identical to the FLAIR and dual-echo images. In addition, 3D T1-weighted images were also acquired. Baseline scans from the trial were used in this study.
Our analysis of this anonymized dataset was approved by our Institutional Review Board (IRB). Across all sites within the CombiRx trial, patients were recruited and scanned with IRB approval and written consent was obtained from each patient. All research was performed in accordance with the Declaration of Helsinki.
After image quality assessment31, all images were filtered using anisotropic diffusion to reduce noise32,33, and FLAIR and T1w images were registered with the T2w images. Skull stripping and bias field correction were performed, and image intensities were normalized34. Brain volumes were initially segmented into 4 tissue classes (NAGM, white matter including both NAWM and DAWM, cerebrospinal fluid or CSF, and T2L) using MRI automated processing (MRIAP)35, an automated segmentation pipeline that is based on both parametric and non-parametric techniques. Lesion masks were manually inspected and corrected by imaging experts.
Expert evaluation
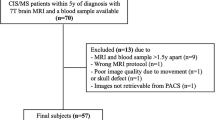
Two neuroimaging experts, a neuroradiologist and an MS neurologist, with 10+ and 15+ years of experience in analyzing MRI of MS, respectively (AK and JAL), independently segmented DAWM volumes from a total of 40 randomly chosen scans. Prior to manual segmentation, the two readers had practice sessions to identify the best MRI contrast types and window setting for DAWM visualization and manual segmentation of DAWM. The manual annotation process began with prior segmentations of the brain and MS lesions as the starting point. Using the ITK-Snap software (version 3.8.0, www.itksnap.org) voxels were reclassified as DAWM based on their appearance on the FLAIR and T2w images (Fig. 1).
Examples of DAWM presentation on FLAIR and T2-weighted images in three MS patients. On zoomed FLAIR images, focal lesions are labeled with red arrows, and DAWM is labeled with yellow arrows. A consensus segmentation by two expert physicians (five tissues—NAGM = grey, NAWM = white, CSF = teal, T2L = salmon pink, DAWM = sky blue) is shown in the middle column.
Special care was taken in areas with possible ambiguity between DAWM and other tissues. Delineation of DAWM from adjacent gray matter structures is especially challenging due to overlap in signal intensity such as in white matter lying along the superior margins of the basal ganglia or in the cortex. Segmentation in these regions was avoided, and the central deep white matter of the brain was taken as the focus of this study. The signal intensity of the large ascending/descending white matter bundles of the brain may also mimic the DAWM signal intensity, such as in the white matter of the corona radiata. To avoid this mimic, only areas with discrete signal changes from the adjacent white matter bundles were segmented.
After initial independent segmentation, the two readers generated a set of consensus segmentations using their previous independent segmentation as a prior. Maps displaying overlap and disagreement of DAWM segmentations were generated, and regions of disagreement were reviewed and either eliminated or accepted to create consensus segmentation maps.
Of the 40 cases assessed by the two readers, 15 were randomly selected for fine-tuning the segmentation model (described in the next section) and the remaining 25 were withheld as an independent test set.
DAWM-Net segmentation
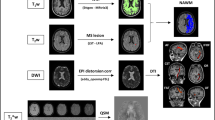
Figure 2 provides a flowchart summarizing the DAWM segmentation process, which consists of three stages. In this study, 2D U-Nets were used as the DL segmentation model in all experiments.
Segmentation approach for DAWM. (Stage 1) Development of heuristic DAWM segmentation based on class score thresholds from a 2D U-Net trained for 4-class segmentation, (Stage 2) generation of imperfect DAWM segmentations for weakly-supervised training of a 2D U-Net, and (Stage 3) fine-tuning on a small set of reader cases to create a final DAWM-Net. Segmentations by algorithm and readers (dark blue) were compared on a withheld test set of 25 patients. Segmentations from an automated intensity thresholding method22 were also tested and their performance was compared to DAWM-Net.
