Abstract
The electronic medical record management system plays a crucial role in clinical practice, optimizing the recording and management of healthcare data. To enhance the functionality of the medical record management system, this paper develops a customized schema designed for ophthalmic diseases. A multi-modal knowledge graph is constructed, which is built upon expert-reviewed and de-identified real-world ophthalmology medical data. Based on this data, we propose an auxiliary diagnostic model based on a contrastive graph attention network (CGAT-ADM), which uses the patient’s diagnostic results as anchor points and achieves auxiliary medical record diagnosis services through graph clustering. By implementing contrastive methods and feature fusion of node types, text, and numerical information in medical records, the CGAT-ADM model achieved an average precision of 0.8563 for the top 20 similar case retrievals, indicating high performance in identifying analogous diagnoses. Our research findings suggest that medical record management systems underpinned by multimodal knowledge graphs significantly enhance the development of AI services. These systems offer a range of benefits, from facilitating assisted diagnosis and addressing similar patient inquiries to delving into potential case connections and disease patterns. This comprehensive approach empowers healthcare professionals to garner deeper insights and make well-informed decisions.
Similar content being viewed by others
Introduction
The emergence of knowledge graphs has prompted many leading researchers around the world to incorporate them into business systems, thereby improving the accuracy and interpretability of recommendations and bringing more comprehensive data analysis1,2,3,4,5,6,7. A knowledge graph, usually built based on graph databases, provides a powerful method for capturing complex relationships between entities by linking the edges of different entities according to information extracted from various heterogeneous data sources. Multi-modal knowledge graphs contain nodes with diverse data types, such as text, image, and voice information, enabling the joint use of multi-modal data. Knowledge graphs, especially multi-modal knowledge graphs, facilitate richer data information for graph-based neural network-based learning.
Ophthalmology, a specialized field dedicated to visual function and eye health, plays a vital role in ensuring the overall well-being of individuals. According to the World Health Organization’s 2019 World Report on Vision, an estimated 2.2 billion people worldwide have vision impairment or blindness, with over 1 billion cases being preventable or treatable conditions that have yet to be addressed8. To improve the clinical serviceability of ophthalmology, we have collected a batch of real desensitized ophthalmic medical records and their matched medical images and reports. This dataset has been reviewed by multiple rounds of experts from Beijing Tongren Hospital and will serve as a standard database for the national medical dataset.
Previous studies on knowledge graph-based medical record management have predominantly focused on demonstrating enhanced visualization capabilities and indexing speeds9,10. Additionally, they have highlighted the efficacy of graph-structured Electronic Medical Records (EMRs) in supervised disease diagnosis through graph embedding techniques11,12,13,14. However, these studies have often overlooked the inherent capacity of graph-based data management to facilitate learning potential associations between medical records with little effort. Specifically, the graph structure inherently connects cases through feature nodes, thereby providing implicit feature correlations that AI algorithms can leverage to perform patient feature clustering without the need to construct specific patient cohorts or incorporate external knowledge vectors.
This capability is particularly advantageous as it contrasts with the complexities associated with traditional relational database management of EMRs, where significant discrepancies exist in database designs across different countries and even within various hospitals9. Information within a single medical record is often dispersed across numerous tables, making unified data structuring challenging. A more coherent data structure schema can be established by adopting a knowledge graph approach aligned with clinical logic, thereby paving the way for more sophisticated AI services.
Inspired by this, we propose a schema to manage EMRs using a graph database, which can construct a multi-modal knowledge graph of audited medical records to enhance their management capabilities. Furthermore, we propose a novel contrastive graph attention network-based auxiliary diagnostic model named CGAT-ADM. This method achieves diagnostic clustering based on graph-structured medical data. It provides the ability to make similar diagnostic EMR recommendations, a fundamental aspect of electronic medical record management. The best part is that CGAT-ADM seamlessly integrates with existing data structures, eliminating the need for additional work while providing AI-based medical record recommendations. Our work demonstrates that medical information based on knowledge graphs has better AI service development capabilities. Moreover, in the supplementary documents, we compared the differences between building EMR management systems based on relational and graph databases, including the differences in data visualization capabilities and query time.
Our study contributes to the field in the following ways:
-
1.
Taking ophthalmology as an example, we propose an application paradigm and graph schema that can transform structured medical records into a multi-modal knowledge graph that matches the required diagnostic features.
-
2.
We propose an auxiliary diagnostic method based on graph contrastive methods and demonstrate the effectiveness of building AI applications using multi-model knowledge graphs as the data foundation.
-
3.
