Abstract
Hypoxic-ischemic encephalopathy (HIE) is a diffuse brain tissue injury caused by acute ischemia and hypoxia, and it is most commonly found in newborn infants but can also occur in adults. Mesenchymal stem cell (MSC) therapies have showed improved outcomes for treating HIE-induced neuronal defects. However, many key issues associated with poor cell viability and tolerance of grafted MSCs after HIE remain to be resolved. Genetic engineering could endow MSCs with more robust regenerative capacities. Our research, along with that of other scientists, has found that the expression of intracellular erythropoietin (EPO) in human umbilical cord MSCs (hUC-MSCs) increases proportionally with the duration of hypoxia exposure. Furthermore, we observed that EPO, when introduced into the EPO gene-modified hUC-MSCs, can be secreted into the extracellular space. However, the underlying mechanisms that support the neuroprotective effects of EPO-MSCs remain unclear. EPO-MSCs, hUC-MSCs, and NC-MSCs were identified by flow cytometry, osteogenic, and adipogenic differentiation assays. The oxygen-glucose deprivation (OGD)-induced SH-SY5Y cell-line was established, and five groups were set up: control, 24-h ischemia-hypoxia, co-cultured with hUC-MSCs, NC-MSCs, and EPO-MSCs after hypoxia. LEGENDplex™ multi-factor flow cytometry was used to detect the secretion of inflammatory factors in cell supernatants and cerebrospinal fluid. Chromosome-targeted excision and tagging (CUT&Tag) sequencing was applied to detect genomic H3K4me2 modifications, and conjoint analysis with transcriptome sequencing (RNA-seq) was performed. Lentiviral vector infection was used to construct SH-SY5Y cells with stable knockdown of RE1-silencing transcription factor (REST), and flow cytometry was used to detect alterations in apoptosis. Finally, the molecular mechanism underlying the neuroprotective and anti-apoptotic effects of EPO-MSCs was investigated using RNA sequencing, qRT-PCR, and western blot assays. Our results suggest that EPO-MSCs are genetically engineered to secrete significantly more EPO. EPO-MSCs treatment has anti-apoptotic properties and offers neuronal protection during ischemic-hypoxic injury. Furthermore, RNA-seq results suggest that multiple inflammation-related genes were down-regulated after EPO-MSCs treatment. Application of RNA-seq and CUT&Tag combined analysis found that the expressions of REST were significantly up-regulated. Lentiviral vector infection to construct REST knockdown SH-SY5Y failed to rescue apoptosis after hypoxia and co-culture with EPO-MSCs, and SETD2-mediated H3K36me3 protein level expression was reduced. EPO-MSCs may promote neuronal survival by affecting H3K4me2 and thus activating the expression of REST and TET3. EPO-MSCs also upregulated the modification level of SETD2-mediated H3K36me3 and regulated the expression of inflammation-related genes such as PLCG2, as well as apoptosis genes BCL2A1. To investigate the neuroprotective effects of EPO-modified hUC-MSCs and the underlying epigenetic regulatory mechanisms, this study aims to provide a theoretical foundation for the potential application of EPO gene-modified hUC-MSCs in the treatment of HIE.
Similar content being viewed by others
Introduction
Hypoxic-ischemic encephalopathy (HIE) is a diffuse brain tissue injury caused by acute ischemia and hypoxia, when the metabolic demands of the brain tissue cannot be met because of respiratory and cardiac arrests, electric shock injuries, poisonings, certain severe craniocerebral injuries, and/or persistent status epilepticus. It is most commonly found in newborn infants, but also occurs in adults1,2. Multiple studies have provided evidence that HIE leads to a delay in cell death through various mechanisms, including oxidative stress, inflammation, apoptosis, and excitotoxicity3. One of the current treatment at the level of in vivo experiments is hypothermia, where the use of 32–34 °C hypothermia within the first 6 h of the primary injury reduces the sequelae caused by oligodendrocyte injury4. However, the neuroprotective effects of hypothermia are limited by the time of onset and severity of encephalopathy5,6. It has been found that the expression of ped-1β, ped L-6 and TNF-a in the cerebrospinal fluid of patients with ischemic-hypoxic encephalopathy increases with the degree of HIE injury, and it has been demonstrated that the concentration of cytokines, such as IL-1α, IL-1β, IL-6, and TNF-α in the cerebrospinal fluid of the cerebral spinal fluid of a rat model of HIE significantly improves with treatment in the group of hypothermia combined with mesenchymal stem cell therapy for ischaemic hypoxic encephalopathy. Therefore, HIE requires systematic exploration of altered inflammatory states as well as new therapeutic measures. Meanwhile, clinically, we measured expression levels of inflammatory factors in cerebrospinal fluid supernatants from HIE patients and control patients, looking forward to discovering the therapeutic potential of mesenchymal stromal cells for hypoxic ischemic encephalopathy.
In recent years, it has been found that both umbilical cord mesenchymal stem cells (hUC-MSCs) and erythropoietin (EPO) have demonstrated good therapeutic effects in the treatment of HIE, but there are limitations7,8. Research has found that the utilization of hUC-MSCs therapy diminishes the magnitude of the injury and enhances the behavioral consequences subsequent to neonatal hypoxic-ischemic brain injury, both in animal models and human subjects9,10. hUC-MSCs can migrate to sites of inflammation and exert anti-inflammatory effects, but are limited by their viability as well as their homing effects11. EPO and its classical receptor (EPOR) have been found to be expressed in both human neurons and astrocytes12. Inflammation-induced EPO signaling is essential for inflammation resolution, and EPO is an important endogenous pro-resolving molecule13. Under normal circumstances, only a mere 1% of the EPO in circulation has demonstrated the ability to traverse the blood-brain barrier (BBB)14. Through diverse technical methods including genetic engineering, surface modification, and tissue engineering, the engineering of these cells or vesicles can enhance their homing abilities and other innate characteristics. This can facilitate targeted delivery to specific tissues or organs, thereby augmenting their therapeutic efficacy15.