In the first development stage, the heuristic U-Net-based segmentation algorithm was developed based on the observations that DAWM has signal intensity (1) intermediate to NAWM and T2L and (2) similar to that of NAGM. First, a 4-class (4 tissues and background) U-Net was trained for brain segmentation based on the validated MRIAP segmentation on the CombiRx anonymized imaging dataset. Based on the histogram of the U-Net class scores, heuristic thresholds were identified such that voxels meeting observations (1) and (2) were reclassified as DAWM36. Additional information on this technique can be found in the Supplementary Material.
In the second stage, the brain segmentation maps, now augmented with the heuristic DAWM segmentation, were used as the target labels to train another 5-class U-Net with weak supervision, with a similar architecture to that of the 4-class U-Net.
In the third stage, a subset of 15 cases with corresponding consensus segmentations by the two expert readers were used for further fine-tuning of the U-Net (10 cases for training, 5 cases for validation). This resulted in the final trained network referred to as DAWM-Net, which segments the brain into background and 5 tissue classes: NAGM, NAWM, CSF, T2L, and DAWM.
To assess the contribution of different image contrasts (FLAIR, T2w, T1w, or proton-density weighted) to the segmentation performance, a sensitivity analysis was performed. In this experiment, a separate set of models were trained with omission of one image contrast and their segmentation performance was evaluated on the test set.
Finally, an intensity thresholding method for automated DAWM and T2L segmentation was applied to the test set for comparison22. In that method, an initial tissue segmentation of the brain was performed, and DAWM and focal lesions were detected using two intensity thresholds based on the mode of the NAWM signal on the T2w images. Our implementation of this technique is described in the Supplementary Materials.
Statistical analysis
DAWM volumes were extracted, and the mean and standard deviation were calculated across the 25 patients in the held-out test set. Inter-reader agreement was assessed using the Dice Similarity Coefficient (DSC) and Spearman correlation of DAWM volumes. Segmentation performance of the proposed DAWM-Net and the automated intensity thresholding method was assessed against consensus segmentations using DSC and Spearman correlation. DSC scores were compared between segmentation methods using a paired t-test. Finally, Bland–Altman analysis was used to determine bias and the 95% confidence limits of agreement (95% LOA) between the segmentation methods37. Descriptive statistics, t-tests, and correlations were calculated using the Python NumPy module (version 1.19.5). Two-sided p values below 0.05 were considered statistically significant.
Results
In the withheld test set of 25 patients, the average DAWM volume detected by DAWM-Net was 10.0 ± 8.1 cm3 (median [range]: 7.4 [1.7–34 cm3]), and the average DAWM volume delineated the two readers was 11.5 ± 5.6 cm3 (median [range]: 11.4 [3.6–26 cm3]). The automated intensity thresholding method overestimated the DAWM volume, yielding an average of 29 ± 7.1 cm3 (median [range]: 28 [16–44 cm3]). Examples of input images and DAWM segmentation are shown in Figs. 3, demonstrating the good segmentation quality obtained with DAWM-Net.
The DSC and Spearman correlation comparing readers and the automated segmentation methods are reported in Table 1. Correlation plots and Bland–Altman plots are shown in Fig. 4. Compared to the consensus segmentations, the DSC of the detected DAWM was 0.49 ± 0.12 for DAWM-Net, significantly higher than DSC of 0.26 ± 0.08 with the automated intensity thresholding method (P < 0.0001, paired t-test). DAWM-Net also segmented T2L, obtaining a DSC of 0.81 ± 0.08, which is also higher than the DSC = 0.30 ± 0.12 obtained with the automated intensity thresholding method (P < 0.0001, paired t-test).
Correlation (a,c,e) and Bland–Altman plots (b,d,f) comparing the following DAWM volumes: DAWM-Net vs. the reader consensus (a,b), the automated intensity thresholding method vs. the reader consensus (c,d), and the automated intensity thresholding method vs. DAWM-Net (e,f). The dotted red line is the line of identity.
When compared to the expert consensus segmentation, Spearman correlation of extracted DAWM volumes from DAWM-Net segmentations was moderate (ρ = 0.52, P = 0.009). This correlation was not significant for the automated intensity thresholding method (ρ = 0.23, P = 0.27). Bland–Altman analysis showed DAWM-Net to have a small negative bias (bias: − 0.76 cm3, 95% LOA: − 6.2, 4.7 cm3). The automated intensity thresholding method had a larger, positive bias and wider 95% LOA (bias: 8.7 cm3, 95% LOA: 1.6, 15.8 cm3). Sensitivity analysis revealed that model performance was most impacted by the omission of the FLAIR image series. Additional details are reported in the Supplementary Material.
