Abstract
Cervical cancer (CC) was considered to be the most common gynaecological cancer, with an estimated 342,000 deaths worldwide each year, as the majority of patients were diagnosed at an advanced stage of the disease. The purpose of this study was to evaluate the predictive value of multi-locus methylation assay for the early detection of CC. The cervical exfoliated cell samples from 492 HPV-positive females with cervical lesions were collected and subjected to methylation detection of gene FAM19A4, EPB41L3 and PAX1 after bisulfite conversion. The levels of gene methylation in patients with different severity of cervical lesions were evaluated and compared. The receiver-operating characteristic (ROC) curve was established and efficacy indexes such as sensitivity, specificity and area under the curve (AUC) were calculated to assess the diagnostic value of DNA methylation detection at multiple gene loci for CC. The methylation levels of FAM19A4, EPB41L3 and PAX1 were significantly increased with the grade of cervical squamous intraepithelial lesions. The sensitivities of FAM19A4, EPB41L3 and PAX1 alone for high-grade squamous intraepithelial lesion (HSIL) and CC diagnosis were 84.6%, 86.3% and 88.0%, respectively; when three markers were combined by a logistic regression model, the sensitivity was 88.0%, with a high specificity of 97.7% and AUC of 0.957 (95% CI 0.937–0.977). Methylation status of FAM19A4, EPB41L3 and PAX1 were highly specific and effective for monitoring the progression of cervical lesions and the tri-gene methylation assay could be used as a triage tool for CC early screening.
Similar content being viewed by others
Introduction
Over the past few decades, cervical cancer (CC), with the increasing morbidity, has been seriously harming human health and becomes a leading cause of female death worldwide in modern society1. The persistent infection of high-risk human papillomavirus (hrHPV) is an important factor in the development and progression of CC, and almost all patients with CC are HPV-positive2. Currently, cervical liquid-based cytology examination and hrHPV testing have been proposed as the main CC screening methods. However, the sensitivity and specificity of these methods are limited, leading to an increase in colposcopy referrals, thereby increasing the physical and mental burden on women3,4. Therefore, a cost-benefical screening strategy with high accuracy and feasibility is urgently needed to predict the occurrence of CC in high-risk patients.
Recent studies have shown that genomic and epigenetic abnormalities are present in the early stages of tumorigenesis. DNA methylation is an epigenetic mechanism that results in heritable silence of genes without changes to their coding sequences5. The occurrence of a variety of tumors is associated with methylation changes in genomic DNA, and these tumor-specific changes are often detected at an early stage. It has been proved that hypermethylation modification of the promoter regions of the tumor suppressor genes is positively correlated with the degree of cervical lesions, making DNA methylation detection a promising tool for CC diagnosis6,7.
The methylation status of cytosine (C) in CpG dinucleotides in the promoter regions of human genes such as FAM19A4, EPB41L3 or PAX1 has been found to be significantly different in cancerous cervical exfoliated cells, compared to normal cervical exfoliated cells8,9,10. However, previous studies usually had small sample sizes and mostly focused on a single gene, resulting in low sensitivity and specificity for diagnosing CC. In a previous background study, we collected and analyzed a large number of 450k methylation array data from public databases, including 259 tissue samples with different degrees of cervical lesions, and finally screened genes of FAM19A4, EPB41L3 and PAX1 by differences in methylation levels and disease correlation. In addition, we collected 53 clinical samples for 450k methylation array testing and validation. This exploratory study aimed to develop a methylation-detection panel containing three genes (FAM19A4, EPB41L3 and PAX1) and investigate the predictive value of polygenic methylation assay in CC screening, so as to provide more references for early diagnosis and treatment.
