Abstract
This study aims to estimate the familial risks of pterygium and assess its relative contributions to environmental and genetic factors using the 2000–2017 Taiwan National Health Insurance Research Database. The marginal Cox's model and the polygenic liability model were made. In Taiwan, the prevalence rate of pterygium in 2017 was 1.64% for individuals with affected first-degree relatives, higher than the general population (1.34%). The adjusted relative risk (RR) for pterygium was highest for twins of the same sex (15.54), followed by siblings of the same sex (4.69), offsprings (3.39), siblings of the different sex (2.88), spouse (2.12), parents (1.86), twins of the different sex (1.57), respectively. The phenotypic variance of pterygium was 21.6% from additive genetic variance, 24.3% from common environmental factors shared by family members, and 54.1% from non-shared environmental factors, respectively. Sensitivity analysis by restricting those with surgical pterygium reveals that aRRs and the three components were similar to those of the overall pterygium. In summary, the prevalence rate of pterygium was higher for individuals with affected first-degree relatives than for the general population. The non-shared environmental factors account for half of the phenotypic variance of pterygium; genetic and shared environmental factors explain the rest.
Similar content being viewed by others
Introduction
Pterygium is a common external eye disease characterized by a wing-shaped fibrovascular growth of the conjunctiva across the limbus onto the cornea1. Patients with pterygium tend to have problems of ocular irritation, corneal astigmatism, visual deterioration, cosmetic concern, or other complications. The prevalence of pterygium lies between 1 and 30% or more, with a pooled rate of around 10%1,2. The prevalence of pterygium in Taiwan was 2.14% in the overall population and 3.48% in the population aged 40 years or older3.
However, biological and pathogenic mechanisms causing pterygium are not fully understood, and etiological theories do not clarify the nature and origin of the lesion4. Older age, male, having outdoor jobs, low education, rural residence, low income, darker skin complexion, and cigarette smoking are risk factors of pterygium1. A very high incidence of pterygia has been reported in certain families over several generations, suggesting heredity may predispose the conjunctiva to react abnormally to atmospheric environmental stimuli4. It was suggested that heredity is crucial for a pterygium to develop, and sunlight triggers chronic inflammation and promotes pterygium enlargement5.
In genetics, the phenotype is the set of observable characteristics or traits of an organism. The ACE model decomposes complex phenotypic variance of quantitative traits or diseases into three categories: additive genetic (A), common (or shared) environmental factors (C), and specific (or non-shared) environmental factors plus measurement error (E). The additive genetic variance assumes that genes act additively with each other both within and between loci6,7.
In this study, we conducted a nationwide study using the Taiwan National Health Insurance Research Database (NHIRD) and the ACE model to estimate the degree of familial aggregation and the extent to which heritability and a shared familial environment of pterygium in this Eastern Asian country.
Materials and methods
Study design and ethical approval
We constructed a nationwide cross-sectional study using the Taiwan National Health Insurance (NIH) database. In 1995, Taiwan launched a compulsory single-payer NHI program, which covered 99.6% of the population and contracted with 93% of the hospitals8. All claim and registration files were available for research in the Health and Welfare Data Science Center. To protect privacy, all claim and registration files can be linked with scrambled IDs, and only summary results can be released from the center. All the methods described were compliant with the Declaration of Helsinki. Furthermore, the Institutional Review Board (IRB) of the Chang Gung Medical Foundation (202002373B1) waived the need for informed consent and ethical approval because of the lowest risk and delink of personal information.
Ascertainment of family relationship
According to the Taiwan National Health Insurance Act9, beneficiaries included the insured (who pays the NHI fee) and their unemployed first-degree relatives (spouse, lineal blood parents, and children). Hence, we could retrieve the parent–offspring and spouse relationships over the 2000–2017 files of registered beneficiaries, a subset of the Taiwan National Health Insurance database, to construct the family relationship (parents, offspring, siblings, twin, spouse) for the study populations. In addition, full siblings were identified as individuals who shared the same parents, and twins were full siblings with the same birth date (± 1 day). As a result, among the 29,253,526 beneficiary records in the 2000–2017 registry, we could identify 4,144,508 families with a mean family size of 4.94 for the 2017 population (N = 23,373,209).
