Abstract
Damage to the development of porcine gametes and embryos caused by high temperatures (HT) is one of the main reasons for the decline in the economic benefits of the livestock industry. Zygotic genome activation (ZGA) marks the beginning of gene expression programs in mammalian pre-implantation embryos. In pigs, ZGA occurs at the 4-cell (4 C) stage, indicating that correct gene expression at this stage plays an important regulatory role in embryonic development. However, the effect of the HT environment on early porcine embryonic development and the RNA expression profile of ZGA remain unclear. In this study, we compared the RNA transcription patterns of porcine 4 C embryos under normal and HT conditions using RNA-seq and identified 326 differentially expressed genes (DEGs). These changes were mainly related to DNA polymerase activity, DNA replication, and nucleotidyltransferase activity. In addition, entries for reverse transcription and endonuclease activity were enriched, indicating that ZGA interfered under HT conditions. Further comparison of the experimental results with the porcine ZGA gene revealed 39 ZGA genes among the DEGs. KEGG and GSEA analysis showed that the oxidative phosphorylation pathway was significantly enriched and signaling pathways related to energy metabolism were significantly downregulated. We also found that NDUFA6 and CDKN1A were located at the center of the protein-protein interaction network diagram of the DEGs. In summary, HT conditions affect mitochondrial function and oxidative phosphorylation levels, and lead to changes in the expression pattern of ZGA in early porcine embryos, with its hub genes NDUFA6 and CDKN1A.
Similar content being viewed by others
Introduction
High temperatures (HT) can lead to seasonal infertility in livestock, which is particularly evident in the pig breeding industry. HT manifests as reduced ovulation, conception, and pregnancy maintenance rates1,2. In addition, HT induces an ovarian heat shock protein response and increases apoptotic signal transduction in granulosa cells3,4. Therefore, understanding the effects of HT on porcine reproduction is an increasingly important area of concern.
Mitochondria are one of the main organelles affected by HT, and their integrity and homeostasis are disrupted under HT conditions. Research has shown that HT experienced by oocytes during maturation can persist during embryonic development, alter nuclear structure, damage the cytoskeleton, cause mitochondrial swelling, and induce organelle damage5. Mitochondrial membrane potential, an important marker for detecting the developmental ability of preimplantation embryos, is reduced in HT embryos6. HT also affects embryonic development by damaging mitochondrial DNA transcription, replication, and expression of genes encoding adenosine 5’ triphosphate (ATP) to produce oxidative phosphorylation complexes7. In addition, the mitochondrial electron transport chain (ETC) is sensitive to heat stress signals. However, research on this topic in embryos is limited. In somatic cells, the core subunits of mitochondrial complex I, NDUFS8 and NDUFV2, are downregulated under HT conditions to regulate the oxidative stress response8. HT oxidizes thiol groups in mitochondrial complexes I, II, IV, and V, thereby reducing the mitochondrial ETC activity9. These findings indicate that mitochondria play an important role in early embryonic response to HT.
Life begins with a transition of genetic control from the maternal to the embryonic genome during zygotic genome activation (ZGA). The genome of the fertilized zygote is initially in transcriptional inertia, but the zygotic gene begins to rapidly transcribe and initiates ZGA after several divisions in early mammalian embryos10,11. In many species, ZGA is not a single biological event, but rather a process of gradual transcriptional activation that consists of two main phases: minor and major ZGA12. The onset and duration of ZGA vary considerably among species; however, within the same species, the process is tightly controlled. In mice, ZGA occurs in the 2-cell (2 C) embryos13, whereas in humans and pigs, it occurs during the 4 C period14,15. ZGA is the driving force for early embryonic regulation of its own development, which is essential for the establishment of embryonic developmental potential and pluripotency. Aberrant ZGA activation, influenced by genetic and environmental factors, leads to defects in preimplantation embryonic development16,17,18,19. During the embryonic stage, from oocytes to ZGA, the embryos are susceptible to HT; this leads to a decrease in developmental ability. In addition, late-stage embryos, such as morulae or blastocysts, acquire HT resistance. The mechanism of temperature-dependent developmental changes is believed to involve the accumulation of antioxidants in embryos in response to HT-induced production of reactive oxygen species20. However, the differences in the RNA transcriptome expression profiles between normal and HT conditions during the ZGA stage of early porcine embryonic development remains unclear.
In this study, we selected porcine 4 C embryos as the research object and studied the effects of HT on the transcriptome characteristics of ZGA in early porcine embryos at the transcriptome level. RNA-seq was used to screen for differentially expressed genes (DEGs) using gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG)21,22,23, gene set enrichment analysis (GSEA), and other analysis methods. Our results indicate that HT substantially affects pathways related to genetic information integration, oxidative phosphorylation, mitochondrial function, and the cell cycle in early porcine embryos, and has different mRNA expression profiles for ZGA. Among them, NDUFA6 and CDKN1A served as hub genes for DGEs.