Comparison between automated techniques (DAWM-Net VS automated intensity thresholding methods) showed the following: DAWM DSC of 0.22, a Spearman correlation of DAWM volumes of 0.53 (P = 0.007) and a Bland–Altman bias and 95% LOA (bias: 9.5 cm3, 95% LOA: 1.8, 17.2 cm3) similar to the comparison between the consensus VS automated intensity thresholding method.
Inter-reader analysis showed DAWM DSC of 0.34 ± 0.16 between the two independent readers, with a weak Spearman correlation of DAWM volumes (ρ = 0.23, P = 0.026) and Bland–Altman bias [95% LOA] of − 1.6 [− 8.9, 5.6] cm3.
Discussion
In this work, we developed a DL approach using semi-supervised training to segment DAWM, focal lesions, and all brain tissues in MS patients from multiparametric MRI. By training with a combined large dataset of imperfect priors and a small dataset of expert segmentations, the DAWM-Net segmentations showed spatial overlap and volume correlation that were comparable to human readers. This tool could support further studies to investigate the clinical significance of DAWM in MS and its contribution to disease burden and progression.
For focal pathologies in neuroradiology, such as lesions or tumors, DSCs from 0.6 to 0.8 are frequently reported29,38,39,40. DAWM-Net was able to segment DAWM and T2L with DSCs of 0.49 and 0.81, respectively, which was more accurate compared to the automated intensity thresholding method that attained DSCs of 0.26 and 0.30, respectively. However, our implementation of the automated intensity thresholding method was optimized for DAWM segmentation, which may have affected the T2L segmentation accuracy.
DAWM segmentation presents unique challenges when compared to focal pathologies. Segmentation performance depends on the size of the segmented tissue and the level of image contrast relative to adjacent tissue38. DAWM segmentation faces some specific difficulties: diffuse appearance with weak and variable contrast relative to its surroundings, confounders such as partial volume artifacts, differential appearance of white matter across levels of the brain (such as hyperintensity of white matter bundles in the centrum semiovale), and spatial inhomogeneities inherent in MRI. Therefore, it was expected that the inter-reader variability of DAWM segmentations would be higher than that of focal, high-contrast T2L. The low inter-reader DSC and weak inter-reader correlation of DAWM volumes in our study reflect these challenges. This challenge of accurate and consistent DAWM segmentation was to some extent ameliorated through the consensus segmentation. Both measures of segmentation performance (spatial overlap and volume correlation) showed better agreement between DAWM-Net and the consensus segmentation than the agreement between the two independent readers.
Assessing inter-reader variability in DAWM segmentation was another goal of this study. A previous study by Maranzano et al. using a semi-automated method reported inter-reader DSC of 0.72–0.79. However, unlike our study, there was no comparison to manual segmentation of DAWM. Instead, that study assessed reproducibility of segmentation by automated intensity thresholding to separate DAWM and T2L from NAWM22. These differences in technique might underly the bias that was observed in Bland–Altman analysis for the automated intensity thresholding method in our study. Using global T2w signal intensity thresholds to separate DAWM across the entire brain volume may introduce errors in regions with the aforementioned confounders. In particular, this strategy may produce false positives in large ascending/descending white matter bundles, which may explain the over-segmentation observed as a positive bias in the Band-Altman analysis. In contrast, readers in the current study performed manual DAWM segmentation on FLAIR images. DAWM-Net showed a small negative bias in comparison to consensus segmentations. While DAWM-Net considers slices individually, it can learn a spatially varying threshold which depends on local image context. This likely led to better performance of DAWM-Net in terms of spatial overlap and volume correlation, which was comparable to that of human readers.