Based on available data, we chose the human neuroblastoma cell line (SH-SY5Y) that exhibits comparable morphological, neurochemical, and electrophysiological traits to typical neurons. The distinctive features of this cell line include its limited differentiation, pyramidal structure, and clearly discernible axon structure16. EPO-MSCs were successfully constructed, and it was found that EPO-MSCs significantly reduced the apoptosis of the oxygen-glucose deprivation (OGD)-induced SH-SY5Y cells, but the specific neuroprotective mechanism was unknown. Research has shown that epigenetic modifications play a crucial role in the process of genomic reprogramming, the expression of genes specific to certain tissues, and higher-order cognitive abilities such as learning and memory. Additionally, these modifications also influence the survival of neurons in the developing brain. Consequently, alterations in the epigenome can serve as a significant mechanism for enduring transcriptional modifications within mature neurons17,18. Hence, investigating the potential neuroprotective mechanism of EPO-MSCs against ischemic-hypoxic SH-SY5Y cells and its associated epigenetic mechanism will offer novel insights and a solid theoretical foundation for the management of HIE.
Materials and methods
Enrolled cases
Clinical data: Twenty patients with HIE (12 male and 8 female; mean age: 47.75 ± 17.14 years; age range: 22–70 years) admitted to Zhengzhou Central Hospital affiliated with Zhengzhou University from April 2021 to December 2022 were selected for the study. The inclusion criteria were as follows: (1) Craniocerebral CT or MRI examinations clearly showed substantial brain injury. (2) History of diseases causing cerebral ischemia and hypoxia, such as post-cardiopulmonary resuscitation, electric shock, and cerebral hemorrhage. (3) Abnormal neurological symptoms such as impaired consciousness, lethargy, and coma, changes in limb muscular strength and muscle tone, and abnormal neurological reflexes. The exclusion criteria were as follows: (1) Severe mental disorder. (2) Severe heart, lung, liver, kidney, or any other organ dysfunction. Eight patients (5 male and 3 female; mean age: 55.38 ± 16.54 years) with increased intracranial pressure and negative cerebrospinal fluid delivery results were selected as the control group. Comparing the sex and age of the patients in the two groups, the difference was not significant (P > 0.05). The lumbar puncture method was used to collect 2–5 mL of cerebrospinal fluid from HIE patients and the control group, which was then centrifuged (1,380×g, 4 °C, 5 min), and the supernatant was stored at -80 °C until bulk cytokine analysis was carried out. The study received approval from the Ethics Committee of Zhengzhou Central Hospital, Zhengzhou University (approval number: 202171), all research was performed in accordance with relevant guidelines/regulations. In addition, all participants in the study provided their informed consent by signing a consent form, all research have been performed in accordance with the Declaration of Helsinki.
Isolation of mesenchymal stem-like cells
Under aseptic conditions, the umbilical cord of a full-term fetus delivered by cesarean section at Zhengzhou Central Hospital affiliated with Zhengzhou University was collected. A 4–5-cm segment of the umbilical cord was taken and washed thoroughly with D-Hanks solution to remove any residual blood in the blood vessels. The Wharton’s jelly tissue of the umbilical cord was then separated and removed. The umbilical cord was further sheared as much as possible and digested with 0.1% collagenase type I for 4 h at 37 °C. After digestion, the umbilical cord was filtered through a 200-mesh mesh filter. The filtrate was collected and centrifuged at 400×g for 8 min. The resulting cell precipitate was resuspended and inoculated into 75cm2 culture flasks. The low-glucose Dulbecco’s modified Eagle’s medium (DMEM; Corning Inc.) supplemented with 10% FBS (Corning Inc.), 1% nonessential amino acids (Gibco), and 1% penicillin/streptomycin (Gibco) (12 mL) was added, and the cells were cultured in a 37 °C, 5% CO2 cell culture incubator. The medium was changed for the first time after 5–6 days, and fresh medium was replaced every 2 days thereafter. When the cells reached approximately 85% confluency, they were digested with 0.25% trypsin solution and passaged in a 1:3 ratio. This process was repeated until the cells reached the third generation, at which point they were collected for phenotypic identification. Based on the research conducted by the International Society for Cellular Therapy, mesenchymal stem-like cells possess three distinctive characteristics. First, they demonstrate adhesive properties to plastic surfaces. Second, they exhibit the expression of surface markers such as CD105, CD73, and CD90, while lacking the expression of CD45, CD34, CD14, CD11b, CD79α, CD19, and HLA-DR. Third, they possess the capability to undergo in vitro differentiation into osteoblasts, adipocytes, and chondroblasts. Flow cytometry, along with assays focusing on osteogenic and adipogenic differentiation were used for the analysis of hUCB-MSCs.
Flow cytometric analysis
Non-adherent hUC-MSCs and EPO-MSCs cells were fixed in 4% paraformaldehyde after being washed twice with ice-cold PBS and centrifuged. Afterwards, the cells underwent a 20-min incubation with mouse anti-human CD45-FITC, CD34-FITC, CD11b-FITC, CD19-FITC, CD29-FITC, CD73-PE, CD105-APC-A750, CD90-APC, and HLA-DR-FITC (all from BioLegend, SanDiego, CA, USA) in the dark. Flow cytometry was carried out using a flow cytometer (Beckman Coulter GmbH, Krefeld, Germany). For cell cycle analysis, cells were fixed overnight at 4°C with 70% ethanol, followed by washing and treatment with propidium iodide (Sigma; P4170; 50 µg/mL) and RNase A (Thermo Fisher; R1253) at 37 °C for 30 min. Subsequently, a minimum of 1 × 104 cells were analyzed using a flow cytometer from BD Biosciences. The Annexin V-FITC/PI Apoptosis Detection Kit (Sigma; APOAF-60TST) was used for apoptosis detection. Signal cells were washed twice with PBS and then stained with Annexin V-FITC and PI for 15 min at room temperature in the dark, following the manufacturer’s instructions. Flow cytometry was employed to capture the signals from the cells, and all data were quantified using the CytExpert Software v1.2 (Beckman Coulter, Brea, CA).
Osteogenic and adipogenic differentiation
To investigate the osteogenic and adipogenic differentiation potential of human umbilical cord blood-derived mesenchymal stem cells (hUC-MSCs), cells from the third passage were seeded at a density of 3 × 103 cells/cm2 and cultured in osteogenic and adipogenic medium (Sigma-Aldrich) for a duration of 3 weeks, with medium changes performed twice/week. Osteogenic differentiation: Cells with a logarithmic growth stage of the P2 generation were selected. The medium in the cell culture flasks was discarded and carefully washed twice with D-Hanks at 37 °C. The medium was then replaced with the osteogenic induction medium, and the medium was changed for the first time after 24 h, and then every 48–60 h thereafter. After 30 days of cultivation, the medium was discarded, and the cells were washed twice with D-Hanks solution. The cells were then fixed with paraformaldehyde, washed with an isopropyl alcohol working solution, and stained with 0.2% Alizarin red.