Materials and methods
Unless otherwise specified, all the chemicals were purchased from Millipore (Burlington, Massachusetts, USA). All experimental operations were conducted on a constant temperature heating table set at 38.5℃ to ensure the stability and accuracy of the experimental environment.
Ethics statement
This study was approved and performed in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of Chungbuk National University Laboratory Animal Center, Cheongju, Korea. All procedures did not involve live animal experiments in our work and all porcine ovaries were purchased from local slaughterhouses. All animal studies were approved and performed according to committee guidelines.
Porcine oocyte collection and in vitro maturation
Fresh ovaries were collected from local slaughterhouses and placed in 37 ℃ constant temperature saline and sent to the laboratory immediately. Aspirate follicles with a diameter of 3–6 mm and select high-quality cumulus-oocyte complexes (COCs). After thrice washing with operating solution, the COCs were transferred to in vitro maturation (IVM) medium and covered with mineral oil. IVM refers to the addition of 10 ng/mL epidermal growth factor, 0.5 µg/mL luteinizing hormone, 0.1 g/L sodium pyruvate, 0.6 mmol/L L-cysteine, 10% (v/v) porcine follicular fluid, and 10 IU/mL follicle-stimulating hormone. Place approximately 30–50 COCs were added to each well and were cultured at 38.5 °C in a humidified 5% CO2 incubator for 44 h. After IVM, COCs were denuded of cumulus cells by continuous pipetting about 30–50 times in a 1 mg/mL hyaluronidase solution.
Parthenogenetic activation and in vitro culture
The denuded oocytes were put in 297 mmol/L mannitol containing 0.05 mM MgSO4, 0.1 mM CaCl2, 0.5 mM HEPES, and 0.01% (w/V) polyvinyl alcohol (PVA), and the pH was adjusted to 7.2. Under these conditions, oocytes were treated with two direct-current pulses of 120 V for 60 µs. To suppress pseudo extrusion of the second polar body, 7.5 µg/mL of cytochalasin B and 4 mg/mL of bovine serum albumin (BSA) were added to bicarbonate-buffered porcine zygote medium 5 (PZM-5), and oocytes were cultured in this medium for 3 h. Add 4 mg/mL BSA to PZM5 to prepare in vitro culture (IVC) medium and wash the activated oocytes thrice. Divide the oocytes into control and HT groups, then culture them in IVC medium at 38.5 °C and 41.5 °C to 4 C stage, respectively.
RNA-seq analysis
Following the manufacturer’s protocol, about 300 samples (4 C) in the lysis buffer were used to construct sequencing libraries with the SMART-Seq® v4 Ultra® Low Input RNA Kit for Illumina. This protocol was composed of first-strand cDNA synthesis, amplify full-length ds cDNA, end repair, dA tailing, ligation of the indexing adapters, and PCR enrichment in sequence to create a final cDNA library. Libraries were quantified by qPCR according to the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms) and qualified using an Agilent Technologies 4200 TapeStation (Agilent Technologies, Waldbronn, Germany). Paired-end sequencing reads were generated using an Illumina NovaSeq sequencing platform (Illumina, San Diego, CA, USA). Filtering out low quality sequences and joint sequences, the Clean Reads were analyzed. Gene- and transcript-level quantification were calculated as the raw read count, fragments per kilobase of transcript per million mapped reads (FPKM), and transcript per million mapped reads (TPM). RNA library sequencing was performed on the Illumina HiseqTM 2500/4000 by Gene Denovo Biotechnology Co., Ltd (Guangzhou, China).
Real-time quantitative PCR
We collected 30 4 C embryos from each group and subsequently extracted mRNA according to the instructions of the Dynabeads mRNA Direct Purification Kit (61012, Thermo Fisher Scientific, Waltham, USA). cDNA was obtained by reverse transcription of mRNA using oligo (dT) 20 primers and SuperScript III reverse transcriptase (Thermo Fisher Scientific). Real-time quantitative PCR (qPCR) was performed using the WizPure qPCR Master Kit and the amplification was performed at 95 °C for 3 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 25 s, 72 °C for 10 s, and a final extension at 72 °C for 5 min. The mRNA quantification data were analyzed using the 2−ΔΔCt method. Sequences of all primers are provided in Table 1.
Statistical analysis
Each experiment was repeated at least three times and statistical comparisons were performed using independent sample t-tests. Statistical significance was set at p < 0.05. The gene expression level of the control group was arbitrarily set to 1 and the HT group was expressed relative to the control group and the results are reported as mean ± SEM. Statistical analysis was performed using GraphPad Prism software (version 5.0; GraphPad Inc., San Diego, CA, USA).