Ascertainment of pterygium
Pterygium was defined if their ICD-9-CM = 372.4 or ICD-10-CM = H11.0 with at least two outpatient visits to eye clinics or one hospital admission. Surgical removal of pterygium was identified based on the procedure code of 85203C, 85204C, 87212C, and 87213C.
Ascertainment of covariates
The demographic variables included age, sex, monthly income, occupation, and residence based on the ___location of the climate zone or the urbanization degree. Income level was based on the payroll-related insurance data for each patient and categorized by quintile plus no income. Occupations were allocated into five categories: (1) dependent, i.e., financially supported by an insured family member; (2) civil servants with government or affiliated positions; (3) non-manual workers working in privately owned institutions; (4) manual workers, including blue-collar workers, farmers, and fishermen; and (5) others, including veterans, members of low-income families, and substitute service draftees. Because the Tropic of Cancer runs through the center of Taiwan, we divided the residence area into three climate zones: subtropical, intermediate, and tropical. Urbanization degree of residence was categorized as urban, suburban, rural, or unknown based on the population density, percentage of residents with college or higher education, percentage of residents ≥ 65 years, percentage of residents who are agricultural workers, and the number of physicians per 100,000 people in 369 towns or districts in Taiwan10.
Statistical analysis
The 1-sample z-test and goodness of fit were made to compare the mean age and the distribution of other demographic variables between the general population and those with affected first-degree relatives of pterygium, where appropriate. The prevalence rate of pterygium for the general population and affected first-degree relatives in 2017 was computed as the number of patients who visited outpatient or inpatient for pterygium during 2017 divided by the total number of NHI beneficiaries registered in 2017 and people with affected first-degree relatives, respectively. The adjusted relative ratio (aRR) was obtained using the marginal Cox proportional hazard model with an equal follow-up time for all individuals11. Furthermore, the robust sandwich estimate was made in the Cox model to resolve the violation of independence in the same family12. The Cox model included the status of first-degree relatives with pterygium (such as a sibling or spouse), age, the climate zone of residence, the urbanization of residence, income levels, occupation, and family size. The aRRs for siblings and spouses were used in the polygenic liability model.
In Falconer’s polygenic liability model, ‘liability’ is a single measure that reflects an individual who is more or less likely to develop the disease with the contribution of genetic and environmental factors. The liabilities of individuals in a population form a continuous variable with normal distribution. The disease status of an individual depends on whether their liability value is below or above a given threshold (unaffected and affected, respectively)13. Under this model, it becomes possible to estimate disease heritability from its prevalence in the general population and relatives of affected individuals. Heritability is the proportion of liability variance ascribable to additive genetic variance based on the phenotypic correlation between relatives. Later on, Reich et al.14,15 and Yang et al.16 developed the equation to estimate heritability and family transmission, as below:
where T0 = \({\Phi }^{-1}\left(1-p\right)\); Ts = \({\Phi }^{-1}\left(1-spouse RR\times p\right)\); T1 = \({\Phi }^{-1}(1-sibling RR\times p)\); p = prevalence of pterygium in the general population; aR: the additive genetic relationship between relatives, for a full sibling, aR = 0.5; i = z/p; z is the height of the standard normal curve of pterygium prevalence; and Ф is the standard normal cumulative distribution function13,14,15,16. To account for the contributions of shared environmental factors to phenotypic variance, we used the spouse as a control, assuming that spouses share the adult family environments but not early life and have no close genetic similarity with blood family members17. The sibling RR and spouse RR were obtained from the results of the Cox model above. The polygenic liability model used in this study assumes no assortative mating, genetic liability variance constant across generations, and did not consider the age of onset of pterygium.