Lipogenic differentiation: Cells with a logarithmic growth stage of the P2 generation were selected. The medium in the cell culture flasks was discarded, and the cells were carefully washed twice with D-Hanks at 37 °C. The medium was then replaced with the lipogenic induction medium and changed every 48 h. After 20 days of incubation, the medium was discarded, and the cells were washed with D-Hanks. The cells were then fixed with 2% paraformaldehyde, washed with a 60% isopropanol working solution. The isopropanol working solution was washed off, and the cells were then stained with Oil Red O.
Cell culture and lentiviral infection
The SH-SY5Y cell line was obtained from Wuhan Punosai Life Science and Technology Co. The cells were cultured in DMEM d-glucose medium (GIBCO) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.hUC-MSCs, NC-MSC, and EPO-MSCs cell lines were cultured in Mesenchymal Stem Cell Culture medium (Yocon). All cells were maintained at 37 °C in a 5% CO2 humidified atmosphere. Lentiviruses used in this study were purchased from Genechem (Shanghai, China). Lentiviral infection was performed following the manufacturer’s instructions. Briefly, SH-SY5Y and hUC-MSCs cells were seeded in 6-well plates with 2 mL complete medium and transduced with lentivirus including with EPO-CDS-eGFP gene overexpressed lentivirus(EPO-MSCs) and control lentivirus(NC-MSCs), sh-REST lentivirus(pLKO.1-sh1, pLKO.1-sh2) and control lentivirus(pLKO.1-Vector) at a multiplicity of infection of 15–20 in the presence of the HitransG A reagent (Genechem). Transduced cells were selected using 5 g/mL puromycin (Selleck, Shanghai, China). After approximately 2 weeks of continuous selection, surviving cell colonies were expanded. The efficacy of overexpression and knockdown was confirmed by western blotting (WB).
Antibodies
The primary antibodies utilized for the Cleavage Under Targets and Tagmentation (CUT&Tag) experiments were rabbit anti-H3K4me2 (diluted 1:1000; Cell Signaling Technology #9725) and rabbit anti-H3K36me3 (diluted 1:1000; Cell Signaling Technology #4909). For WB, the primary antibodies used were rabbit anti-EPO (diluted 1:1000 for WB, ThermoFisher PA5-85107); mouse anti-β-actin (diluted 1:5000 for WB, Proteintech #66009-1-Ig); mouse anti-BCL2A1 (diluted 1:2000 for WB, Cell Signaling Technology #15071T); rabbit anti-REST (diluted 1:500 for WB, Proteintech #22242-1-AP); Rabbit IgG (H + L) secondary antibody (diluted 1:1000 for WB, ThermoFisher #31460); and Mouse IgG (H + L) secondary antibody (diluted 1:1000 for WB, ThermoFisher #31430).
ELISA
Supernatants obtained from hUC-MSCs, NC-MSCs and EPO-MSCs cultures were collected and subjected to ELISA analysis using the Cloud-Clone AEA028Ra kit, as per the manufacturer’s instructions. The optical density (OD) was measured at 450 nm using a standard ELISA plate reader. These experiments were performed in triplicate.
The establishment of oxydextrose deprivation SH-SY5Y model
SH-SY5Y cells were inoculated with 5 × 106 cells/well in 6-well plates and cultured for 24 h. When the cell density reached 60–80%, the cells were replaced with sugar-free and serum-free DMEM medium, and then cultured in a triple-gas incubator (1% O2, 5% CO2, and 94% N2, by volume) at 37 °C for 24 h. The 6-well plates were co-cultured with hUC-MSCs, NC-MSCs, and EPO-MSCs transwell plates that were prepared 1 day in advance for 48 h. The 6-well plates were co-cultured with hUC-MSCs, NC-MSCs, and EPO-MSCs transwell plates laid out 1 day beforehand for 48 h. The cells and supernatants were collected and divided into the following five groups: SH-SY5Y group before hypoxia, SH-SY5Y group after hypoxia, hUC-MSCs co-culture group after hypoxia, NC-MSCs co-culture group after hypoxia, and EPO-MSCs co-culture group after hypoxia.
Cerebrospinal fluid and cellular supernatant cytokine level profiling
Expression levels of inflammatory factors in cerebrospinal fluid supernatants from HIE patients and control patients, as well as in cell supernatants before and after co-culture, were measured using the BioLegend® LEGENGplex™ Human Inflammation Panel 1 Assay Kit and analyzed according to the manufacturer’s instructions for the following 13 factors: IL-1β, MCP-1, IL-23, IL-17 A, IL-18, IFN-α2, IFN-γ, IL-6, IL-12p70, IL-8, IL-10, TNF-α, and IL-33. The experiment was repeated thrice to ensure loading criteria. Assays were performed on-line with a Cytoflex flow cytometer, and data were analyzed by the official online analysis tool and mass concentrations were specified in pg/mL.
RNA expression profiling by RNA sequencing
Transcriptome sequencing was conducted on SH-SY5Y samples, both treated and untreated with hUC-MSCs, NC-MSCs, and EPO-MSCs. Total RNA was extracted using the Trizol method, and its purity was assessed using a spectrophotometer. RNA integrity was evaluated through agarose gel electrophoresis and Agilent 2100 BioAnalyzer. The Illumina NEBNext® UltraTM RNA Library Prep Kit was employed for library construction. Subsequently, the Illumina platform was utilized for library sequencing, generating 150 bp paired terminal reads to obtain sequence information of the target fragments. After quality control and alignment to the reference genome, DESeq2 software was employed to analyze the differentially expressed genes (DEGs) between the two groups. Finally, the DEGs underwent gene enrichment analysis based on gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)19.
CUT&Tag assay
The experimental procedure was conducted following the instructions provided in the Novozymes Hyperactive Universal CUT&Tag Assay Kit for Illumina TD903 kit (Vazyme Biotech, #TD903)20. Nuclei were extracted from SH-SY5Y cells in the co-culture group of NC-MSCs and EPO-MSCs after hypoxia. The target DNA sequence was identified using a primary anti-H3K4me2 rabbit antibody and a secondary goat-anti-rabbit antibody. The target DNA sequence was captured using the pA-Tn5 transposon, and the purified products were amplified by PCR after fragmentation. The concentration of the amplified products was determined. The purified products were then subjected to PCR amplification, and their concentration was determined. Once the CUT&Tag library was successfully constructed, the Illumina HiSeq-X sequencer was employed for subsequent sequencing.