Results
Global transcriptome analysis of control and HT oocytes
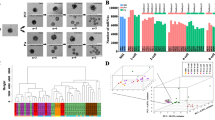
To investigate the differences in gene expression in response to HT, high-throughput RNA-seq analysis was performed on control and HT 4 C stage embryos. The principal component analysis (PCA) showed good intra-group reproducibility, and the two groups were clearly distinguished from each other, as shown in Fig. 1A. By comparing transcript abundances, 326 differentially expressed genes (DEGs) were identified, of which 141 were up-regulated and 185 were down-regulated, as shown in Fig. 1B and C. To verify the accuracy of the RNA-Seq data, we randomly selected 11 genes for qRT-PCR. All genes showed expression patterns consistent with the RNA-Seq data, verifying the reliability and accuracy of the sequencing analysis, as shown in Fig. 1D.
Global transcriptome analysis of control and HT oocytes. (A) PCA showing the reproducibility between repetitive samples and the difference between groups. (B) Heatmap of mRNA expression profiles in Control and HT embryos, showing changes in a subset of genes in response to HT. The color key (from blue to red) of Z-score value (− 2, − 2) indicates low to high expression levels. (C) DEGs between control and HT embryos were shown in the volcano map. Genes that expressed higher (up-regulated) in HT embryos are shown in red, and genes that were lower expressed (down-regulated) are shown in yellow. (D) Validation of RNA-seq data by qPCR. *p < 0.05; ns means not significantly.
HT interferes with genetic information in porcine embryos
To better understand the roles of the DEGs during the HT process, the DEGs were enriched into Gene Ontology (GO) terminology. The enrichment difference bubble chart showed that most genes were enriched in the GO terms contained in the cellular component category, whereas the terms contained in the molecular function category were the most significantly enriched, as shown in Fig. 2A. The top 15 enriched GO terms classified by –log10(Q value) are shown in Fig. 2B. The top enriched terms in molecular function were DNA polymerase activity and nucleotidyltransferase activity, whereas the most enriched terms in biological processes were reverse transcription and DNA replication. In addition, we found that after HT exposure, the expression of GMNC, FBXO4 and RPA3 related to DNA replication, decreased, while the expression of TOP1MT and POLM increased, which together with downregulated POLE2 are related to DNA polymerase activity, as shown in Fig. 2C.
HT interferes with genetic information in porcine embryos. (A) The enrichment difference bubble chart of GO enrichment analysis on DEGs. The abscissa of the image represents the normalized ratio of up-regulated genes to down-regulated genes. Greater than 0 means that more genes in this pathway are up-regulated. (B) The top 15 enriched items analyzed by GO. (C) The heat map for gene expression levels associated with DNA replication and DNA polymerase activity in enriched GO terms.
HT interferes with ZGA in porcine embryos
In pigs, ZGA occurs during the 4 C phase, during which the embryo undergoes intracellular maternal mRNA and protein degradation and transcriptional initiation of its own genome. HT led to the enrichment of DEGs in these terms, indicating that porcine embryonic ZGA was disturbed. Therefore, we compared the expression of the porcine ZGA gene24 in the control and HT groups and found 39 ZGA genes among the DEG, as shown in Fig. 3A. Thereafter, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed to explore the pathways affected by these ZGA genes. As shown in Fig. 3B, these ZGA genes are significantly enriched in TGF-beta signaling, HIF-1 signaling, and selenocompound metabolism and cell cycle pathways. Furthermore, GO analysis of these ZGA genes revealed significant enrichment of terms related to cycling binding, as shown in Fig. 3C. The expression levels of these ZGA genes in control and HT embryos are shown in Fig. 3D. A total of 23 ZGA genes, including CDKN1A, BTF3, NDUFA4, and CDC23 were down-regulated in the HT group embryos.
HT interferes with ZGA in porcine embryos. (A) Venn diagram shows that 39 DEGs are overlapped with porcine ZGA genes. (B) GO enrichment analysis on the ZGA genes in DEGs showing the top 5 enriched KEGG pathways. (C) GO enrichment analysis on the ZGA genes in DEGs showing the top 5 enriched GO items. (D) The heat map for gene expression levels associated with ZGA.
HT interferes with the energy metabolism of porcine embryos
Subsequently, we explored the impact of HT-induced DEGs on signaling pathways in embryos using KEGG pathway enrichment analysis. The top ten enriched pathways are shown in Fig. 4A. We found that oxidative phosphorylation was a significantly enriched pathway, with a lower expression tendency in the HT group. Metabolic reprogramming of the main energy supply pathway, from glycolysis to oxidative phosphorylation, is another key event in the development of porcine preimplantation embryos. As shown in Fig. 4B, the expression levels of the oxidative phosphorylation genes, ATP5MC3, ATP5MC3, NDUFA6, etc. were lower in the HT group.