Materials and methods
Study participants and samples
The cervical exfoliated cell samples from participants used in the experiment were all from Obstetrics and Gynecology Hospital Affiliated to Fudan University, Shanghai, China from March 2023 to May 2023, with a total of 492 consecutive cases. According to the WHO 2020 pathological classification criteria for CC and 2020 American Cancer Society guidelines, the clinical samples used in the trial were divided into control group (151 cases), low-grade squamous intraepithelial lesion (LSIL) group (107 cases), high-grade squamous intraepithelial lesion (HSIL) group (121 cases), and CC group (113 cases). The control group was diagnosed with a normal or inflammatory cervix by HPV screening and thinprep cytologic test (TCT). The histopathological report was verified by two professional pathologists with associate senior titles and above. Specimens are collected by a stationary physician and baseline clinical characteristics were obtained from all participants at the time of admission. This study complied with the Declaration of Helsinki and was approved by the Institutional Ethics Committee of Obstetrics and Gynecology Hospital Affiliated to Fudan University. All participants provided written informed consent.
Inclusion criteria for all subjects were as follows: (a) Patients with HPV infection who were not pregnant and had not received vaginal medication or vaginal irrigation within 7 days prior to visit; (b) Patients with cervical inflammation or intraepithelial neoplasia or CC; (c) Patients and their families agreed to participate in the study and signed an informed consent. Exclusion criteria for all subjects were as follows: (a) Patients with a history of pelvic chemoradiotherapy; (b) Patients with a history of cervical and uterine surgery; (c) Patients treated with antibiotics in the last 1 week; (d) Patients who were menstruating at the time of enrollment; (e) Patients with HPV infection and vaginal lesions but no cervical inflammation.
DNA extraction and bisulfite transformation
Samples were collected using a disposable cervical sampling swab and stored in cell preservation solution (PreservCyt Solution, Hologic, National Machinery: 20140197) at − 80℃. Genomic DNA extraction was performed from cervical exfoliated cells using UE Genomic DNA Miniprep Kit (Suzhou Youyi Landi Biotechnology, catalog number: UE-MN-MS-GDNA-250 ). 40–1000 ng of genomic DNA was taken and transformed into bisulfite-converted DNA (bisDNA) according to the instructions of the recommended bisulfite conversion kit EZ DNA Methylation-Gold (ZYMO RESEARCH, catalog number: D5006), and finally bisDNA was eluted with 20 µl of ultrapure water and stored at − 20 ℃ until analysis. The gene methylation assay, as previously described11, firstly used bisulfite to treat CpG dinucleotides on genomic DNA, and bisulfite treatment would produce different transformation effects on CpG dinucleotides in different methylation states: if the cytosine (C) in CpG dinucleotide was methylated, it would remain unchanged after treatment; If the cytosine (C) in the CpG dinucleotide was not methylated, it would be converted to uracil (U) after treatment. Subsequently, the methylation status was quantitatively analyzed using the fluorescent probes that specifically identified untransformed CpG dinucleotides.
Methylation-specific PCR (MSP) analysis
The bisDNA from the previous step was used as a template for methylation-specific PCR (MSP) amplification using Hongshi PCR Analysis System (SLAN-96 S), and the specific primers could only amplify the methylated genes, so as to detect the methylation status of the three genes FAM19A4, EPB41L3 and PAX1. ACTB was set as the internal reference gene to monitor the extraction effect of each sample and the amplification of the PCR reaction, in order to effectively avoid false negatives. When the Ct value of the ACTB gene was ≤ 32.0, the sample test result was valid. The Ct value was defined as 45.0 when the gene was not amplified. Calculated the ΔCt value for each target gene, where the target gene ΔCt = Ct (FAM19A4/EPB41L3/PAX1)-Ct (ACTB). MSP program setting were as follows: 94℃ for 30 s in stage 1, 94 ℃ for 5 s and 60 ℃ for 30 s in stage 2 (stage 2 for 45 cycles). A blank control and a positive control were included in each assay.