This study demonstrated the process, structure, and characteristics of establishing a graphical data electronic medical record management system using de-identified real-world medical data.
Related work
This section reviews relevant work from two perspectives: (i) the development of multi-modal knowledge graphs and their applications in medical information analysis and (ii) auxiliary diagnostic methods using EMRs.
Multi-modal knowledge graph in medical research
The Multi-modal Knowledge Graph integrates information from multiple modalities, enriches the representation of entities and relationships by labeling images and other forms, and supports multi-modal reasoning. Furthermore, incorporating multi-modal data as additional features can assist in resolving information gaps in some natural language tasks1.
Significant research has been conducted on integrating EMR information with knowledge graphs, which involves converting patient-related information into a graph structure for analysis. For example, Hasan et al. built a knowledge graph from the registration information of cancer patients for various applications, including visualization analysis and patient queue analysis9. Using a graph structure to store patient medical record information has many advantages over relational database storage, including no need for extra code to perform complex searches, faster execution speed, and better connection of databases with different structures. Linfeng et al. present a systematic approach to constructing a medical knowledge graph from EMRs, demonstrating its effectiveness in ranking tasks and graph embedding learning15. Aldwairi et al. proposed a new graph-based data management system for effective information storage, retrieval, and processing using healthcare data10.
However, the application of medical record-driven multi-modal knowledge graphs in clinical systems is relatively limited based on our knowledge. Zheng et al. made an effort to address this gap by constructing a multi-modal knowledge graph that incorporated patients’ lung CT scans, X-rays, and dialogue information between patients and doctors to enhance the diagnostic accuracy of COVID-1912. This study aimed to integrate doctor-patient dialogue and medical imaging into graph structures. However, it is worth noting that the doctor-patient dialogue primarily focused on the patient’s chief complaints, such as fever, dry cough, and muscle pain, without incorporating more extensive patient information.
Auxiliary diagnostic methods using EMRs
In recent years, research based on actual medical information has become increasingly popular, especially with the release of large-scale medical databases such as the MIMIC-III database16, TCGA database, NHANES database, and UK BioBank. Among them, the analysis based on EMRs can be used to build clinical decision support systems to assist physicians in making more accurate clinical decisions, such as patient diagnosis, medication recommendations, surgical recommendations, etc11,17. Assistive diagnostics using healthcare data is a fundamental topic in analyzing EMRs, which aims to provide a scientifically accurate method for providing diagnostic results or clinical recommendations through AI.
For example, Lin et al. constructed a medical knowledge graph from EHRs, ICD-9 ontology, DrugBank, and medical entity descriptions from Wikipedia and developed a patient similarity learning method based on the Siamese CNN model to compute the similarity score between all patient pairs18. Pokharel et al. used a structure called “time trees” to represent EHR information and utilized the doc2vec embedding technique to calculate patient similarity scores19. Sun et al. embedded a publicly available dataset of patient data into hidden representations and dynamically retrieved patients with the same diagnostic results20. Rui Li et al. developed an interpretable and accurate risk prediction model to support EHR data exploration, knowledge graph demonstration, and model interpretation in heart failure risks13. The work in14 constructed a knowledge graph system for EHR and verified that it could improve the utilization efficiency of unused information in clinical routine practice in chronic kidney disease patients.
Previous studies have shown that EMR based on graph management performs well in terms of visualization capability and data query speed9,10. In addition, the potential of using graph-based EMRs for supervised learning was demonstrated12,13,14. This article aims to demonstrate another unexplored capability of EMR based on graph management: to achieve AI-based assisted diagnosis through feature associations between audited medical records without the need for additional data annotation or external knowledge feature vectors. This concept is particularly applicable to medical data analysis, as medical data is often limited in quantity and difficult to obtain annotations. By examining the feature associations between reviewed medical records, a rich data graph can be constructed to serve as the basis for training the model. This method helps improve the accuracy of diagnosis and explores potential case associations and disease patterns, providing more support for clinical decision-making.
Methodology
As we introduced in the related work, past research on EMR data modeling primarily employed two forms: sequential or graphical. Moreover, the majority of the work was conducted based on supervised learning11,17, and classify medical records into two or more categories. In other words, it involves patient queue selection, data processing, and the construction of auxiliary diagnostic or classification models. However, since we can construct knowledge graphs with stronger interpretability based on EMR, we consider using the graph structure of the knowledge graph as the data foundation for providing AI services.