HT interferes with the energy metabolism of porcine embryos. (A) KEGG enrichment analysis on DEGs showing the top 10 enriched KEGG pathways. (B) The heat map for gene expression levels associated with oxidative phosphorylation in enriched KEGG pathways. (C) GSEA results showing the suppression of the “oxidative phosphorylation”, “inner mitochondrial membrane protein complex”, “ATP synthesis coupled proton transport”, and “mitochondrial protein complex” signaling pathway in HT embryos. (D) The heat map for gene expression levels associated with mitochondrial function.
Gene Set Enrichment Analysis (GSEA) captured several gene sets with differential expression during transcriptome changes. GSEA analysis indicated significant down-regulation of the “oxidative phosphorylation”, “inner mitochondrial membrane protein complex”, “ATP synthesis coupled proton transport”, and “mitochondrial protein complex” signaling pathways associated with energy metabolism, as shown in Fig. 4C.
Mitochondria are the main sites of oxidative phosphorylation, and a decrease in oxidative phosphorylation may be due to mitochondrial dysfunction. Based on the GO terms, we identified genes related to mitochondrial function and found that their expression patterns matched this result, as shown in Fig. 4D. Most Genes related to the mitochondria, mitochondrial proton-transporting ATP synthase complex, mitochondrial respiratory chain complex, and ATP binding were down-regulated in the HT group embryos.
NDUFA6 and CDKN1A are the hub genes of the protein interaction network
After determining that HT mainly affected the pathways and genes related to DNA replication and energy metabolism in porcine oocytes, we plotted the regulatory network between the KEGG pathways. The results are shown in Fig. 5A. The cell cycle and oxidative phosphorylation pathways are central to the regulatory network. This further illustrates the important roles of the cell cycle and oxidative phosphorylation pathways in HT-induced porcine embryonic injury. Thereafter, we plotted a protein-protein interaction network diagram of the differentially expressed genes, as shown in Fig. 5B. We found that NDUFA6 and CDKN1A were at the center of the two interaction networks. Interestingly, NDUFA6 and CDKN1A were involved in the oxidative phosphorylation and cell cycle pathways, respectively. Therefore, we believe that NDUFA6 and CDKN1A are hub genes among the differentially expressed genes associated with HT.
NDUFA6andCDKN1Aare the hub genes of the protein interaction network. (A) The string diagram shows the cell cycle and oxidative phosphorylation pathways are in the center of pathway relationship network. (B) The string diagram shows the NDUFA6 and CDKN1A genes are in the center of protein-protein interaction networks.
Discussion
HT are considered one of the most challenging issues in livestock management. The reproductive system is more sensitive to temperature than the other parts of the body and the impact of HT on fertility is persistent. Although the animals are no longer exposed to HT conditions, the low fertility rate present from warm months of the year (June to September) can still be detected in autumn (October to November)25. Studies have described the effects of HT on the reproductive function of domestic animals during gamete and early embryonic development. For example, our previous research showed that HT leads to meiotic division failure of oocytes and a decrease in the proportion of embryos developing into blastocysts26,27. The pre-implantation embryos in the early stages are highly sensitive to HT damage, and exposure to temperatures only 1.5–2.0 °C higher than normal body temperature can affect the development of mouse and bovine 1-cell embryos28,29. Furthermore, some studies have exposed embryos to temperature fluctuations for 24 h to mimic the rectal temperature of cows exposed to heat stress. It was found that embryonic development was impaired after 8 days in this environment, but the effect was not significant after only one day of exposure, indicating that embryonic development may be interrupted by short-term severe or long-term mild heat shock30. However, the molecular mechanisms underlying this process need to be elucidated and improved upon. Our data indicated that there are multiple differences in RNA expression during early porcine embryonic development under HT conditions.