Integration of CUT&Tag and RNA-seq data
Differential analysis of H3K4me2 CUT&Tag data and RNA was performed using the “edgeR” package in the R software, to obtain a matrix of differential chromatin regions and differential gene expression, and then to integrate the target genes regulated by the differential chromatin states for visualization analysis. Target genes with attenuated silencer signal intensity and up-regulated expression and target genes with enhanced silencer signal intensity and down-regulated expression were counted and analyzed for Reactome, GO, and KEGG enrichment.
Western blotting
Proteins were extracted from samples using RIPA buffer containing proteinase inhibitors. The protein concentrations were determined using the BCA Protein Assay Kit (Beyotime, Shanghai, China) and separated by SDS-PAGE gel electrophoresis (Solarbio, Beijing). Subsequently, they were transferred onto a PVDF membrane. The membranes were then incubated overnight at 4 °C with anti-EPO, anti-REST, anti-Bcl-2, anti-H3K36me3, anti-β-actin, and anti-GAPDH antibodies, followed by incubation with secondary antibodies for 1 h at room temperature. The blots were visualized using an ECL chemiluminescence kit (enhanced), and the band intensity was quantified using ImageJ software.
qRT-PCR
RNA extraction was carried out using Trizol reagent (Invitrogen, USA) from EPO-MSCs or SH-SY5Y cells. Reverse transcription of each RNA sample was performed using the PrimeScript RT reagent kits (Takara, Japan) as per the manufacturer’s guidelines. For the PCR reaction, specific primers and SYBR Premix Ex Taq II (Takara, Japan) were utilized in a final volume of 20 µL. The PCR cycling conditions were as follows: pre-denaturation at 95 °C for 30 s, denaturation at 95 °C for 5 s, annealing at 60 °C for 30 s, 40 cycles, and elongation at 60 °C for 30 s. The relative gene expressions were analyzed using the 2 − ΔΔCT method with GAPDH serving as the internal control. Primer sequences are listed in Table 1.
Statistical analysis
Statistical analysis was conducted using SPSS software (version 20.0; IBM Corporation, Armonk, NY, USA). All values are presented as mean ± s.d.unless otherwise stated. Two independent samples t-tests were employed to analyze the data between two groups, while one-way analysis of variance (ANOVA) was utilized for comparisons among multiple groups. Statistical significance was defined as P < 0.05.
Results
EPO-MSCs were effectively engineered to preserve their inherent biological characteristics and secrete EPO into the extracellular milieu
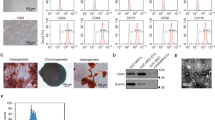
Under standard culture conditions, hUC-MSCs, NC-MSCs, and EPO-MSCs exhibited adherent growth in plastic bottles, displaying distinct cell morphology characterized by epithelioid or spindle arrangement within 48 h post lentiviral transfection. Notably, clear green fluorescence expression was observed under a fluorescence microscope, and the transfection efficiency was quantified using flow cytometry (Fig. 1A, B). Upon induction with a commercially available stem cell differentiation kit for osteoblasts, and adipoblasts, hUC-MSCs successfully differentiated into osteoblasts and adipocytes (Supplementary Fig. 1C, D). The expression of stem cell surface markers CD73, CD90, and CD105 were detected in 98.5%, 99.9%, and 97.6% of hUC-MSCs, respectively (Fig. 1e). Furthermore, compared to the control group, both protein expression and mRNA levels of erythropoietin (EPO) were significantly elevated (Fig. 1C, D). No significant differences were observed in terms of cell proliferation, apoptosis rate, and cell cycle among the hUC-MSCs, NC-MSCs, and EPO-MSCs (Supplementary Fig. 1A, B, E). Additionally, EPO levels in both the supernatant and cytosol were significantly increased after EPO overexpression (Fig. 1F, G).
Identification and transfection of EPO-MSCs. (a,b) The transfected EPO-MSCs and NC-MSCs show green fluorescence under a 10X microscope and flow cytometry for GFP expression efficiency. (c) Relative expression of EPO mRNA level in the hUC-MSCs group, the NC-MSCs group, and the EPO-MSCs group. (d) Immunoblotting analysis and semiquantitative analysis of EPO in the hUC-MSCs group, the NC-MSCs group, and the EPO-MSCs group. (e) Identification of specific antigenic markers in EPO-MSCs by flow cytometry. (f) EPO secretion from hUC-MSCs, NC-MSCs, and EPO-MSCs cell supernatants. (g) Intracellular EPO secretion in hUC-MSCs, NC-MSCs, and EPO-MSCs cells. Data are shown as the means + SDs (*p < 0.05, **p < 0.01, ***p < 0.001), Scale bar = 200 μm.
EPO-MSCs exert a protective effect on ischemic-hypoxic SH-SY5Y by releasing EPO and inhibiting the inflammatory environment partly through the release of MCP-1 and IL-6
To explore the protective mechanism of EPO-MSCs on neurons under OGD conditions, we constructed an in vitro OGD model using SH-SY5Y cells and co-cultured them with hUC-MSCs, NC-MSCs, and EPO-MSCs at the beginning of the reoxygenation period. Flow cytometry was used to detect cell apoptosis, and the results indicated that OGD induced the apoptosis of SH-SY5Y cells. After 48 h of co-culture with hUC-MSCs and NC-MSCs, SH-SY5Y cell apoptosis was significantly reduced (Fig. 2A, B), and further improved after 48 h of co-culture with EPO-MSCs.
EPO-MSCs are protective against ischemic hypoxia in SH-SY5Y cells. (a) Apoptosis in the Blank group, the Hypoxia group, the hUC-MSCs group, the NC-MSCs group, and the EPO-MSCs group. (b) Early, late, and total apoptosis. (c) Secretion of inflammatory factors in the cellular supernatant (pg/ml). (d) Expression of inflammatory factors in the cerebrospinal fluid supernatant of patients with HIE (pg/ml). Data are presented as means ± SDs (*p < 0.05, **p < 0.01, ***p < 0.001).