The primers and probes were designed by us and not derived from other publications or patents. The primers and probes were both synthesized by Shanghai Sangon Bioengineering and these specific sequences were summarized in Table 1. All primers were lyophilized powder and dissolved in TE buffer at PH8.0. PAGE electrophoresis did not contain miscellaneous bands. Determined with a visible-ultraviolet spectrophotometer, the OD260 nm/OD280 nm was between 1.6 and 2.0. All probes were lyophilized powder and stored in brown tubes, with a purity of HPLC grade. The probes were also dissolved with TE buffer at PH8.0. Visible-ultraviolet spectrophotometer determined that OD260nm/OD280nm was between 1.6 and 2.0. The probes had a specific absorption peak at the fluorescein excitation wavelength of 494 nm (FAM fluorophore) or 576 nm (ROX fluorophore). We selected PerfectStart™ II Probe qPCR SuperMix (TransGen Biotech, catalog number: AQ711) with the best amplification efficiency and reaction stability as the raw material for the MSP reaction.
Statistic analysis
All acquired data were analyzed and visualized using SPSS 17.0 and GraphPad Prism 8 software. For continuous variables, normal distribution data presented as means ± standard deviation were compared using t-test between two groups or one-way analysis of variance (ANOVA) among multiple groups. Skewed distribution data expressed as median value (interquartile interval) were analyzed using the Mann-Whitney U test between two groups or Kruskal–Wallis H test among multiple groups. Categorical variables were expressed as percentage and evaluated using Fisher’s exact test. Receiver-operating characteristic (ROC) curves were established and logistic regression analysis was performed to assess the clinical predictive value of polygenic methylation assay for HSIL and CC. A two-tailed p value < 0.05 was considered to be statistically significant.
The ΔCt data of each target gene were analyzed separately using SPSS 17.0, and the software automatically generated a ROC curve. According to the coordinate point data from SPSS 17.0, excel was used to calculate “sensitivity + specificity”, and the point with the largest value was the optimal cut-off value.
Results
Characteristics of study participants
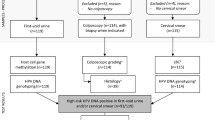
According to the inclusion criteria, a total of 505 subjects were selected, and according to the exclusion criteria, 492 subjects were finally recruited. They were divided into four groups according to the results of pathological tests from the hospital: control group (151 cases), LSIL group (107 cases), HSIL group (121 cases) and CC group (113 cases) (Fig. 1). The age distribution of the four groups was normal and there were no significant differences in these groups (P > 0.05, Table 2). The age range for the control group was 20–62 years old (38.00 ± 15.36). For the LSIL group, the age range was 31–64 years old (40.25 ± 12.28). The HSIL group ranged in age from 26 to 74 years old (41.88 ± 11.64), while the CC group ranged in age from 28 to 78 years old (40.50 ± 12.47). It could be concluded that the incidence of cervical intraepithelial neoplasia and CC is tending towards a younger age group, with the majority of each group being under 35 years old. Obviously, both the HSIL and CC groups had a higher proportion of hrHPV16/18 infection compared to the control and LSIL groups (p < 0.05, Table 2).
Relationship between gene methylation and cervical lesions
The methylation degree of FAM19A4 (Fig. 2A), EPB41L3 (Fig. 2B) or PAX1 (Fig. 2C) was further compared in the four groups. According to the principle of MSP, a smaller ΔCt value usually indicated a higher methylation level of the target gene. As shown in Fig. 2, the methylation levels of all three genes were significantly higher in the HSIL and CC groups than in the control and LSIL groups (p < 0.05), and the CC group had higher gene methylation degree compared to the HSIL group (p < 0.05). However, it showed no significant difference between control group and LSIL group (p > 0.05). It could be seen that the higher the methylation level of the target gene, the greater the risk of cervical malignancy, especially HSIL and CC, indicating a positive correlation between the degree of gene methylation and the severity of cervical lesions. In particular, the CC group included two sample types, squamous cell carcinoma (SCC) and adenocarcinoma (AC), and there was no significant difference in the degree of methylation of the three genes between the two cancer types, as shown in supplementary Fig. 1 (p > 0.05).