First, we compared and analyzed dynamic changes in global gene expression in porcine embryos under normal and HT conditions. Principal component analysis showed good intra group reproducibility between the control and HT group, indicating that the RNA-Seq data were of high quality. Subsequently, we identified 326 DEGs between the two groups and validated their expression, further demonstrating the reliability of the RNA-seq analysis. To elucidate the biological functions of HT during early porcine embryonic development at the molecular level, the DEGs were subjected to functional enrichment analyses using GO. These results indicated that these terms were mainly related to the integration of genetic information. Previous studies using animal models have shown that exposure to HT during fertilization can alter the expression of DNA repair genes during early embryonic development31. HT caused by cryptorchidism can lead to a decrease in the expression of repair genes that play a role in the final stage of DNA repair, such as DNA polymerase beta, which promotes the recruitment of DNA ligase III32. Multiple studies have indicated that GMNC play an important role in initiating DNA replication by interacting with Cdc45 and CDK2/cyclin E. GMNC depletion impairs DNA replication in Xenopus laevis oocytes33. In addition, it has been identified that the F-box protein FBXO4 binds to phospho-cyclin D1Thr286 for targeted degradation in response to DNA damage and MAPK signals34,35,36. The replication protein A (RPA) complex, composed of RPA1, RPA2, and RPA3, is the main single-stranded DNA (ssDNA)-binding heterotrimer complex and is involved in various DNA metabolic pathways, including DNA replication, recombination, repair, and DNA damage checkpoints37,38. The absence of RPA in oocytes triggers canonical DNA damage response mechanisms and pathways such as the activation of ATM and P53. RPA deficiency leads to chromosomal translocation in MI and MII oocytes, ultimately resulting in female infertility39. Furthermore, POLE2 (DNA polymerase epsilon 2) is involved in DNA replication, repair, and cell cycle control40. In our study, the expression of GMNC, POLE2, and RPA3 was significantly downregulated in early porcine embryos treated with HT, which supports this point.
ZGA involves a series of dynamic epigenetic changes, including degradation of maternal factors, activation of the zygotic genome, and chromatin recombination, thereby enabling the embryo to achieve pluripotency41,42. The thermal sensitivity of embryos is phased out, and early stage embryos are more susceptible to HT infection29. ZGA in bovine embryos begins after 8 C, and HT treatment at 42℃ affects the development from 1 C to 8 C stages. However, it had almost no effect at the morula and blastocyst stages. Even if early stage embryos survive and continue to develop, the quality of their blastocysts will significantly decrease29,30,43 which suggesting that the normal progression of ZGA is crucial for the embryo to acquire the ability to resist stress. As for which contribution of sperm or oocyte to embryo heat tolerance is more important, studies have compared the differences between Brahman cattle (embryos are less affected by heat stress) and Holstein cattle. It has been found that after fertilization of Holstein cow oocytes with Brahman or Angus breed bull semen (both types of semen have no difference in heat stress response44), there is no breed × temperature interaction, indicating that oocytes play a more important role than sperm in resisting the harmful effects of heat shock in embryos45. Therefore, we compared the RNA-seq data with the pig ZGA gene library and found that 39 DEGs overlapped with the porcine ZGA gene. This indicates that under HT conditions, ZGA of early porcine embryos was significantly affected.
Mitochondria produce cellular ATP via oxidative phosphorylation (OXPHOS), which is crucial for the maintenance of cellular homeostasis. Mitochondria are one of the main organelles affected by heat stress, and in vitro heat damage can induce electron transfer chain (ETC) dysfunction and inhibit mitochondrial ATP synthesis46. There is a tight regulatory connection between the folding of oxidative proteins within the endoplasmic reticulum (ER) and mitochondrial oxidative phosphorylation (OXPHOS). The accumulation of unfolded proteins induces ER stress, leading to the production of reactive oxygen species (ROS) and the activation of cellular signaling mechanisms to counteract the accumulation of unfolded proteins, known as “unfolded protein response” (UPR). One of the three main sensors of UPR is the activating transcription factor 6 (ATF6). Our previous studies have also shown that AFT6 regulates the homeostasis of the ER, Golgi apparatus, and mitochondria, as well as regulates early porcine embryo damage through AIFM2 mediated apoptosis under HT conditions27. In this study, we demonstrated that the OXPHOS pathway was enriched in early porcine embryos treated with HT, and that genes related to mitochondrial ATP synthesis and ETC function were significantly altered. Proteomic analysis of cows has demonstrated that HT leads to reduced expression of proteins involved in OXPHOS and mitochondrial energy pathways, such as NDUFS8 and NDUFV2, which are the core subunits of complex I8. In addition, complex I inactivates and dissociates under HT conditions, slowing down electron flow and reducing oxygen uptake in the ETC, leading to the depletion of ATP synthesis and an imbalance in mitochondrial division/fusion dynamics, inducing mitochondrial dysfunction47. Under acute stress, changes in Ndufa6 transcripts have been detected in the hippocampi of mice using RNA-Seq analysis48. In addition, analysis of multiple myeloma (MM) using single-cell RNA-seq and bulk RNA-seq data indicated that NDUFA6 is a hub gene for MM and can serve as a novel prognostic biomarker for MM49. For the cell cycle, a wide range of stress stimuli, such as oxidants, irradiation, and genotoxins can induce upregulation of CDKN1A, leading to cell aging mediated by upregulation of CDKN1A, which is also a form of cell cycle arrest50,51,52. Previous studies have shown that CDKN1A expression is detected at all stages of mouse pre-implantation embryonic development, and CDKN1A mRNA gradually increases from the 4-cell stage and reaches its peak at the morula stage before decreasing at the blastocyst stage. The expression level of CDKN1A was significantly upregulated when embryos are exposed to heat stress. Knocking down CDKN1A in heat stressed embryos can alleviate developmental delay and reduced blastocyst formation caused by heat stress53. CDKN1A could also mediate cell cycle arrest during the morula/blastocyst stage and apoptosis after the inner cell mass blastocyst stage. The increase in CDKN1A expression level was related to the arrest before and after the morula stage54,55. These findings indicated that NDUFA6 and CDKN1A play crucial regulatory roles in early porcine embryonic development by actively responding to HT signals.