Inflammatory stimuli have been shown to induce cell apoptosis. Therefore, it is of interest to investigate the impact of EPO-MSCs on the inflammatory microenvironment. Therefore, we analyzed the secretion of inflammatory cytokines into the supernatant of all five groups using the bead-based LEGENDplex™ Human Inflammation Panel immunoassay. The results showed that the 24-h ischemia-hypoxia SH-SY5Y model group had significantly increased expression levels of MCP-1 and IL-6 in the cell supernatant compared with the control group before hypoxia (MCP-1: 711.24 ± 79.38 vs. 0.00 ± 0.00; IL-6: 482.43 ± 2.84 vs. 0.00 ± 0.00, P < 0.01). The expression levels of MCP-1 and IL-6 in the hUC-MSCs co-culture group were significantly reduced compared with the model group (MCP-1: 347.78 ± 6.54 vs. 711.24 ± 79.38; IL-6: 86.74 ± 4.92 vs. 482.43 ± 2.84, both P < 0.01). The expression levels of MCP-1 and IL-6 in the NC-MSC co-culture group and hUC-MSCs co-culture group did not significantly differ (P > 0.05). The MCP-1 and IL-6 expression levels were significantly reduced in the EPO-MSCs co-culture group compared with the NC-MSCs co-culture group (MCP-1: 144.38 ± 9.21 vs. 332.76 ± 9.07; IL-6: 24.61 ± 1.32 vs. 95.23 ± 3.10, all P < 0.01) (Fig. 2C). These results suggested that EPO-MSCs improved the inflammatory microenvironment by down-regulating the release of MCP-1 and IL-6 in the ischemic-hypoxic SH-SY5Y model.
Hypoxic-ischemic encephalopathy is associated with neuronal death that occurs through multiple mechanisms such as oxidative stress, inflammation, apoptosis, and excitotoxicity. Furthermore, to explore the expression level of inflammatory factors in HIE, we utilized the LEGENGplex™ multifactor flow assay to assess the secretion of inflammatory factors in the cerebrospinal fluid supernatant. Our findings revealed significantly elevated levels of HIE inflammation compared to the control group (P < 0.01, Fig. 2D). IL-8 expression levels in the HIE group were higher than those in the control group (P < 0.05, Fig. 2d). IL-10, INF-γ, IL-12p70, IL-33, and IL-17 A were not statistically significant in the HIE and control groups (P > 0.05, Supplementary Fig. 2A).
EPO-MSCs exert neuroprotective effects through epigenetic regulation
To investigate the specific mechanism of apoptosis inhibition and regulation of MCP-1 and IL-6 in EPO-MSCs, we performed transcriptome sequencing in three groups: SH-SY5Y, NC-MSCs, and EPO-MSCs. A total of 9,160 different genes were identified in the NC-MSCs group compared to the SH-SY5Y group. Among these genes, 4351 were up-regulated and 4,809 were down-regulated (Supplementary Fig. 3A, B, C, D). The results of the NC-MSCs group suggest that apoptosis of ischemic-hypoxic SY5Y cells was reduced, and the KEGG pathway was enriched and aggregated in the MAPK pathway leading to reduced apoptosis (Supplementary Fig. 3E), suggesting that hUC-MSCs have a neuroprotective function, which is in line with previous reports7.
RNA-seq analysis identified 405 differentially expressed genes (DEGs), among which 288 genes were up-regulated and 117 genes were down-regulated in the EPO-MSCs group compared to the NC-MSCs group (Fig. 3A, B). Seven of the genes screened for significant down-regulation were associated with inflammation (Table 2). GO functional enrichment analysis revealed the main enrichment of the DEGs in “histone H3-K4 monomethylation,” “transcription regulator complex,” “MAP kinase activity,” “DNA-binding transcription factor activity,” and “histone methyltransferase activity” items (Fig. 3C), this suggests that epigenetic regulation maybe very important in the neuroprotective effects of EPO-MSCs. Additionally, the differentially expressed genes between the two groups were mainly enriched in the Wnt signaling and MAPK signaling pathways (Fig. 3D).
Combined RNA-seq and CUT&Tag analysis reveals the involvement of REST in epigenetic remodeling functions
As a context-dependent epigenetic mark, H3K4me2 can act as an activating mark. Therefore, we used CUT&Tag to detect genomic H3K4me2 modifications, analyzed the possible transcriptional activation genes, and explored the neuroprotective mechanism of EPO-MSCs. Principal component analysis and Spearman correlation analysis showed a highly significant correlation between the three biological replicates of EPO-MSCs treatment and control groups (Fig. 4A, B). After co-cultured with EPO-MSCs, the number of peaks for H3K4me2 changed dramatically, with 29,884 lost peaks, 9,308 shared peaks, and 10,442 gained peaks (Fig. 4C). CUT&Tag signal heatmaps showed intense peak signals for H3K4me3 in the vicinity of transcription start sites within a 3Kb region, and the peak signals decreased significantly after co-cultured with EPO-MSCs (Fig. 4D). Genomic annotation results showed that the lost CUT&Tag peaks for H3K4me2were mainly enriched in promoter regions (Fig. 4E). Furthermore, bioinformatics analysis found abundant H3K4me2 signals in the promoter region of REST and TET3, suggesting that REST and TET3 might be regulated by histone modification (Table 3). All differentially expressed genes were analyzed using the Reactome Pathway database (REACTOME) for enrichment analysis. The results suggest that regulatory regions are highly enriched in transcriptional regulation, as well as in chromatin organization and chromatin-modifying enzymes (Fig. 4F).
Combined RNA-seq and CUT&tag analysis: (a,b) Principal Component Analysis (PCA) diagram. (c) Venn diagram. (d) A heatmap was generated to visualize the enrichment of H3K4me2 CUT&tag peaks in proximity to gene transcription start sites (TSS) and transcription end sites (TES). (e) Distribution of H3K4me2 on the genome. (f) Reactome enrichment analysis.
REST deficiency leads to increased neuronal apoptosis and downregulation of H3K36me3
REST, a transcriptional repressor, plays a significant role in regulating neural development and differentiation. To elucidate the neuroprotective mechanism of EPO-MSCs by regulating REST, SH-SY5Y cells were stably transfected with a lentiviral vector for REST knockdown or the corresponding empty vector control. After 10 days, approximately 99.24% and 99.33% of REST lentivirus-transfected SH-SY5Y cells were GFP + as shown by flow cytometry analysis, suggesting high transduction efficiency (Fig. 5A, B). Quantitative PCR and western blot analyses showed that REST mRNA and protein levels were markedly inhibited after infection in SH-SY5Y cells (Fig. 5C, D).