The ACE model decomposed the phenotypic variance of a multifactorial disease (\(\sigma_{P}^{2}\)) into three components: \(\sigma_{A}^{2}\) the additive genetic variance, \(\sigma_{C}^{2}\) the variance of common environmental factors shared by family members, and \(\sigma_{E}^{2}\) the variance of non-shared environmental factors. In other words, the ACE model is \(\sigma_{P}^{2} = \sigma_{A}^{2} + \sigma_{C}^{2} + \sigma_{E}^{2}\). In the polygenic liability model, heritability (H) is the proportion of the additive genetic variance among the phenotypic variance \(\left( {H = {{\sigma_{A}^{2} } \mathord{\left/ {\vphantom {{\sigma_{A}^{2} } {\sigma_{P}^{2} }}} \right. \kern-0pt} {\sigma_{P}^{2} }}} \right)\). Familial transmission (FT) is the sum of the additive genetic variance and the variance of common environmental factors shared by family members divided by the phenotypic variance \({(}FT = ({{\sigma_{A}^{2} + \sigma_{C}^{2} )} \mathord{\left/ {\vphantom {{\sigma_{A}^{2} + \sigma_{C}^{2} )} {\sigma_{P}^{2} }}} \right. \kern-0pt} {\sigma_{P}^{2} }}{)}\). Hence, the variance of common environmental factors shared by family members divided by the phenotypic variance of a multifactorial disease is familial transmission minus heritability (\(\sigma_{C}^{2}\)/\(\sigma_{P}^{2}\) = FT – H). Moreover, the variance of non-shared environmental factors divided by the phenotypic variance of a multifactorial disease is one minus familial transmission (\(\sigma_{E}^{2}\)/\(\sigma_{P}^{2}\) = 1-FT).
We did a sensitivity analysis by adding extra criteria of pterygium requiring surgical removal to determine how robust the main results (aRRs of pterygium for those with affected first-degree relatives and the three components in the ACE model) apply to a rigid definition of pterygium.
Results
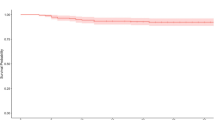
Among the general population, 171,813 females and 141,438 males had a medical visit for their pterygium in 2017, equivalent to the prevalence rate of 1.46% and 1.22%, respectively. The age-specific prevalence rates of pterygium for males and females were similar, remaining low at a young age, increasing at 40–49 years, continuing to increase until age 70–79, and declining afterward (Fig. 1). There were 399,134 people (1.71%) had first-degree relatives with pterygium. Individuals with the affected first-degree relative had significantly higher prevalence rates of pterygium (1.67% and 1.61% for females and males, respectively) than the general population. Again, the age-specific prevalence rates of pterygium among those with the affected first-degree relative are higher than those in the general population (Fig. 1). Those with affected first-degree relatives lived more in rural areas, had a higher income, worked more as non-manual workers, and lived more in tropical than subtropical areas than the general population for both sexes (Table 1).
The adjusted relative risk (aRR), genetic and environmental contributions
The aRRs (95% CI) of pterygium were 2.48 (2.41–2.56) for those with affected first-degree relatives, 1.86 (1.79–1.93) for parents, 3.39 (3.28–3.50) for offsprings, 4.69 (4.12–5.33) for siblings of the same sex, 2.88 (2.50–3.31) for siblings of the different sex, 15.54 (7.09–34.03) for twins of the same sex, 1.57 (0.70–3.51) for twins of the different sex, and 2.12 (2.08–2.16) for spouses, respectively. When stratified by sex, the aRR and 95% CI of pterygium for those with affected first-degree relatives were similar between females and males (Fig. 2). The contributions to the phenotypic variance of pterygium were 21.6% for additive genetic variance, 24.3% for common environmental factors shared by family members, and 54.1% for non-shared environmental factors, respectively (Fig. 3).
The adjusted relative risks (aRR) of pterygium in those with affected first-degree relatives. The aRR for the status of first-degree relatives with pterygium was obtained using the Cox model with a robust sandwich estimate, which also included age, the climate zone of residence, the urbanization of residence, income levels, occupation, and family size.