There is currently no standard method to measure DAWM volume. Several prior studies have relied on qualitative assessment of DAWM in lieu of volume quantitation6,9,20. Other studies have used a region of interest approach to sample DAWM but did not assess its volume10,41. Previous reports relying on visual assessment have seen DAWM in more than a quarter of MS patients6,9. In our test set of 25 baseline scans, randomly selected from the CombiRx trial, DAWM was segmented in all patients (minimum DAWM volume: 3.6 cm3). The reported prevalence of DAWM in previous studies may be affected by a subjective threshold considering what volume of DAWM is substantial enough to be considered a visual indication of DAWM presence. On the other hand, with automated, voxel-wise segmentation techniques such as those considered here, DAWM is detected regardless of its volume22. Thus, comparing prevalence of DAWM between studies with differing techniques is not straightforward. The 25-patient set in this study is not sufficiently powered to measure the prevalence of DAWM. We will investigate the prevalence of DAWM in an upcoming study with a large patient population.
Only a few previous studies have estimated DAWM volumes. In consecutive studies applying the automated intensity thresholding technique, DAWM averaged around 20 cm3 in volume for MS patients (range: 1.82–43.87 cm3)18,19,22. This is similar to our results from the T2w image automated intensity thresholding technique. Although the automated segmentation methods showed moderate correlation, our measurements from the reader consensus and DAWM-Net were closer to 10 cm3. This difference is likely due to the more conservative manual segmentation strategy used by our readers, which was imparted to DAWM-Net through semi-supervised training. Another possible reason for this difference may be due to the different primary image contrast used (T2w vs. FLAIR), warranting further investigation. DAWM-Net included both T2w and FLAIR images as inputs, but its performance was shown to be most strongly dependent on the FLAIR series in the sensitivity analysis.
One limitation of our study is that the CombiRx dataset only included early relapsing–remitting MS patients. Generalization of the DAWM-Net model to other MS phenotypes should be explored in the future. A second limitation of DAWM-Net is the requirement for normalized, multiparametric data as input, which may not be immediately available. We evaluated our method in a relatively small withheld set of 25 MRI volumes. Validation in larger external datasets, possibly with different imaging parameters, will be required to fully determine the generalizability of the proposed algorithm. Data used in this study were acquired in a large multi-site trial, thus are not highly specific and contribute to a wider range of a-priori information for the training of the DL pipeline, which would potentially be in favor of the system’s generalizability. To improve DAWM segmentation consistency, readers avoided ambiguous white matter regions, focusing on the central deep white matter of the brain. This focus was likely imparted to DAWM-Net during training and could potentially result in underestimation of DAWM volume. A final limitation is that our manual segmentation data, used for both development and evaluation, is generated by only two expert readers. Addition of more expert readers could potentially improve segmentation performance and better assess inter-reader agreement. An independent study would most accurately and efficiently evaluate the efficacy of DAWM-Net in the face of these limitations.
In conclusion, we have demonstrated that a DL model combined with semi-supervised training can perform comprehensive segmentation of all brain tissues, including DAWM, on data from a large multi-center study of RRMS. Accurate and automated segmentation of DAWM will facilitate research efforts focused on enhancing our understanding of DAWM and its potential as an imaging biomarker of disease progression.
Data availability
Supporting code and the trained DAWM-Net model will be available at the following ___location: https://github.com/uthmri.
References
Walton, C. et al. Rising prevalence of multiple sclerosis worldwide: Insights from the Atlas of MS. Mult. Scler. 26, 1816–1821 (2020).
Thompson, A. J. et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 17, 162–173 (2018).
Cahalane, A. M., Kearney, H., Purcell, Y. M., McGuigan, C. & Killeen, R. P. MRI and multiple sclerosis- the evolving role of MRI in the diagnosis and management of MS: The radiologist’s perspective. Ir. J. Med. Sci. 187, 781–787 (2018).
Zhao, G., Li, D. K. B., Wang, X. Y., Cheng, Y. & Paty, D. W. MRI dirty-appearing white matter in MS. Neurology 54, A121 (2000).
Zhao, G. et al. Possible prognostic significance of dirty-appearing white matter on MRI in multiple sclerosis. Mult. Scler. 9, S61 (2003).
Vertinsky, A. T. et al. Diffusely abnormal white matter, T2 burden of disease, and brain volume in relapsing-remitting multiple sclerosis. J. Neuroimaging 29, 151–159 (2019).