Driven by this, we propose a multi-modal knowledge graph-driven ophthalmic medical record management schema, and based on this, we have developed an AI-assisted application that requires no additional data processing. Our study was approved by the Ethics Committee of Beijing Tongren Hospital, Affiliated with Capital Medical University (Approval No. TRECKY2018-056-GZ(2022)-07). All methods were performed in accordance with the relevant guidelines and regulations. We confirm that informed consent was obtained from all subjects participating in the study, or from their legal guardian(s) in the case of minors.
In this section, we first introduce the methods of data collection and constructing multi-modal knowledge graphs. On this basis, we introduce a novel auxiliary diagnostic model called CGAT-ADM, which combines metapath2vec21, graph contrastive methods22, and Siamese GAT model23. By inputting clinical information without diagnostic results into the model, medical record recommendations that match the basic factual diagnostic results can be returned from the medical record database. The abbreviations used throughout the text are listed in Table 1.
Construction of multi-model knowledge graph from medical records
The information management systems in most hospitals are built on relational databases such as MySQL or SQL Server, and the data source in this study is no exception. A patient’s EMR will be divided and managed across multiple tables in such a structure. For the expert-reviewed and de-identified ophthalmic EMRs, we have developed a set of transformation engines to convert the structured data into a graph structure consisting of heterogeneous nodes and edges.
Table 2 summarizes the data statistics of the graph database. The transformation of medical records into a graph structure is based on two fundamental principles:
-
1.
Different medical records are linked through public nodes such as age, gender, diagnosis results, historical diseases, and other common attributes. This design provides a bridge for potential relationships between different medical records.
-
2.
Each medical record includes separate left and right eye nodes, with associated diagnosis results, exam results, ophthalmology history, and medical images connected to their respective eye nodes.
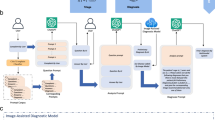
Fig. 1 illustrates the basic data structure of a medical record graph. Gender and age nodes function as public nodes that link each patient’s medical record. Systemic diseases, ophthalmic diseases, abnormal ophthalmic examination items, and patient diagnosis results serve as public nodes to connect different patients. Characteristic nodes retain specific textual descriptions of lifestyle habits and disease history.
These public nodes provide a bridge for associating medical records of different patients, making data management more efficient and providing a basis for modeling relationships between other patients. All graph data is stored in a Neo4j database. The abnormal examination results from medical imaging are also maintained as feature nodes in the knowledge graph. Our supplementary documents introduce the characteristic information of EMRs directly used for diagnosis under the guidance of clinical experts, as well as the process of data collection. Our publicly available code provides the source code for converting structured EMRs into graphical structured data.
Model realization of auxiliary diagnostic search engine
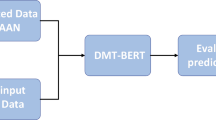
We have incorporated a patient’s medical imaging feature information with their background information on a graph. However, we cannot determine the relationships between patients or disease groups. We developed a medical record graph data engine called Contrastive Graph Attention Network-based Auxiliary Diagnostic Model to achieve this. This module is based on metapath2vec21, graph contrastive methods22, Bert24, and GAT23.
The framework has two layers: Input Layer and Graph Coding Layer. The Input Layer consists of two parts: (1) node-type embedding using the metapath2vec and (2) constructing medical records pairs using contrastive methods. The Graph Coding Layer uses the GAT model as the backbone. The framework of the CGAT-ADM is shown in Fig. 2. The notations and their descriptions in the figure are shown in Table 3.
Input layer
As shown in Fig. 2, the input layer of the CGAT-ADM model consists of two parts. The first part is the node-type embedding based on metapath2vec, and the second part is the construction of positive and negative samples based on contrastive methods.
Node type embedding based on metapath2vec:
Metapath2vec is a graph embedding method used for learning graph embeddings to address the problem of entity representation in heterogeneous graph networks. It produces high-quality entity representations using meta-paths as prior knowledge and employing neural networks for learning. Unlike random walks, the path acquisition process of metapath2vec takes into account the type of entity node and forms an explicit entity type path schema. The Skip-gram model is used for path vector modeling during the training process.
We first constructed a large-scale sampling based on a multi-model medical graph according to the design of the meta-path. The meta-path starts by traversing all information about a patient from the head node of a medical record. Then, it jumps to the following medical record with the same diagnosis result through the diagnosis result node for further traversal. Each medical record was randomly sampled five times, and 10% of non-critical nodes were removed. We performed 100 rounds of medical record sampling from the entire training dataset. Subsequently, the sampling results were trained using the Skip-gram.