Conclusion
HT affects mitochondrial function, oxidative phosphorylation pathways, DNA polymerase activity, DNA replication, and cell cycle in early porcine embryos. In addition, the expression patterns of ZGA genes in porcine embryos changed under HT conditions. Hub genes are NDUFA6 and CDKN1A.
Data availability
The original contributions presented in the study are included in the article/Supplementary Material. The original data GEO Submission (GSE271158) (NCBI tracking system #24709464) could be found at the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE271158.
References
Love, R. J., Evans, G. & Klupiec, C. Seasonal effects on fertility in gilts and sows. J. Reprod. Fertil. Suppl.48, 191–206 (1993).
Xue, J. L., Dial, G. D., Marsh, W. E. & Davies, P. R. Multiple manifestations of season on reproductive performance of commercial swine. J. Am. Vet. Med. Assoc.204, 1486–1489 (1994).
Seibert, J. T. et al. Differentiating between the effects of heat stress and lipopolysaccharide on the porcine ovarian heat shock protein response1. J. Anim. Sci.97, 4965–4973. https://doi.org/10.1093/jas/skz343 (2019).
Luo, M., Li, L., Xiao, C., Sun, Y. & Wang, G. L. Heat stress impairs mice granulosa cell function by diminishing steroids production and inducing apoptosis. Mol. Cell. Biochem.412, 81–90. https://doi.org/10.1007/s11010-015-2610-0 (2016).
Pohland, R. et al. Influence of long-term thermal stress on the in vitro maturation on embryo development and heat shock protein abundance in zebu cattle. Anim. Reprod.17, e20190085. https://doi.org/10.1590/1984-3143-AR2019-0085 (2020).
Brad, A. M., Hendricks, K. E. & Hansen, P. J. The block to apoptosis in bovine two-cell embryos involves inhibition of caspase-9 activation and caspase-mediated DNA damage. Reproduction. 134, 789–797. https://doi.org/10.1530/REP-07-0146 (2007).
Roth, Z. Symposium review: reduction in oocyte developmental competence by stress is associated with alterations in mitochondrial function. J. Dairy. Sci.101, 3642–3654. https://doi.org/10.3168/jds.2017-13389 (2018).
Skibiel, A. L., Zachut, M., do Amaral, B. C., Levin, Y. & Dahl, G. E. Liver proteomic analysis of postpartum holstein cows exposed to heat stress or cooling conditions during the dry period. J. Dairy. Sci.101, 705–716. https://doi.org/10.3168/jds.2017-13258 (2018).
England, K., O’Driscoll, C. & Cotter, T. G. Carbonylation of glycolytic proteins is a key response to drug-induced oxidative stress and apoptosis. Cell. Death Differ.11, 252–260. https://doi.org/10.1038/sj.cdd.4401338 (2004).
Aoki, F., Worrad, D. M. & Schultz, R. M. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol.181, 296–307. https://doi.org/10.1006/dbio.1996.8466 (1997).
Li, L., Lu, X. & Dean, J. The maternal to zygotic transition in mammals. Mol. Aspects Med.34, 919–938. https://doi.org/10.1016/j.mam.2013.01.003 (2013).
Vastenhouw, N. L., Cao, W. X. & Lipshitz, H. D. The maternal-to-zygotic transition revisited. Development. 146https://doi.org/10.1242/dev.161471 (2019).
Hamatani, T., Carter, M. G., Sharov, A. A. & Ko, M. S. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell.6, 117–131. https://doi.org/10.1016/s1534-5807(03)00373-3 (2004).
Tomanek, M., Kopecny, V. & Kanka, J. Genome reactivation in developing early pig embryos: an ultrastructural and autoradiographic analysis. Anat. Embryol. (Berl). 180, 309–316. https://doi.org/10.1007/BF00315889 (1989).
Jukam, D., Shariati, S. A. M. & Skotheim, J. M. Zygotic genome activation in vertebrates. Dev. Cell.42, 316–332. https://doi.org/10.1016/j.devcel.2017.07.026 (2017).
Zenk, F. et al. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science. 357, 212–216. https://doi.org/10.1126/science.aam5339 (2017).