REST deficiency leads to increased neuronal apoptosis and downregulation of H3K36me3. (a,b) The transfected pLKO.1-sh1 and pLKO.1-sh2 show green fluorescence under a 10× microscope and flow cytometry for GFP expression efficiency. (c) Relative expression of REST mRNA level in the hUC-MSCs group, the pLKO.1-sh1 group, and the pLKO.1-sh2 group. (d) Immunoblotting analysis and semiquantitative analysis of REST in the pLKO.1-sh1 group and the pLKO.1-sh2 group. (e) Apoptosis in the pLKO.1-sh1 group and the pLKO.1-sh2 group. (f) Early, late, and total apoptosis. (g,h) Immunoblotting analysis and semiquantitative analysis of H3K36me3 in the pLKO.1-sh1 group and the pLKO.1-sh2 group. Data are shown as the means + SDs (*p < 0.05, **p < 0.01, ***p < 0.001).
To explore the mechanism of the apoptosis-protective effects of the transcription factor REST in SH-SY5Y cells with ischemic-hypoxic injury, cells from the pLKO.1-Vector and pLKO.1-sh1 groups were OGD for 24 h and then co-cultured with EPO-MSCs for 48 h. At 48 h, the early apoptosis rate, late apoptosis rate, and total apoptosis rate of the pLKO.1-sh1 group were significantly increased compared to the control group (P < 0.01, Fig. 5E, F). The expression level of H3K36me3 was decreased in the pLKO.1-sh1 group compared with the pLKO.1-Vector group, also shown by the western blotting results (P < 0.01, Fig. 5G, H).
The gene expression profiles of the SH-SY5Y group with knockdown of REST after ischemic hypoxia treatment and the SH-SY5Y group co-cultured with EPO-MSCs were analyzed by transcriptome sequencing
The RNA-seq results demonstrated that there were 95 genes that exhibited differential expression between the REST knockdown and control groups. Out of these genes, 58 were down-regulated, while 37 were up-regulated (Fig. 6A, B). KEGG enrichment analysis indicated a significant up-regulation in the biological function of apoptotic signaling pathways (Fig. 6C), with only one gene BCL2A1 related to the pathway, a member of the BCL-2 family (Fig. 6D). Reactome pathway analysis also indicated a significant down-regulation in sevral erythropoietin pathway (Fig. 6E), such as erythropoietin activates phospholipase C gamma (PLCG), erythrocytes take up carbon dioxide and release oxygen, signaling by erythropoietin, with only one gene PLCG2 related to these pathways, which has been proven to be a key downstream gene of EPO, and it is related to the occurrence of inflammation (Fig. 6F). At the same time, we also noticed that the expression of SETD2, the only methyltransferase that catalyzes the formation of the H3K36me3 modificationwas, was significantly downregulated in the absence of REST (Fig. 6G), these results of three genes including BCL2A1, PLCG2 and SED2 were also validated by qRT-PCR (Fig. 6h, i, j). This suggested that REST mediates the targeting of TET3 to DNA, and subsequently SETD2 mediates the production of H3K36me3, which leads to chromatin remodelling and transcriptional activation of neuronal genes.
Transcriptome sequencing results analysis. (a) Principal component analysis (PCA) diagram. (b) Volcano map of pLKO.1-Vector, pLKO.1-sh2 differentially expressed genes. (c) KEGG function enrichment analysis. (d) TPM(Transcripts Per Million) values for BCL2A1 of the pLKO.1-Vector, pLKO.1-sh1 groups. (e) Reactome pathway analysis. (f) TPM values for PLCG2 of the pLKO.1-Vector, pLKO.1-sh1 groups. (g) TPM values for SETD2 of the pLKO.1-Vector, pLKO.1-sh1 groups. The data presented in this study are displayed as means with standard deviations (SDs). (h–j) Quantitative analysis of the relative mRNA expression level of BCL2A1, PLCG2 and SETD2 from three independent experiments. Statistical significance is indicated by asterisks, with * representing p < 0.05, ** representing p < 0.01, and *** representing p < 0.001.
Discussion
hUC-MSCs and EPO have each demonstrated short-term benefits with delayed administration, and combination therapy may provide the most benefit. hUC-MSCs have the characteristics of immune regulation and homing20. Engineering these cells or vesicles through different technical means such as genetic engineering, surface modification, and tissue engineering can enhance their homing abilities and other inherent characteristics. This enables them to specifically target particular tissues or organs, thereby improving their therapeutic efficacy21. In this study, we successfully generated umbilical cord-derived MSCs that overexpress EPO. These EPO-MSCs exhibit the characteristic properties of stem cells. We utilized SH-SY5Y cells to establish an in vitro model of OGD cell injury, aiming to mimic HIE. Apoptosis was significantly reduced in the OGD-treated SH-SY5Y cells co-cultured with EPO-MSCs compared to the co-cultured group with NC-MSCs as well as hUC-MSCs. However, the exact mechanism underlying this phenomenon is still not completely understood.
To investigate the mechanism by which apoptosis is inhibited in EPO-MSCs, this study conducted transcriptome sequencing of SH-SY5Y cells after 24 h of OGD, as well as after co-cultivation with NC-MSCs and EPO-MSCs. The results revealed a significant down-regulation of several genes associated with the onset of inflammation, such as AMH, HSF4, PDIA2, and MIR4435-2HG in the EPO-MSCs group compared to the NC-MSCs group. Previous studies have demonstrated that hypoxic-ischemic injury induces neuronal cell death through increased apoptosis and triggers an inflammatory response, resulting in neuronal loss and axonal degeneration22. Inflammatory factors such as IL-1β, TNF-α, IL-6, IL-8, IL-10, and MCP-1 are significantly upregulated in brain tissue following ischemic-hypoxic brain injury23. The release of danger-associated molecular pattern molecules (DAMPs) from neurons activates M1-type macrophages, leading to the generation of reactive oxygen species and pro-inflammatory cytokines24. Activated microglia secrete numerous pro-inflammatory factors, contributing to neuronal cell death25.
In this study, we systematically examined the secretion of 13 inflammatory factors in the supernatant of cerebrospinal fluid of patients with HIE by LEGENDplex™ multifactor flow assay and found that IL-1β, IL-6, IFN-α2, MCP-1, IL-8, IL-18, and IL-23 production were significantly increased in the HIE group compared with the control group. Among them, the elevation of MCP-1 and IL-6 were involved in the inflammatory response to neurological damage26. It has been found that under normal conditions, immune cells in the CNS are mostly in a quiescent state and do not express MCP-1. However, when pathogens invade the CNS, several neural activation and immune cells are activated, resulting in the secretion and production of MCP-127. The increase in MCP-1 further exacerbates inflammatory responses and increases the occurrence of a variety of encephalopathies such as hydrocephalus27. The chemokine MCP-1 and its receptor CCR2 play an important role in mediating the p38 MAPK signaling pathway, leading to the disruption of the BBB28. IL-6 acts as an activation signal for NF-κB. NF-κB, as a eukaryotic transcription-associated factor, belongs to the convergence point of the signaling pathway and is capable of modulating the level of expression of many genes, especially inflammation-related genes. In particular, NF-κB has a key role in the expression of inflammation-related genes29,30.