The Skip-gram model comprises an input layer, a hidden layer, and an output layer. The input layer receives the unique heat vector of the node, and the output layer gives the conditional probability of each node category under the given central node in the form of the probability distribution. The hidden layer is a fully connected neural network layer responsible for learning the relationship between nodes. After metapath2vec, we obtained 768-dimensional embedding vectors for all node types. These vectors are used to create a node-type vector query table with a size of node-type number \(\times\) 768.
Constructing positive and negative samples based on contrastive methods:
Contrastive learning was first widely used in Computer Vision25,26,27. It involves data augmentation, such as rotating, cropping, and adding noise, to obtain different views of the target sample (image) and take these different views of the image as positive samples. This approach relies on prior knowledge that the augmentation will not affect the image’s label. Graph contrastive learning is a standard data augmentation method for graphs that involves randomly deleting nodes to generate positive and negative samples22.
Inspired by these studies, we have expanded the existing medical record graphs and generated pairs of medical record samples and their similarity index using contrastive methods. In a patient’s medical record information, the diagnosis result is a core feature. If the diagnosis results of two patients are similar, there is a high potential correlation between them. We utilize the diagnostic results as a monitoring signal to quantify the similarity between the two cases. Considering the potential for partial overlap among cases with multiple diseases, we employ the F1 score as a measure of similarity in negative sample construction and high correlation negative samples.
The F1 score is calculated based on the number of true positives (TP), false positives (FP), and false negatives (FN). TP represents the number of consistent items in the diagnostic results of two patients, while FP+FN represents the sum of different diagnostic results of two patients.
The process involves applying three algorithms to construct positive, negative, and high-correlation negative samples. We have detailed descriptions of it in the supplementary document and open-source code.
Graphic coding layer
As illustrated in Fig. 2, the Graphic Coding Layer retrieves data from the Data Input Layer. It performs encoding to capture the global information of the medical records for achieving multi-modal fusion. We aim to generate a high-dimensional spatial clustering of patients with various diseases at a macro level and obtain spatial coordinates for patients sharing the same disease. Concurrently, these coordinates aim to reflect potential associations between patients, encompassing systemic disease information, specific lesion manifestations, and more. In our proposed CGAT-ADM method, we encode three distinct information types from patient medical records: text mode, numerical mode, and node category mode.
In the text mode, we initially establish the principle of text concatenation for each node type and encode the concatenated text information for each node input in the medical record. The text of each node in the graph is fed into MacBERT-base, a Chinese pre-training model based on the BERT-base model28. We utilize the 768-dimensional vector of the first token as the text vector representation for the node. MacBERT-base consists of a 12-layer transformer encoder specifically designed for Chinese language processing.
Regarding the numerical mode, we utilize a 3-layer fully connected layer with a Sigmoid activation function to convert the numerical information into a 768-dimensional numerical vector representation. We only encode the node types that contain numerical information, such as the patient’s unaided vision, optimal corrected vision, intraocular pressure, and axial length.
For each node in the graph, we retrieve the node category from the table obtained through metapath2vec and concatenate it with the corresponding node category vector, which has a size of node count \(\times\) 768.
We implemented three matrices of size node count \(\times\) 768. By concatenating these matrices, we obtained a resulting matrix of size 3 \(\times\) node count \(\times\) 768. We applied a fully connected layer and Softmax to simplify the input graph into a node count \(\times\) 768 matrices, representing the node features of the input graph.
In Fig. 2, the graph encoding layer is depicted as a Siamese GAT model with shared parameters. The GAT backbone model consists of a three-layer GAT layer, each with 12 heads of Multi-Head Attention. The node coding obtained from the node encoding layer and the graph’s adjacency matrix are inputs, and the first token is selected as the overall vector representation.
We employ the cosine similarity of their respective graph vectors to calculate the similarity between pairs of cases. The established similarity metric is then utilized to guide the Mean Squared Error Loss (MSE). The MSE formula used in our study is as follows:
Here, n represents the number of samples, \(y_{\text {pred}_i}\) denotes the predicted value, and \(y_{\text {true}_i}\) represents the corresponding actual value. This formula calculates the squared differences between predictions and actual values, averages them over all samples, and yields the MSE value. As previously stated, CGAT-ADM aims to optimize the extraction of medical record information, thereby enhancing the utilization of medical data.