Sensitivity analysis
When limiting the pterygium that received surgical removal, the 2017 prevalence rates were 0.59% for the general population and 0.72% for those with affected first-degree relatives (Supplementary Table S1). The age-specific prevalence of surgical pterygium for the general population and individuals whose first-degree relatives had surgical pterygium are presented in Supplemental Fig. 1. It is reasonable to observe the lower prevalence of surgical pterygium than that of overall pterygium. The aRRs (95% CI) of surgical pterygium became 2.89 (2.68–3.11) for those with affected first-degree relatives, 2.10 (1.93–2.29) for parents, 4.12 (3.80–4.46) for offsprings, 4.61 (3.39–6.29) for siblings of the same sex, 3.30 (2.43–4.49) for siblings of the different sex, 18.94 (5.37–66.73) for twins of the same sex, 1.54 (0.23–10.17) for twins of the different sex, and 2.51 (2.36–2.66) for spouses, respectively (Supplemental Fig. 2). The percentage of additive genetic variance, common environmental factors shared by family members, and non-shared environmental factors for surgical pterygium became 14.5%, 24.6%, and 60.9%, respectively (Supplemental Fig. 3). In brief, aRRs and the three components of the ACE model for the surgical pterygium were similar to those of the overall pterygium.
Discussion
This nationwide study using the Taiwan National Health Insurance database reveals that the non-shared environmental factors account for half of the phenotypic variance of pterygium; genetic and shared environmental factors explain the rest. The aRRs of overall (or clinically diagnosed) pterygium in those with affected first-degree relatives was 2.48 (2.41–2.56), and that of surgical pterygium was 2.89 (2.68–3.11). The aRRs of overall pterygium in a spouse was 2.12 (2.08–2.16), and that of surgical pterygium was 2.51 (2.36–2.66).
The prevalence of pterygium varies widely from 1.4 to 33% globally, depending on the study area, study setting, and age coverage2. The rates of pterygium for aged 15–84 years old in China was 9.84%, varied with age, sex and geographic ___location18. The prevalence rate of pterygium in southern Taiwan was 25.2% among adults over 40 years in a community complex health screening study from Sept. 2006 to Oct. 200719. In this study, the 2017 prevalence rate of pterygium (1.34%) in Taiwan was similar to a previous study, which also used the NHIRD to estimate the prevalence of "clinically diagnosed" pterygium in Taiwan3. They reported that the annual incidence of pterygium in the whole population decreased from 3.7% in 2000 to 2.0% in 2011 in Taiwan3. In this study, we saw a higher prevalence of pterygium for those with affected first-degree relatives than for the general population.
In this study, we used "clinically diagnosed" or "surgical" pterygium to define this disease because they represent a noteworthy condition needing medical or surgical care. The grading of pterygium severity was typically based on its thickness and size20. For the proportion of pterygium from grade I (mild) to IV (severe) in the community-based studies, grade II pterygium was most common (44.00%), followed by grade I (26.70%) in Gambella, Ethiopia, and also in other countries21. In this study, most patients should have high grades of pterygium because they require medical or surgical treatment.
The indications for pterygium surgery are visual disturbance secondary to growth over the pupil or induced astigmatism, restriction in eye movements, chronic redness and foreign body sensation, or cosmetic concerns22. The severity of "surgical" pterygium is clinically more severe, though not precisely more severe in its thickness or size, among our "clinically diagnosed" pterygium subjects. We analyzed aRRs and the three components of the ACE model of "surgical" pterygium as a sensitivity analysis to compare with those of "clinically diagnosed" pterygium. In brief, aRRs and the three components of the ACE model were similar for both types of pterygia, implying that our conclusion may hold constant in different severity of pterygium, such as those in community-based studies.
Our study confirmed familial aggregation of pterygium by demonstrating its greater prevalence and aRR among individuals with affected family members compared to the general population. However, biological relatives tend to share similar environmental and lifestyle risk factors in addition to genes; both contribute to familial aggregation. Therefore, we examined the risk of pterygium for those whose spouse had pterygium, assuming that any increased risk from environmental and lifestyle factors common to family members. Our findings showed that genetic factors play a minor role compared to environmental factors in the pathogenesis of pterygium.
Previous reports also revealed a high incidence of pterygium in certain families over generations, and various models of inheritance have been described, including autosomal dominant with reduced penetrance, polygenic, multifactorial, and non-Mendelian inheritance. Several genes23, microRNAs, and polymorphisms of vascular endothelial growth factors have some role in the pathogenesis of pterygium24,25. Although many genes and pathways were proposed to be involved in the inheritance of pterygium, the genetic basis remains to be elucidated.