Ge, Y., Grossman, R. I., Babb, J. S., He, J. & Mannon, L. J. Dirty-appearing white matter in multiple sclerosis: Volumetric MR imaging and magnetization transfer ratio histogram analysis. AJNR Am. J. Neuroradiol. 24, 1935–1940 (2003).
Filippi, M. & Rocca, M. A. Dirty-appearing white matter: A disregarded entity in multiple sclerosis. AJNR Am. J. Neuroradiol. 31, 390–391 (2010).
Vavasour, I. M. et al. Diffusely abnormal white matter in clinically isolated syndrome is associated with parenchymal loss and elevated neurofilament levels. Mult. Scler. Relat. Disord. 57, 103422 (2022).
West, J. et al. Normal appearing and diffusely abnormal white matter in patients with multiple sclerosis assessed with quantitative MR. PLoS ONE 9, e95161 (2014).
Laule, C. et al. Pathological basis of diffusely abnormal white matter: insights from magnetic resonance imaging and histology. Mult. Scler. 17, 144–150 (2011).
Laule, C. et al. Diffusely abnormal white matter in multiple sclerosis: Further histologic studies provide evidence for a primary lipid abnormality with neurodegeneration. J. Neuropathol. Exp. Neurol. 72, 42–52 (2013).
Seewann, A. et al. Diffusely abnormal white matter in chronic multiple sclerosis: Imaging and histopathologic analysis. Arch. Neurol. 66, 601–609 (2009).
Kitzler, H. H. et al. Deficient MWF mapping in multiple sclerosis using 3D whole-brain multi-component relaxation MRI. Neuroimage 59, 2670–2677 (2012).
Ropele, S. et al. A comparison of magnetization transfer ratio, magnetization transfer rate, and the native relaxation time of water protons related to relapsing-remitting multiple sclerosis. AJNR Am. J. Neuroradiol. 21, 1885–1891 (2000).
Karampekios, S. et al. Quantification of magnetization transfer rate and native T1 relaxation time of the brain: correlation with magnetization transfer ratio measurements in patients with multiple sclerosis. Neuroradiology 47, 189–196 (2005).
Vos, C. M. et al. Blood-brain barrier alterations in both focal and diffuse abnormalities on postmortem MRI in multiple sclerosis. Neurobiol. Dis. 20, 953–960 (2005).
Dadar, M., Narayanan, S., Arnold, D. L., Collins, D. L. & Maranzano, J. Conversion of diffusely abnormal white matter to focal lesions is linked to progression in secondary progressive multiple sclerosis. Mult. Scler. J. 27, 208–219 (2021).
Dadar, M. et al. Diffusely abnormal white matter converts to T2 lesion volume in the absence of MRI-detectable acute inflammation. Brain 145, 2008–2017 (2022).
Holmes, R. D. et al. Nonlesional diffusely abnormal appearing white matter in clinically isolated syndrome: Prevalence, association with clinical and MRI features, and risk for conversion to multiple sclerosis. J. Neuroimaging 31, 981–994 (2021).
Papadaki, E. et al. T2 relaxometry evidence of microstructural changes in diffusely abnormal white matter in relapsing–remitting multiple sclerosis and clinically isolated syndrome: Impact on visuomotor performance. J. Magn. Reson. Imaging 54, 1077–1087 (2021).
Maranzano, J. et al. Automated separation of diffusely abnormal white matter from focal white matter lesions on MRI in multiple sclerosis. Neuroimage 213, 116690 (2020).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference. 234–241 (2015).
Brosch, T. et al. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans. Med. Imaging 35, 1229–1239 (2016).
Valverde, S. et al. Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage 155, 159–168 (2017).
Gabr, R. E. et al. Brain and lesion segmentation in multiple sclerosis using fully convolutional neural networks: A large-scale study. Mult. Scler. J. 26, 1217–1226 (2020).
Krishnan, A. P. et al. Joint MRI T1 unenhancing and contrast-enhancing multiple sclerosis lesion segmentation with deep learning in OPERA trials. Radiology 302, 662–673 (2022).
Coronado, I., Gabr, R. E. & Narayana, P. A. Deep learning segmentation of gadolinium-enhancing lesions in multiple sclerosis. Mult. Scler. J. 27, 519–527 (2021).