Results
Dataset
Our data were collected from Beijing Tongren Hospital from 2022 to 2023. The diagnosis results of patients included seven categories: Choroid Melanoma (CM), Retinal Vein Occlusion (RVO), Age-related Macular Degeneration (AMD), Diabetic Retinopathy (DR), Retinal Detachment (RD), Refractive Error (RE), and Pathological Myopia (PM). Most patients suffer from one disease, while others suffer from multiple diseases simultaneously. In addition to the diagnostic results, the surgical and medication information provided by the doctor is also recorded. Most of the medical records in our dataset contain fundus images centered on the macula and OCT images of the macula area in both eyes of the patient. The fundus image was acquired using the Topcon non-contact fundus camera, while the OCT images were collected using the Heidelberg OCT scanner. To protect patient privacy, sensitive information such as patient names, birthdays, and contact information were removed from the real medical records used in our study. 10,124 patients have completed three rounds of medical expert review, data de-identification, and data structuring. This includes multiple visit records for the same patient. In this study, we only retained the medical records of the first visit for each patient, resulting in a total of 5,367 patients. Among them are 224 patients with the diagnosis label CM, 1,146 patients with RVO, 1,159 patients with AMD, 1,696 patients with DR, 530 patients with RD, 426 patients with RE, and 448 patients with PM. The majority of patients have one type of disease, while some patients are diagnosed with multiple diseases at the same time. There are a total of 2,990 female patients, accounting for 55.72%. The median age of all patients is 59, ranging from 1 to 98. For more information about our workflow, please refer to the supplementary document.
To verify the capability of CGAT-ADM, we used a 7:1:2 split method to divide the dataset into a training set, a validation set, and a test set. Fig. 3 illustrates the workflow of data assignment and the distribution of disease labels among patients.
Baseline methods
In this study, we have implemented a pipeline to provide diagnostic assistance in the form of medical record recommendations using a graph data structure. To evaluate the effectiveness of the proposed CGAT-ADM, we compared it with the following baseline methods.
Principal Component Analysis (PCA): An unsupervised algorithm for dimension reduction and feature extraction. We use PCA to map the original high-dimensional data through linear transformation to a low-dimensional space. The number of components is set to 16. Then, we use the cosine distance to measure the similarity between two medical records.
Code Sum based Matching (CSM): A method proposed by Choi et al. This method converts ICD diagnostic codes into medical concept vectors. Then, the patient similarity is determined by calculating the cosine distance between patient vectors29. In our experiment, the patient node vector obtained through metapath2vec is used as the medical concept vector, and the similarity of patients is determined by calculating the cosine distance between patients.
Graph Neural Networks (GNN): GNN is a versatile model for processing graph data, characterized by effective graph modeling, capturing connectivity and dependencies, and leveraging message passing for contextual information exchange among nodes30. GNN obtains a comprehensive representation of the entire graph by treating it as a whole, making it particularly suitable for handling non-Euclidean data. GNN’s ability to propagate and aggregate information among nodes enables it to capture the graph’s relational dependencies and global patterns.
Graph Convolutional Network (GCN): GCN is a deep learning method that utilizes the underlying graph structure. It primarily employs neighborhood aggregation to perform convolution operations on node features and can dynamically adjust weights through the learning process31. GCN extracts and encodes neighborhood information for each node like convolution, allowing for information aggregation from adjacent nodes.
Graph Attention Network (GAT): GAT is a graph neural network that leverages the attention mechanism. It incorporates weighted aggregation based on the relationship weight between a node and its neighbors, thereby enhancing the perception of local information23. In GAT, each node assigns a weight coefficient to its neighboring nodes, which is learned and reflects the importance of each neighbor to the node. These weights are then used to aggregate the features of the neighboring nodes, resulting in a more refined representation that preserves local information.
Evaluation metric
For each patient in the test dataset, we calculate the cosine similarity between that patient and all other patients and retrieve the top k most similar patients. The diagnostic similarity between the input patients and their top k most similar patients is evaluated using the precision metric. We denote MP@k as the mean average precision of the top k most similar patients to the query patient of the validation dataset. Since we are building real medical records for ophthalmic clinical practice, which means that a patient can diagnose multiple diseases simultaneously, this is not a multi-classification or multi-label task. We use MP@k to evaluate the quality of search results, which is a commonly used metric used to measure the average accuracy of information retrieval systems in ranking the top k in the returned results. In machine learning, precision refers to the ratio of correctly classified samples by the classifier to the total number of samples classified by the classifier.
To better evaluate the performance of different models, we set k = 1, 5, 10, and 20, respectively, to retrieve the top 1, 5, 10, and 20 most similar patients in the training database for each medical record graph in the test dataset.