The area between 37° north and south of the equator is named the "pterygium belt" because of its high prevalence of pterygium and high UV radiation26. Hence, exposure to ultraviolet (UV) light is the major environmental risk factor for pterygium, whereas hereditary is an underlying factor for its occurrence. It is proposed that UV light may damage DNA in predisposed individuals with many genes involved. UV light may damage limbal stem cells, alter the function of stromal fibroblasts, or induce inflammatory responses to develop pterygium1. However, the molecular processes underlying human health and disease are highly complex. Often, genetic and environmental factors contribute to a given disease in a non-additive manner27. Large non-shared environmental influences may include un-estimated genes and environment interaction. Taiwan lies in the "pterygium belt," and the Tropic of Cancer runs through the island. Our results show that the risk of pterygium was higher for inhabitants of the tropical area than for those in the other areas in Taiwan, indicating a dosage effect of UV light on the pathogenesis of pterygium. Therefore, preventions for UV light exposure, such as staying in the shade, wearing a wide-brimmed hat or sunglasses, etc., are advised to prevent pterygium development, especially for those with a family history.
There are several limitations to the study. First, it was confined to Taiwan, so the results may not be generalizable to other settings. Second, the NHIRD is primarily a health insurance database containing limited diagnosis criteria and disease severity information. The results of laboratory testing and examinations are not available in NHIRD. Third, our data from the NHIRD did not include patients with mild pterygium who did not seek medical help or did not attract physicians' attention for a diagnosis. In other words, the prevalence rate reported here may be underestimated because we might miss some cases with mild pterygium or receiving treatment before 2000. On the other hand, some diagnosed cases of pterygium may be misclassified. Hence, we did a sensitivity analysis on the surgical pterygium for more assurance. Fourth, we did not have data on potential confounding factors, such as sunlight exposure due to occupation or outdoor activity. Thus, we cannot test the interactions between family history and other confounders and their independent contributions to the risk of pterygium. Fifth, our analysis of relative genetic and environmental contributions was based on the multifactorial liability model, and our results are subject to assumptions, so they should be interpreted cautiously. However, the published data on other diseases, such as gout, atrial fibrillation, nasopharyngeal carcinoma, type I DM, aortic dissection, thyroid cancer, and glaucoma, support the validity of this model17,28,29,30,31,32,33. Finally, we cannot account for the effects of assortative mating, whereby spouses are more similar for a phenotype than they would be if mating occurred randomly in the population. A biased estimation of relative genetic and environmental contributions may occur if this assortment is not negligible. The rate of consanguineous marriage in Taiwan is unknown34 but should be very small because marriage between close relatives is prohibited according to Article 983 of the Civil Code in Taiwan35.
Our main strengths include data from the entire population of approximately 23 million individuals and systematic methods to identify and ascertain first-degree relatives, which allow a reasonable estimation of the prevalence and RRs of pterygium with minimal selection bias. In addition, identifying pterygium cases and using consistent case definitions for individuals at risk and their relatives ensured the minimal problem of information bias.
In conclusion, this study is the first to provide quantitative estimates of familial aRR of pterygium. Our results confirm the clinical belief that pterygium clusters within families. The non-shared environmental factors account for half of the phenotypic variance of pterygium; genetic and shared environmental factors explain the rest.
Data availability
The data for this study were sourced from the National Health Insurance Research Database Taiwan (NHIRD), which is housed exclusively in the Health and Welfare Data Science Center. Due to data sharing restrictions, we cannot make the research data publicly available.
References
Shahraki, T., Arabi, A. & Feizi, S. Pterygium: An update on pathophysiology, clinical features, and management. Ther Adv Ophthalmol 13, 25158414211020150. https://doi.org/10.1177/25158414211020152 (2021).
Liu, L., Wu, J., Geng, J., Yuan, Z. & Huang, D. Geographical prevalence and risk factors for pterygium: A systematic review and meta-analysis. BMJ Open. 3(11), e003787. https://doi.org/10.1136/bmjopen-2013-003787 (2013).