Commowick, O., Cervenansky, F., Cotton, F. & Dojat, M. MSSEG-2 challenge proceedings: Multiple sclerosis new lesions segmentation challenge using a data management and processing infrastructure. In MICCAI 2021–24th International Conference on Medical Image Computing and Computer Assisted Intervention 126 (2021).
Lublin, F. D. et al. Randomized study combining interferon and glatiramer acetate in multiple sclerosis. Ann. Neurol. 73, 327–340 (2013).
Narayana, P. A. et al. Regional cortical thickness in relapsing remitting multiple sclerosis: A multi-center study. Neuroimage Clin. 2, 120–131 (2012).
Perona, P. & Malik, J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 12, 629–639 (1990).
Gerig, G., Kubler, O., Kikinis, R. & Jolesz, F. A. Nonlinear anisotropic filtering of MRI data. IEEE Trans. Med. Imaging 11, 221–232 (1992).
Nyul, L. G., Udupa, J. K. & Zhang, X. New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging 19, 143–150 (2000).
Sajja, B. R. et al. Unified approach for multiple sclerosis lesion segmentation on brain MRI. Ann. Biomed. Eng. 34, 142–151 (2006).
Gabr, R. E. & Narayana, P. A. Automatic segmentation of diffusely abnormal white matter in MS using deep neural network. In 2021 Annual Meeting of the Internation Society of Magnetic Resonance in Medicine. Abstract #2804 (2021).
Bland, M. J. & Altman, D. G. Statistical methods for assessing agreement between two methods of clinical measurement. The Lancet 327, 307–310 (1986).
Cao, Y. et al. Automatic detection and segmentation of multiple brain metastases on magnetic resonance image using asymmetric UNet architecture. Phys. Med. Biol. 66, 015003 (2021).
Narayana, P. A. et al. Are multi-contrast magnetic resonance images necessary for segmenting multiple sclerosis brains? A large cohort study based on deep learning. Magn. Reson. Imaging 65, 8–14 (2020).
Khademi, A. et al. Segmentation of white matter lesions in multicentre FLAIR MRI. Neuroimage Rep. 1, 100044 (2021).
Kitzler, H. H. et al. Multi-component relaxation in clinically isolated syndrome: Lesion myelination may predict multiple sclerosis conversion. NeuroImage Clin. 20, 61–70 (2018).
Acknowledgements
This work was funded by an NIH National Institute of Neurological Disorders and Stroke R21 Grant (5R21NS118320-02). We would also like to thank Professor Fred D. Lublin for providing access to the CombiRx dataset used in this work. The CombiRx study (NCT00211887) was funded by the NIH, National Institute of Neurological Disorders and Stroke (phase III study: UO1NS045719).
Author information
Authors and Affiliations
Contributions
B.C.M.: Methodology, Formal analysis, Software, Investigation, Data Curation, Writing—Original Draft, Visualization. R.E.G.: Methodology, Software, Formal analysis, Investigation, Resources, Data, Writing—Original Draft, Writing—Review and Editing Curation, Visualization, Supervision, Project administration, Funding acquisition. Y.Y.: Software, Investigation, Writing—Review and Editing, Visualization. A.K.: Methodology, Validation, Writing—Review and Editing. J.A.L.: Methodology, Validation, Writing—Review and Editing. M.A.J.: Resources, Writing—Review and Editing. V.L.: Methodology, Software, Writing—Review and Editing. XL—Methodology, Software, Writing—Review and Editing. F.D.L.: Resources, Writing—Review and Editing. J.S.W.: Resources, Writing—Review and Editing. P.A.N.: Methodology, Resources, Writing—Review and Editing, Funding acquisition. K.M.H.: Methodology, Formal analysis, Resources, Writing—Review and Editing, Visualization, Supervision, Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Musall, B.C., Gabr, R.E., Yang, Y. et al. Detection of diffusely abnormal white matter in multiple sclerosis on multiparametric brain MRI using semi-supervised deep learning. Sci Rep 14, 17157 (2024). https://doi.org/10.1038/s41598-024-67722-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-67722-2