Results
Table 4 shows the performance comparison results of the models. our CGAT-ADM achieved the highest precision of 0.8940 in MP@1, 0.8742 in MP@5, 0.8686 in MP@10 and 0.8563 in MP@20. Contrastive methods are used to construct medical record pairs, and significant performance improvements have been achieved by incorporating GAT, GCN, and GNN on top of the metapath2vec method. This demonstrates that the graph contrastive strategy can greatly enhance the capability of medical record analysis based on a multi-model knowledge graph. The CGAT-ADM approach significantly improves performance by incorporating more comprehensive information, including medication, medical history, intraocular pressure, and visual acuity. This integration yields a remarkable improvement of over 13.19% in MP@1, 13.25% in MP@5, 13.73% in MP@10, and 12.08% in MP@20 compared to the GAT method. Fig. 4 displays the high-dimensional results of all the EMRs in our dataset obtained through CGAT-ADM, which were then reduced to 2 dimensions using t-SNE and visualized in a scatter plot. In summary, the research findings showcase the potential of CGAT-ADM as a physician-assisted tool in real-world and intricate clinical settings. It also highlights its efficacy in identifying potential disease patterns by fusing medical records with multiple data sources.
Ablation study
Generally, patient similarity learning based on diagnostic results using EMRs is centered on doing semantic similarity studies with medical text information32,33,34 or converting patients’ medical data into specific codes or graphs for similarity learning13,20,35. Compared to traditional research, we suggest encoding and integrating different types of information separately to improve the overall performance of medical record similarity analysis. Based on the ablation experiment results in Table 5,
The GAT model based on node types achieved an MP@1 of 0.7898, MP@5 of 0.7719, MP@10 of 0.7637, and MP@20 of 0.7640. Based on node text information, the GAT model achieved an MP@1 of 0.8508, MP@5 of 0.8444, MP@10 of 0.8387, and MP@20 of 0.8318. By fusing text, node types, and numerical information, CGAT-ADM achieved an MP@1 of 0.8940, MP@5 of 0.8742, MP@10 of 0.8686, and MP@20 of 0.8563. The improvement in system performance by adding numerical information is relatively small, mainly due to the limited number of numerical nodes of these medical records. However, we still retain the design of the numerical information channel to enhance the model’s scalability of integrating data for more potential examination indicators.
Programming architecture
The CGAT-ADM model is trained using PyTorch. The CGAT-ADM model processes all healthcare data to construct their high-dimensional representations stored in the Milvus vector database. The underlying data structure is stored in the Neo4j graph database, while the medical imaging records of patients are saved as URLs in the graph database. This study demonstrates a clustering service that stores multimodal EMR information in a graph structure. We have built a framework for online querying and visualization of record recommendations. The back end of the application is invoked using Node.js. The front end is developed using Vue.js. Doctors or healthcare professionals can input partial or complete patient information through API to retrieve the most similar patients from the database using the CGAT-ADM engine. It is essential to highlight that the data is stored in the Neo4j graph database, while the vector database engine maintains high-dimensional positional vectors for all the records.
Discussion
This study introduces a schema for converting existing EMRs into graph-structured data. Based on this schema, diagnostic results can be clustered with less further data processing. The CGAT-ADM model we propose is a multi-modal knowledge graph-based medical record-assisted diagnosis and recommendation method. In the field of ophthalmology, doctors primarily rely on patient communication and the observation of medical images to make diagnoses. This study aimed to simulate clinical reasoning by leveraging AI techniques and medical data in a graph structure. Using multi-modal knowledge graphs for medical record management demonstrations improved capabilities for deploying AI applications in this ___domain. This particularly benefits regions with limited medical resources, scarce doctors, or relatively inexperienced medical professionals. The quality of medical services can be improved by establishing a standardized medical record system that is desensitized and reviewed by experts and utilizing AI methods. In addition, we also presented a visual display of medical records based on graph databases and a comparison of query time between graph data management medical records and relational databases in the supplementary documents.
The CGAT-ADM method is defined as an auxiliary diagnostic model because the anchor of the strategy of the graph contrast method is the coarse diagnosis and fine diagnosis results of patients, thus achieving clustering based on diagnostic results. This model can automatically recommend patients similar to their condition based on partial or comprehensive patient information, thereby providing potential diagnostic results and recommendations for similar patients. Through the above work, we demonstrate that managing electronic medical record information in a multi-modal knowledge graph possesses advantages that relational databases do not have, namely the ability to provide AI services with less additional data work. The development of the vast majority of auxiliary diagnostic systems typically involves several necessary processes, including defining classification labels, collecting clinical data, and building classification models. This work is generally supervised learning and requires manual screening of clinical data to construct sequences, graphs, or based on organized medical images. The business logic we demonstrate directly builds auxiliary diagnostics through the underlying data structure of EMR management systems. Previous research has emphasized the advantages of knowledge graph management of medical data, including faster indexing speeds and better visualization capabilities9,10,13,14. Conversely, this study showcases the potential of building medical AI services based on the graph structure. It is worth noting that although some studies have proposed using tabular data for classification36,37, graph structures, due to their unique triple structure, have a distinct advantage when building recommendation systems, which is currently difficult for tabular data to achieve.