Diagnostic potential of target gene methylation for HSIL and CC
ROC analysis was conducted to evaluate the predictive value of gene methylation levels including FAM19A4, EPB41L3 and PAX1 to diagnose HSIL and CC patients. To differentiate the HSIL + CC cases from the control + LSIL cases (Fig. 3B), cut-off value of ΔCt was 10.445 (sensitivity, 84.6%; specificity, 96.1%) for FAM19A4, 9.08 (sensitivity, 86.3%; specificity, 95.3%) for EPB41L3, and 10.015 (sensitivity, 88.0%; specificity, 97.7%) for PAX1. To differentiate the CC cases from the HSIL cases (Fig. 3C), cut-off value of ΔCt was 5.375 (sensitivity, 68.1%; specificity, 66.9%) for FAM19A4, 2.665 (sensitivity, 54.9%; specificity, 81.8%) for EPB41L3, and 4.955 (sensitivity, 81.4%; specificity, 58.7%) for PAX1. Consistent with the previous result that there was no significant difference in gene methylation degree between control group and LSIL group, it was also of low value in the diagnosis of LSIL (Fig. 3A, p > 0.05). Meanwhile, it could be seen that the methylation levels of the three genes had higher diagnostic value for HSIL + CC (Fig. 3B), but their specificity and sensitivity in differentiating HSIL from CC were relatively low (Fig. 3C). All area under the curve (AUC) and p values were shown in Fig. 3D.
ROC curve of FAM19A4, EPB41L3 and PAX1 for predictive values to differentiate the LSIL cases from control (A), the HSIL + CC cases from the control + LSIL cases (B) and the CC cases from the HSIL cases (C). All AUC and p values are shown in (D), and additional information, such as sensitivity and specificity, can be found in the "Results" section.
We further performed logistic regression analysis to determine the scoring weights of the three target genes in the model, and established a combined scoring formula based on the ΔCt values of the three target genes. Then, the optimal scoring threshold were determined based on the principle of the largest Youden index. After analysis, the scoring formula for value at risk (VAR) of the logistic regression model differentiating the HSIL + CC cases from the control + LSIL cases was: VAR = 6.498 − 0.107*ΔCt (FAM19A4)-0.167*ΔCt (EPB41L3)-0.305*ΔCt (PAX1). As shown in Fig. 4A, VAR cut-off value was 0.599 with 88% sensitivity and 97.7% specificity (AUC, 0.957; 95%CI, 0.937–0.977; p < 0.0001). However, the scoring formula differentiating the CC cases from the HSIL cases was: VAR = 1.475 − 0.105*ΔCt (EPB41L3) -0.157*ΔCt (PAX1). As shown in Fig. 4B, VAR cut-off value was 0.257 with 78.8% sensitivity and 62% specificity (AUC, 0.752; 95%CI, 0.691–0.814; p < 0.0001). Apparently, compared with the single target gene prediction model, the combined tri-gene methylation assay had higher sensitivity, specificity and AUC for predicting HSIL + CC cases (Fig. 4A). It was worth noting that in the logistic regression model of tri-gene methylation assay that differentiated HSIL from CC, the sensitivity, specificity, and AUC were relatively low, and the role of gene FAM19A4 even did not reach a statistically significant level which was not included in the above formula (p > 0.05, Fig. 4B).
ROC curve of logistic regression analysis for predictive values to differentiate the HSIL + CC cases from the control + LSIL cases (A) and the CC cases from the HSIL cases (B). AUC is 0.957 (95% CI, 0.937–0.977) for A (88% sensitivity, 97.7% specificity, p < 0.0001) and 0.752 (95%CI, 0.691–0.814) for B (78.8% sensitivity, 62% specificity, p < 0.0001).