Menezo, Y., Clement, P., Dale, B. & Elder, K. Modulating oxidative stress and epigenetic homeostasis in preimplantation IVF embryos. Zygote. 30, 149–158. https://doi.org/10.1017/S0967199421000356 (2022).
Wu, S. L. et al. Exposure to acrylamide induces zygotic genome activation defects of mouse embryos. Food Chem. Toxicol.175, 113753. https://doi.org/10.1016/j.fct.2023.113753 (2023).
Zhang, T. et al. Arginine regulates zygotic genome activation in porcine embryos under Nutrition Restriction. Front. Vet. Sci.9, 921406. https://doi.org/10.3389/fvets.2022.921406 (2022).
Sakatani, M. Effects of heat stress on bovine preimplantation embryos produced in vitro. J. Reprod. Dev.63, 347–352. https://doi.org/10.1262/jrd.2017-045 (2017).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51, D587–D592. https://doi.org/10.1093/nar/gkac963 (2023).
Bu, G. et al. Coordination of zygotic genome activation entry and exit by H3K4me3 and H3K27me3 in porcine early embryos. Genome Res.32, 1487–1501. https://doi.org/10.1101/gr.276207.121 (2022).
Badinga, L., Collier, R. J., Thatcher, W. W. & Wilcox, C. J. Effects of climatic and management factors on conception rate of dairy cattle in subtropical environment. J. Dairy. Sci.68, 78–85. https://doi.org/10.3168/jds.S0022-0302(85)80800-6 (1985).
Sun, M. H. et al. High Temperature-Induced m6A epigenetic changes affect early porcine embryonic developmental competence in pigs. Microsc Microanal. 29, 2174–2183. https://doi.org/10.1093/micmic/ozad131 (2023).
Sun, M. H. et al. ATF6 aggravates apoptosis in early porcine embryonic development by regulating organelle homeostasis under high-temperature conditions. Zool. Res.44, 848–859. https://doi.org/10.24272/j.issn.2095-8137.2023.080 (2023).
Gwazdauskas, F. C., McCaffrey, C., McEvoy, T. G. & Sreenan, J. M. In vitro preimplantation mouse embryo development with incubation temperatures of 37 and 39 degrees C. J. Assist. Reprod. Genet.9, 149–154. https://doi.org/10.1007/BF01203755 (1992).
Sakatani, M., Alvarez, N. V., Takahashi, M. & Hansen, P. J. Consequences of physiological heat shock beginning at the zygote stage on embryonic development and expression of stress response genes in cattle. J. Dairy. Sci.95, 3080–3091. https://doi.org/10.3168/jds.2011-4986 (2012).
Rivera, R. M. & Hansen, P. J. Development of cultured bovine embryos after exposure to high temperatures in the physiological range. Reproduction. 121, 107–115 (2001).
Harrouk, W., Codrington, A., Vinson, R., Robaire, B. & Hales, B. F. Paternal exposure to cyclophosphamide induces DNA damage and alters the expression of DNA repair genes in the rat preimplantation embryo. Mutat. Res.461, 229–241. https://doi.org/10.1016/s0921-8777(00)00053-7 (2000).
Tramontano, F., Malanga, M., Farina, B., Jones, R. & Quesada, P. Heat stress reduces poly(ADPR)polymerase expression in rat testis. Mol. Hum. Reprod.6, 575–581. https://doi.org/10.1093/molehr/6.7.575 (2000).
Balestrini, A., Cosentino, C., Errico, A., Garner, E. & Costanzo, V. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat. Cell. Biol.12, 484–491. https://doi.org/10.1038/ncb2050 (2010).
Lin, D. I. et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol. Cell.24, 355–366. https://doi.org/10.1016/j.molcel.2006.09.007 (2006).
Pontano, L. L. et al. Genotoxic stress-induced cyclin D1 phosphorylation and proteolysis are required for genomic stability. Mol. Cell. Biol.28, 7245–7258. https://doi.org/10.1128/MCB.01085-08 (2008).
Jia, L. & Sun, Y. F-box proteins FBXO31 and FBX4 in regulation of cyclin D1 degradation upon DNA damage. Pigment Cell. Melanoma Res.22, 518–519. https://doi.org/10.1111/j.1755-148X.2009.00611.x (2009).
Zou, Y., Liu, Y., Wu, X. & Shell, S. M. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol.208, 267–273. https://doi.org/10.1002/jcp.20622 (2006).
Wold, M. S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem.66, 61–92. https://doi.org/10.1146/annurev.biochem.66.1.61 (1997).
Miao, X. et al. Replication protein A1 is essential for DNA damage repair during mammalian oogenesis. bioRxiv. https://doi.org/10.1101/2023.07.04.547725 (2023).