We are fully aware of the limitations of this study. Firstly, this study integrates over 30 different abnormal results obtained from medical imaging platforms when constructing graph-based EMRs. In the future, we will consider building visual extractors to extract feature values from medical images directly. Moreover, this research is based on ophthalmic medical record data, and different medical disciplines may have different graph data construction schemas. This involves the medical disciplines’ diagnostic processes, clinical challenges, and data modeling methods. Similar to the table design of relational databases, the structure design of graphs also requires in-depth discussions with medical experts and software developers. Our data has undergone expert review, anonymization, and informed consent from patients. However, in real-world applications, especially when using knowledge graphs as the foundation for data management to build artificial intelligence applications, it is necessary to consider more issues, such as patient privacy, especially when using medical data to build artificial intelligence services.
Furthermore, this study is based on data that experts have reviewed, and more prospective experiments, including clinical applications, will be beneficial in demonstrating the superiority of our approach. This will be the focus of our next phase of work. We are also considering further exploration of the integration of artificial intelligence and multi-modal knowledge graphs, including integrating external variable datasets such as sequencing information and drug data. This integration will provide new opportunities for advanced clinical analysis, such as determining clinical trial eligibility, evaluating drug treatment efficacy, analyzing patient demographic variables, and identifying risk factors.
Data Availibility Statement
The desensitization dataset used in this study can be obtained from the corresponding author upon reasonable request. More data samples can be found in the supplementary documents.
References
Zhu, X. et al. Multi-modal knowledge graph construction and application: A survey. IEEE Transactions on Knowledge and Data Engineering (2022).
Zhong, W. et al. Multi-dimensional quality-driven service recommendation with privacy-preservation in mobile edge environment. Computer Communications 157, 116–123 (2020).
Chi, X., Yan, C., Wang, H., Rafique, W. & Qi, L. Amplified locality-sensitive hashing-based recommender systems with privacy protection. Concurrency and Computation: Practice and Experience 34, e5681 (2022).
Liu, H., Kou, H., Yan, C., Qi, L. et al. Keywords-driven and popularity-aware paper recommendation based on undirected paper citation graph. Complexity 2020 (2020).
Qi, L. et al. Data-driven web apis recommendation for building web applications. IEEE transactions on big data 8, 685–698 (2020).
Liu, H., Kou, H., Yan, C. & Qi, L. Link prediction in paper citation network to construct paper correlation graph. EURASIP Journal on Wireless Communications and Networking 2019, 1–12 (2019).
Zheng, S. et al. Pharmkg: a dedicated knowledge graph benchmark for bomedical data mining. Briefings in bioinformatics 22, bbaa344 (2021).
Burton, M. J. et al. The lancet global health commission on global eye health: vision beyond 2020. The Lancet Global Health 9, e489–e551 (2021).
Hasan, S. S. et al. Knowledge graph-enabled cancer data analytics. IEEE journal of biomedical and health informatics 24, 1952–1967 (2020).
Aldwairi, M., Jarrah, M., Mahasneh, N. & Al-khateeb, B. Graph-based data management system for efficient information storage, retrieval and processing. Information Processing & Management 60, 103165 (2023).
Si, Y. et al. Deep representation learning of patient data from electronic health records (ehr): A systematic review. Journal of biomedical informatics 115, 103671 (2021).
Zheng, W. et al. Pay attention to doctor-patient dialogues: multi-modal knowledge graph attention image-text embedding for covid-19 diagnosis. Information Fusion 75, 168–185 (2021).
Li, R., Yin, C., Yang, S., Qian, B. & Zhang, P. Marrying medical ___domain knowledge with deep learning on electronic health records: a deep visual analytics approach. Journal of medical Internet research 22, e20645 (2020).
Shang, Y. et al. Ehr-oriented knowledge graph system: toward efficient utilization of non-used information buried in routine clinical practice. IEEE Journal of Biomedical and Health Informatics 25, 2463–2475 (2021).
Li, L. et al. Real-world data medical knowledge graph: construction and applications. Artificial intelligence in medicine 103, 101817 (2020).
Miotto, R., Li, L., Kidd, B. A. & Dudley, J. T. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Scientific reports 6, 1–10 (2016).