In this study, the LEGENDplex™ multifactor flow assay was utilized to analyze the cell supernatants before and after co-culture. The results revealed that the group treated with EPO-MSCs exhibited a significant reduction in the expression levels of MCP-1 and IL-6 in ischemic-hypoxic SH-SY5Y cells.
To investigate the specific mechanism by which EPO-MSCs regulate MCP-1 and IL-6, this study conducted transcriptome sequencing and analyzed GO functional enrichment clustering. The results revealed the presence of biological process enrichment analysis for H3K4 methylation and post-transcriptional regulation of genes. H3K4me2 serves as an epigenetic regulatory signal for the transcription activation, indicating dynamic changes in nucleosome positioning at the genomic level. Specifically, it can indicate the translocation of nucleosomes that occur in response to external stimuli. When nucleosomes are displaced in regions where transcription factors bind, the genomic DNA becomes exposed, leading to increased accessibility of chromatin and facilitating the binding of transcription factors31. These relocated nucleosomes move upstream and downstream of the binding region, resulting in an increased density of nucleosomes on the flanking side of the wing. Therefore, the regions with enhanced nucleosome signals on both sides and decreased nucleosome signals in the middle can be better identified by labeling H3K4me232. It has been demonstrated that the occupancy of H3K4me2 and the expression levels of genes in close proximity to REST-associated Donana Biological Reserve are augmented following treatment with EPO. REST serves as a downstream mediator of EPO signaling33. In this study, the target molecule selected was H3K4me2, and the CUT&Tag technique was employed to detect genomic modifications of H3K4me234. The results revealed that H3K4me2 specifically binds to the promoter regions of the REST and TET3 genes. REST, a highly expressed epigenetic modifier in the developing nervous system, plays a crucial role in regulating the maturation, function, and survival of neuronal cells17. Increased expression of REST is neuroprotective35,36, and TET3 is a direct downstream target gene of REST37.
A lentivirus was used to construct an SH-SY5Y cell line with REST knocked down to study its specific mechanism. The results showed a significant increase in the rate of apoptosis following the knockdown of REST, when compared to the control group. It has been shown that REST mediates the targeting of TET3 to DNA, which promotes the generation of 5hmC, and subsequently NSD3 mediates the production of H3K36me3, which leads to chromatin remodelling and transcriptional activation of neuronal genes37,38. Through transcriptome sequencing, this study further revealed the mechanisms involved and found that the expression of the apoptosis gene BCL2A1 was up-regulated, this is consistent with the results of our co cultivation. We also found PLCG2, a gene that responds to EPO signal stimulation, among significantly downregulated genes, Accumulating evidence suggests that the modulation of the TREM2/PLCG2 signaling pathway is a potential therapeutic candidate for AD(Alzheimer’s disease)39, here we demonstrate for the first time that PLCG2 is directly regulated by REST to protect neurons from apoptosis.
Conclusions
EPO-MSCs may promote neuronal survival by affecting H3K4me2 and thus activating the expression of REST and TET3. EPO-MSCs also upregulated the modification level of SETD2-mediated H3K36me3 and regulated the expression of inflammation-related genes such as PLCG2, as well as apoptosis genes BCL2A1. This study presents a theoretical framework supporting the potential use of EPO-MSCs for the treatment of HIE. However, the number of cases in the control group and the number of cerebrospinal fluid samples of HIE patients in this study is relatively small, which were likely also accompanied by differences in cerebrospinal fluid collection procedures and the kits used. The SH-SY5Y cell line cannot replace the complex immune microenvironment of the human body, and the fewer inflammatory factors detected are a major limitation for reproducing the disease model. Therefore, the quality and accuracy of the data profile may impact the results.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Wang, W. & Jia, L. Regulatory mechanism of microRNA-30b on neonatal hypoxic-ischemic encephalopathy (HIE). J. Stroke Cerebrovasc. Dis. 30(3), 105553 (2021).
Chen, H. R. et al. Monocytes promote acute neuroinflammation and become pathological microglia in neonatal hypoxic-ischemic brain injury. Theranostics 12(2), 512–529 (2022).
Dixon, B. J. et al. Intranasal administration of interferon beta attenuates neuronal apoptosis via the JAK1/STAT3/BCL-2 pathway in a rat model of neonatal hypoxic-ischemic encephalopathy. ASN Neuro 8, 5 (2016).
Du, T. et al. Cerebrospinal fluid is a significant fluid source for anoxic cerebral oedema. Brain 145, 787 (2021).
Hu, X. et al. Rh-CSF1 attenuates oxidative stress and neuronal apoptosis via the CSF1R/PLCG2/PKA/UCP2 signaling pathway in a rat model of neonatal HIE. Oxid. Med. Cell. Longev. 1, 1 (2020).
Massaro, A. N. et al. Short-term outcomes after perinatal hypoxic ischemic encephalopathy: a report from the children’s hospitals neonatal Consortium HIE focus group. J. Perinatol. 35, 4 (2016).
Li, F. et al. The neuroprotective effect of mesenchymal stem cells is mediated through inhibition of apoptosis in hypoxic ischemic injury. World J. Pediatr. 16, 193 (2020).
Choi, J. W., Kang, S. J., Choi, J. I., Kwack, K. & Kim, M. Role of nuclear-receptor-related 1 in the synergistic neuroprotective effect of umbilical cord blood and erythropoietin combination therapy in hypoxic ischemic encephalopathy. Int. J. Mol. Sci. 23, 5 (2022).
Frenette, P. S. et al. Mesenchymal stem cell: keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annu. Rev. Immunol. 31(1), 285–316 (2013).
Soon, P. W. et al. Hypothermia augments neuroprotective activity of mesenchymal stem cells for neonatal hypoxic-ischemic encephalopathy. PLoS ONE 10(3), e0120893 (2015).
Nassar, W. et al. Umbilical cord mesenchymal stem cells derived extracellular vesicles can safely ameliorate the progression of chronic kidney diseases. Biomater. Res. 20, 21 (2016).