Lin, Y. H. et al. Epidemiologic study of pterygium in Taiwan. Jpn J Ophthalmol. 63(4), 297–303. https://doi.org/10.1007/s10384-019-00670-x (2019).
Romano, V., Steger, B., Kovacova, A., Kaye, S. B. & Willoughby, C. E. Further evidence for heredity of pterygium. Ophthalmic Genet. 37(4), 434–436. https://doi.org/10.3109/13816810.2015.1111911 (2016).
Anguria, P., Kitinya, J., Ntuli, S. & Carmichael, T. The role of heredity in pterygium development. Int J Ophthalmol. 7(3), 563–573. https://doi.org/10.3980/j.issn.2222-3959.2014.03.31 (2014).
Martin, N. G. & Eaves, L. J. The genetical analysis of covariance structure. Heredity. 38(1), 79–95 (1977).
Lyu, X. & Garrison, S. M. Effects of genetic relatedness of kin pairs on univariate ACE model performance. Twin Res. Human Genet. 26(4–5), 257–268 (2023).
Hsieh, C. Y. et al. Taiwan’s national health insurance research database: Past and future. Clin. Epidemiol. 11, 349 (2019).
Ministry of Justice, Taiwan. Laws & regulations database: Regulations governing the deduction and payment of the supplementary insurance premium of the National Health Insurance. https://law.moj.gov.tw/ENG/LawClass/LawAll.aspx?pcode=L0060027. Accessed 9 Jan 2023.
Li, L. A study on the order of the degree of urbanisation among the Lo Chi-Hon Strata of Taiwan counties. Surv Res Method Appl. 15, 5–30 (2004).
Lin, D. Cox regression analysis of multivariate failure time data: The marginal approach. Stat. Med. 13(21), 2233–2247 (1994).
Lin, D. Y. & Wei, L.-J. The robust inference for the Cox proportional hazards model. J. Am. Stat. Assoc. 84(408), 1074–1078 (1989).
Falconer, D. The inheritance of liability to diseases with variable age of onset, with particular reference to diabetes mellitus. Ann. Human Genet. 31(1), 1–20 (1967).
Reich, T., James, J. & Morris, C. The use of multiple thresholds in determining the mode of transmission of semi-continuous traits. Ann. Human Genet. 36(2), 163–184 (1972).
Reich, T., Rice, J., Cloninger, C. R., Wette, R. & James, J. The use of multiple thresholds and segregation analysis in analyzing the phenotypic heterogeneity of multifactorial traits. Ann. Hum. Genet. 42(3), 371–390 (1979).
Yang, J., Visscher, P. M. & Wray, N. R. Sporadic cases are the norm for complex disease. Eur. J. Human Genet. 18(9), 1039–1043 (2010).
Kuo, C. F. et al. Familial aggregation of gout and relative genetic and environmental contributions: A nationwide population study in Taiwan. Ann. Rheum. Diseases. 74, 369–374 (2015).
Song, P., Chang, X., Wang, M. & An, L. Variations of pterygium prevalence by age, gender and geographic characteristics in China: A systematic review and meta-analysis. PLoS One. 12(3), e0174587 (2017).
Chen, C. L. et al. The epidemiology of patients with pterygium in southern Taiwanese adults: The Chiayi survey. Taiwan J. Ophthalmol. 3(2), 58–61. https://doi.org/10.1016/j.tjo.2013.03.001 (2013).
Gazzard, G. et al. Pterygium in Indonesia: Prevalence, severity and risk factors. Br J Ophthalmol. 86(12), 1341–1346. https://doi.org/10.1136/bjo.86.12.1341 (2002).
Fekadu, S. A., Assem, A. S. & Adimassu, N. F. Prevalence of pterygium and its associated factors among adults aged 18 years and above in Gambella town, Southwest Ethiopia, May 2019. PLoS One. 15(9), e0237891. https://doi.org/10.1371/journal.pone.0237891 (2020).
Singh, S. K. Pterygium: Epidemiology prevention and treatment. Community Eye Health. 30(99), S5-s6 (2017).
Yue, X. L. & Gao, Z. Q. Identification of pathogenic genes of pterygium based on the gene expression Omnibus database. Int. J. Ophthalmol. 12(4), 529–535. https://doi.org/10.18240/ijo.2019.04.01 (2019).