Shickel, B., Tighe, P. J., Bihorac, A. & Rashidi, P. Deep ehr: a survey of recent advances in deep learning techniques for electronic health record (ehr) analysis. IEEE journal of biomedical and health informatics 22, 1589–1604 (2017).
Lin, Z., Yang, D. & Yin, X. Patient similarity via joint embeddings of medical knowledge graph and medical entity descriptions. IEEE Access 8, 156663–156676 (2020).
Pokharel, S., Zuccon, G., Li, X., Utomo, C. P. & Li, Y. Temporal tree representation for similarity computation between medical patients. Artificial Intelligence in Medicine 108, 101900 (2020).
Sun, Z., Lu, X., Duan, H. & Li, H. Deep dynamic patient similarity analysis: Model development and validation in icu. Computer Methods and Programs in Biomedicine 225, 107033 (2022).
Dong, Y., Chawla, N. V. & Swami, A. metapath2vec: Scalable representation learning for heterogeneous networks. In Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, 135–144 (2017).
You, Y. et al. Graph contrastive learning with augmentations. Advances in neural information processing systems 33, 5812–5823 (2020).
Veličković, P. et al. Graph attention networks. arXiv preprint[SPACE]arXiv:1710.10903 (2017).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint[SPACE]arXiv:1810.04805 (2018).
Misra, I. & Maaten, L. v. d. Self-supervised learning of pretext-invariant representations. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 6707–6717 (2020).
Chen, T., Kornblith, S., Norouzi, M. & Hinton, G. A simple framework for contrastive learning of visual representations. In International conference on machine learning, 1597–1607 (PMLR, 2020).
Wei, C., Wang, H., Shen, W. & Yuille, A. Co2: Consistent contrast for unsupervised visual representation learning. arXiv preprint[SPACE]arXiv:2010.02217 (2020).
Cui, Y. et al. Revisiting pre-trained models for chinese natural language processing. arXiv preprint[SPACE]arXiv:2004.13922 (2020).
Choi, E., Schuetz, A., Stewart, W. F. & Sun, J. Medical concept representation learning from electronic health records and its application on heart failure prediction. arXiv preprint[SPACE]arXiv:1602.03686 (2016).
Defferrard, M., Bresson, X. & Vandergheynst, P. Convolutional neural networks on graphs with fast localized spectral filtering. Advances in neural information processing systems 29 (2016).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. arXiv preprint[SPACE]arXiv:1609.02907 (2016).
Slater, L. T. et al. Towards similarity-based differential diagnostics for common diseases. Computers in biology and medicine 133, 104360 (2021).
Zhou, X., Li, Y. & Liang, W. Cnn-rnn based intelligent recommendation for online medical pre-diagnosis support. IEEE/ACM Transactions on Computational Biology and Bioinformatics 18, 912–921 (2020).
Li, X. et al. Intelligent diagnosis with chinese electronic medical records based on convolutional neural networks. BMC bioinformatics 20, 1–12 (2019).
Gu, Y. et al. Structure-aware siamese graph neural networks for encounter-level patient similarity learning. Journal of Biomedical Informatics 127, 104027 (2022).
Huang, X., Khetan, A., Cvitkovic, M. & Karnin, Z. Tabtransformer: Tabular data modeling using contextual embeddings. arXiv preprint[SPACE]arXiv:2012.06678 (2020).
Hegselmann, S. et al. Tabllm: Few-shot classification of tabular data with large language models. In International Conference on Artificial Intelligence and Statistics, 5549–5581 (PMLR, 2023).
Acknowledgements
This work was supported in part by the Shenzhen International Science and Technology Information Center and the Shenzhen Bay Laboratory.
Author information
Authors and Affiliations
Contributions
W.G., F.R., L.S., W.W., and L.M. conceived this project. W.G., L.S., F.L., and R.Z. were involved in data collection and organization. W.G., F.R., Z.D., D.X., C.C., and Z.N. conducted the experiments and analyzed the results. W.G. and Z.G. wrote and revised the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Code availability
This study includes three main steps: (1) extracting EMR graph structure from relational databases, (2) metapath2vec and constructing medical record pairs through contrastive methods, (3) CGAT-ADM. You can obtain all these codes at the following address: https://github.com/AI-thpremed/Multi-Modal-KG-EMRs-Management.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, W., Rong, F., Shao, L. et al. Enhancing ophthalmology medical record management with multi-modal knowledge graphs. Sci Rep 14, 23221 (2024). https://doi.org/10.1038/s41598-024-73316-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-73316-9