Zhu, Y., Chen, G., Song, Y., Chen, Z. & Chen, X. POLE2 knockdown reduce tumorigenesis in esophageal squamous cells. Cancer Cell. Int.20, 388. https://doi.org/10.1186/s12935-020-01477-4 (2020).
Eckersley-Maslin, M. A., Alda-Catalinas, C. & Reik, W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell. Biol.19, 436–450. https://doi.org/10.1038/s41580-018-0008-z (2018).
Schulz, K. N. & Harrison, M. M. Mechanisms regulating zygotic genome activation. Nat. Rev. Genet.20, 221–234. https://doi.org/10.1038/s41576-018-0087-x (2019).
Sakatani, M., Kobayashi, S. & Takahashi, M. Effects of heat shock on in vitro development and intracellular oxidative state of bovine preimplantation embryos. Mol. Reprod. Dev.67, 77–82. https://doi.org/10.1002/mrd.20014 (2004).
Chandolia, R. K., Reinertsen, E. M. & Hansen, P. J. Short communication: lack of breed differences in responses of bovine spermatozoa to heat shock. J. Dairy. Sci.82, 2617–2619. https://doi.org/10.3168/jds.S0022-0302(99)75517-7 (1999).
Block, J., Chase, C. C. Jr. & Hansen, P. J. Inheritance of resistance of bovine preimplantation embryos to heat shock: relative importance of the maternal versus paternal contribution. Mol. Reprod. Dev.63, 32–37. https://doi.org/10.1002/mrd.10160 (2002).
Davidson, J. F. & Schiestl, R. H. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol. Cell. Biol.21, 8483–8489. https://doi.org/10.1128/MCB.21.24.8483-8489.2001 (2001).
Chen, K. L. et al. Heat stress induces apoptosis through disruption of dynamic mitochondrial networks in dairy cow mammary epithelial cells. Vitro Cell. Dev. Biol. Anim.56, 322–331. https://doi.org/10.1007/s11626-020-00446-5 (2020).
Sannino, G. et al. Acute stress enhances the expression of neuroprotection- and neurogenesis-associated genes in the hippocampus of a mouse restraint model. Oncotarget. 7, 8455–8465. https://doi.org/10.18632/oncotarget.7225 (2016).
Morita, H., Nishida, Y., Uemura, N. & Hosomi, H. Effect of pentobarbital anesthesia on renal sympathetic nerve activity in the rabbit. J. Auton. Nerv. Syst.20, 57–64. https://doi.org/10.1016/0165-1838(87)90081-6 (1987).
Gorospe, M., Wang, X. & Holbrook, N. J. Functional role of p21 during the cellular response to stress. Gene Expr. 7, 377–385 (1999).
Hoeferlin, L. A., Oleinik, N. V., Krupenko, N. I. & Krupenko, S. A. Activation of p21-Dependent G1/G2 arrest in the absence of DNA damage as an antiapoptotic response to metabolic stress. Genes Cancer. 2, 889–899. https://doi.org/10.1177/1947601911432495 (2011).
Munoz-Espin, D. et al. Programmed cell senescence during mammalian embryonic development. Cell. 155, 1104–1118. https://doi.org/10.1016/j.cell.2013.10.019 (2013).
Ock, S. A., Knott, J. G. & Choi, I. Involvement of CDKN1A (p21) in cellular senescence in response to heat and irradiation stress during preimplantation development. Cell. Stress Chaperones. 25, 503–508. https://doi.org/10.1007/s12192-020-01090-4 (2020).
Choi, I., Carey, T. S., Wilson, C. A. & Knott, J. G. Transcription factor AP-2gamma is a core regulator of tight junction biogenesis and cavity formation during mouse early embryogenesis. Development. 139, 4623–4632. https://doi.org/10.1242/dev.086645 (2012).
Adiga, S. K. et al. p21 provides stage specific DNA damage control to preimplantation embryos. Oncogene. 26, 6141–6149. https://doi.org/10.1038/sj.onc.1210444 (2007).
Funding
This work was supported by the Natural Science Foundation of Chongqing (CSTB2024NSCQ-MSX1284), China; the National Research Foundation (NRF) of Korea grant funded by the Korea government (MSIT) (2022R1A2C300769), Republic of Korea; and the Fundamental Research Funds for the Central Universities (SWU-KQ24014).
Author information
Authors and Affiliations
Contributions
MHS and XSC designed the experiments; MHS performed the majority of the experiments; ZCL, SHL and XHL contributed to the materials; MHS and ZCL wrote the manuscript; XSC revised the manuscript. All authors approved the submission of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
The authors agree with publishing this manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, MH., Zhan, CL., Li, XH. et al. Transcriptome analysis of the effects of high temperature on zygotic genome activation in porcine embryos. Sci Rep 14, 21849 (2024). https://doi.org/10.1038/s41598-024-73166-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-73166-5