Peng, M. L. et al. Vascular endothelial growth factor gene polymorphism and protein expression in the pathogenesis of pterygium. Br J Ophthalmol. 98(4), 556–561. https://doi.org/10.1136/bjophthalmol-2013-303436 (2014).
Chien, K. H. et al. Correlation of microRNA-145 levels and clinical severity of pterygia. Ocul Surf. 11(2), 133–138. https://doi.org/10.1016/j.jtos.2012.12.001 (2013).
Cameron ME. Pterygium throughout the world. Thomas; 1965.
Virolainen, S. J., VonHandorf, A., Viel, K., Weirauch, M. T. & Kottyan, L. C. Gene-environment interactions and their impact on human health. Genes Immun. 24(1), 1–11. https://doi.org/10.1038/s41435-022-00192-6 (2023).
Chang, S. H. et al. Association of a family history of atrial fibrillation with incidence and outcomes of atrial fibrillation: A population-based family cohort study. JAMA Cardiol. 2(8), 863–870. https://doi.org/10.1001/jamacardio.2017.1855 (2017).
Huang, S. F. et al. Familial aggregation of nasopharyngeal carcinoma in Taiwan. Oral Oncol. 73, 10–15. https://doi.org/10.1016/j.oraloncology.2017.07.020 (2017).
Kuo, C. F. et al. Familial aggregation and heritability of type 1 diabetes mellitus and coaggregation of chronic diseases in affected families. Clin. Epidemiol. 10, 1447–1455 (2018).
Chen, S. W. et al. Association of family history with incidence and outcomes of aortic dissection. J Am Coll Cardiol. 76(10), 1181–1192. https://doi.org/10.1016/j.jacc.2020.07.028 (2020).
Lin, H. T. et al. Familial aggregation and heritability of nonmedullary thyroid cancer in an Asian population: A nationwide cohort study. J. Clin. Endocrinol. Metab. 105(7), e2521–e2530. https://doi.org/10.1210/clinem/dgaa191 (2020).
Lee, J. S., Kuo, C. F., Chen, W. M., Lin, K. K. & See, L. C. Genetic and environmental contributions of primary angle-closure glaucoma and primary open-angle glaucoma: A nationwide study in Taiwan. Am. J. Ophthalmol. 258, 99–109. https://doi.org/10.1016/j.ajo.2023.07.001 (2024).
Bittles AH. Chapter 1. The prevalence and outcomes of consanguineous marriage in contemporary societies. vol 28. Cousin marriages: Between tradition, genetic risk and cultural change. Berghahn Books, New York and Oxford; 2015:33–45.
Ministry of Justice, Taiwan: Civil Code article 983, Laws & regulations database. http://law.moj.gov.tw/LawClass/LawAll.aspx?pcode=B0000001. Accessed 10 May 2024.
Acknowledgements
This study was based on the National Health Insurance Research Database provided by the Health and Welfare Data Center (HWDC) from the Ministry of Health and Welfare (MOHW), Taiwan.
Funding
CMRPG3L1591 from Chang Gung Memorial Hospital, Taiwan. The sponsors had no role in the design or conduct of this research.
Author information
Authors and Affiliations
Contributions
Jiahn-Shing Lee and Lai-Chu See have grant support to access the data in Health and Welfare Data Science Center, Taiwan. Jiahn-Shing Lee, Chang-Fu Kuo and Lai-Chu See wrote the manuscript. Chang-Fu Kuo was instrumental in the methodology of constructing the family relationship using NHIRD, ACE model, and polygenic liability model. Lai-Chu See and Wei-Min Chen undertook the task of data processing and statistical analysis. The data curation and interpretation tasks were jointly handled by Jiahn-Shing Lee, Ken-Kuo Lin, Li-Hung Tsai, Chang-Fu Kuo, and Lai-Chu See. All authors made significant contributions to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lee, JS., Chen, WM., Lin, KK. et al. Assessing genetic and environmental components for pterygium: a nationwide study in Taiwan. Sci Rep 14, 18464 (2024). https://doi.org/10.1038/s41598-024-69482-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69482-5