Lauria, G. et al. Erythropoietin in amyotrophic lateral sclerosis: a multicentre, randomised, double blind, placebo controlled, phase III study. J. Neurol. Neurosurg. Psychiatry 86(8), 879–886 (2015).
Luo, B. et al. Phagocyte respiratory burst activates macrophage erythropoietin signalling to promote acute inflammation resolution. Nat. Commun. 7, 12177 (2016).
Urrutia, A. A. et al. Prolyl-4-hydroxylase 2 and 3 coregulate murine erythropoietin in brain pericytes. Blood 128(21), 2550–2560 (2016).
Agostini, F. et al. Nucleofection of adipose mesenchymal stem/stromal cells: improved transfection efficiency for GMP grade applications. Cells 10, 12 (2021).
Perlikowska, R., Silva, J., Alves, C., Susano, P. & Pedrosa, R. The therapeutic potential of naturally occurring peptides in counteracting SH-SY5Y cells injury. Int. J. Mol. Sci. 23, 19 (2022).
Lin, F. L. et al. HADC8 inhibitor WK2-16 therapeutically targets lipopolysaccharide-induced mouse model of neuroinflammation and microglial activation. Int. J. Mol. Sci. 20(2), 410 (2019).
Li, X. et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 20(2), 259–276 (2008).
Kanehisa, M. & Goto, S. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Yang, J. et al. Tropoelastin improves adhesion and migration of intra-articular injected infrapatellar fat pad MSCs and reduces osteoarthritis progression. Bioact Mater. 10, 443–459 (2022).
Al-Amoodi, A. S. et al. Refining the migration and engraftment of short-term and long-term HSCs by enhancing homing-specific adhesion mechanisms. Blood Adv. 6(15), 4373–4391 (2022).
Biouss, G. et al. Experimental necrotizing enterocolitis induces neuroinflammation in the neonatal brain. J. Neuroinflamm. 16(1), 97 (2019).
Arumugam, T. V. et al. Age and energy intake interact to modify cell stress pathways and stroke outcome. Ann. Neurol. 67(1), 41–52 (2010).
Guo, A. et al. Inhibition of connexin hemichannels alleviates neuroinflammation and hyperexcitability in temporal lobe epilepsy. Proc. Natl. Acad. Sci. U.S.A. 119(45), e2213162119 (2022).
Gupta, D. P., Park, S. H., Yang, H. J., Suk, K. & Song, G. J. Neuroprotective and anti-neuroinflammatory effects of a poisonous plant Croton tiglium Linn. extract. Toxins 12(4), 261 (2020).
Kumar, H., Kawai, T. & Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16 (2011).
Singh, S., Dixit, A. & Ravichandiran, V. MCP-1: function, regulation, and involvement in disease. Int. Immunopharmacol. 101, 107598 (2021).
Chen, Z. et al. Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 77(9), 2266–2278 (2017).
Kolattukudy, P. E. & Niu, J. Inflammation, endoplasmic reticulum stress, autophagy, and the monocyte chemoattractant protein-1/CCR2 pathway. Circ. Res. 110(1), 174–189 (2012).
Huang, X. Y. et al. Everolimus inhibits PI3K/Akt/mTOR and NF-kB/IL-6 signaling and protects seizure-induced brain injury in rats. J. Chem. Neuroanat. 114, 101960 (2021).
Nie, Y., Shu, C. & Sun, X. Cooperative binding of transcription factors in the human genome. Genomics 112(5), 3427–3434 (2020).
Wang, S., Meyer, D. H. & Schumacher, B. H3K4me2 regulates the recovery of protein biosynthesis and homeostasis following DNA damage. Nat. Struct. Mol. Biol. 27(12), 1165–1177 (2020).
Sollinger, C. et al. Erythropoietin signaling regulates key epigenetic and transcription networks in fetal neural progenitor cells. Sci. Rep. 7(1), 14381 (2017).
Kaya-Okur et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10(1), 1930 (2019).
Ryan, B. J. et al. REST protects dopaminergic neurons from mitochondrial and α-synuclein oligomer pathology in an alpha synuclein overexpressing BAC-transgenic mouse model. J. Neurosci. 41(16), 3731–3746 (2021).
Lam, X. J., Maniam, S., Cheah, P. S. & Ling, K. H. REST in the road map of brain development. Cell. Mol. Neurobiol. 43, 3417 (2023).
Perera, A. et al. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell. Rep. 11(2), 283–294 (2015).
Sun, Z. et al. H3K36me3, message from chromatin to DNA damage repair. Cell. Biosci. 10, 9 (2020).
Tsai, A. P. et al. PLCG2 is associated with the inflammatory response and is induced by amyloid plaques in Alzheimer’s disease. Genome Med. 14, 17 (2022).
Acknowledgements
We express our gratitude to Professor Jianping Ye for his invaluable guidance throughout our study. We would like to acknowledge the Branch Center of Advanced Medical Research Center, Zhengzhou Central Hospital Affiliated to Zhengzhou University for their support.
Funding
This work was supported by the Henan Province Science and Technology Research and Development Projects (Grant Number LHGJ20191045, LHGJ20220858), the Henan Province Science and Technology Research and Development Projects (Grant Number 222102310032), and the Health Commission of Henan Province (Grant Number SYJS2022150).
Author information
Authors and Affiliations
Contributions
Ruibo Li was responsible for conception and design, collection and assembly of data, and data analysis and writing the manuscript. Yu Jiang was responsible for writing the manuscript. Lei Sun was responsible for conception and design. Ning Kong and Jixiang Tang was responsible for perform Quantitative qRT-PCR and Western bloting assays. Baodong Ma, Ranran Jin and Yiming Shao was responsible for collection data and CUT&TAG assays support. Yueyao Ban and Wenjin Zhang was responsible to collect the information of patients and data analysis. Hui Zhang and Han Yue was responsible for conception and design, data analysis and interpretation, funding acquisition and final approval of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was reviewed and approved by Ethics Committee of Zhengzhou Central Hospital Affiliated to Zhengzhou University (Ethics No. 202171).
Consent for publication
All authors provided their explicit consent for the publication of this research paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, Y., Li, R., Ban, Y. et al. EPO modified MSCs protects SH-SY5Y cells against ischemia/hypoxia-induced apoptosis via REST-dependent epigenetic remodeling. Sci Rep 14, 23252 (2024). https://doi.org/10.1038/s41598-024-74261-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-74261-3