Discussion
For the first time, we simultaneously detected methylation alterations in genes FAM19A4, EPB41L3, and PAX1 in patients with HPV-positive cervical lesions in a large population, and explored their relationship with disease progression. The major new findings of this study were: (i) methylation levels of these three genes were markedly elevated in HSIL and CC patients and could be considered as new biomarkers accurately distinguishing between HSIL + CC cases and control + LSIL cases; (ii) The methylation degree of all three genes had high sensitivity and specificity for the diagnosis of HSIL + CC, especially the diagnostic performance of the tri-gene methylation assay was more prominent; (iii) methylation levels of the three genes were positively correlated with the severity of cervical lesions, and they had significant differences in group HSIL and CC. Although slightly inferior, methylation assay of the three genes still had considerable sensitivity and specificity for distinguishing between HSIL and CC, which could act as novel indicators to monitor disease progression; (iv) Consistent with the findings of previous studies2, although all of the included subjects were HPV-positive, patients in groups HSIL and CC seemed to have a higher proportion of hrHPV16/18 infection.
CC is one of the most common malignant tumors in women with extremely high morbidity and mortality rate12. Although nearly 90% of CC burden worldwide occurs in developing countries, significantly lower screening rates are usually reported among women in these countries than those in developed countries13. To date, 228 genotypes of HPV have been identified. Persistent infection of hrHPV (currently 14 HPV types are termed high-risk, mainly with HPV16 and HPV18, and all HPV tests aim at their detection) usually results in the development of malignancy of cervix14.
Scientific screening, rational triage of risk groups and active interventions are critical to effectively prevent disease progression, improve prognosis and reduce fatality. Cervical liquid-based cytology examination is mainly used to stain the exfoliated cells from the cervix to observe morphological changes and then determine whether the cells are cancerous. The specificity of cytology examination is high, but the sensitivity is low, which is easy to lead to missed diagnosis and treatment. The judgment of cytology results needs to be interpreted by professional pathologists under the microscope, and as a result, results are susceptible to subjective decisions3. HPV testing is an etiological examination to detect whether the cervix is infected with HPV virus through nucleic acid testing for pathogens4. The sensitivity of HPV testing is high, but the specificity is poor. The false-positive rate is relatively high, and it is impossible to distinguish between transient infection and pathogenic infection. Transient HPV infection will not develop into cervical precancerous lesions or CC, which is easy to increase the rate of colposcopy referral and cervical tissue biopsy, causing unnecessary suffering to women15.
Aberrant genomic DNA methylation of cervical exfoliated cells is a common epigenetic alteration in the process of CC. HPV can induce hypermethylation of the promoter of certain tumor suppressor genes in the host, thereby causing gene silencing and participating in the occurrence of CC16. The quantitative detection of DNA methylation of related genes can act as a new means for CC screening, which has important clinical value for monitoring high-risk populations17,18,19,20,21,22.
The FAM19A4 gene, a member of the TAFA family, is expressed at low levels in most normal tissues and slightly higher in brain tissue, and is generally thought to be involved in the immune response as an immunomodulator with the function of preventing pathogen invasion23,24. It was found that the promoter of FAM19A4 gene had a high degree of abnormal methylation in cervical precancerous lesions and CC, and the methylation detection of FAM19A4 gene promoter had a higher specificity than cytology examination and HPV testing in CC screening25. Luttmer et al.26 found that the sensitivity of FAM19A4 methylation assay, cytology examination and HPV16/18-genotyping testing for the detection of cervical lesion (grade ≥ CIN 3) was 75.6%, 85.6% and 72.2%, respectively, and the specificity was 71.1%, 49.8% and 57.4%, respectively. According to another study, patients with a positive FAM19A4 methylation test could be directly referred for colposcopy, while a negative test could predict a transient HPV infection and a low risk of CC27. In our study, methylation levels of FAM19A4 gene promoter had a higher sensitivity and specificity for the diagnosis of HSIL + CC, at 84.6% and 96.1%, respectively. The sensitivity and specificity of further differentiating HSIL from CC were 68.1% and 66.9%, respectively.
EPB41L3 (erythrocyte membrane protein band 4.1 like 3), also known as 4.1B or DAI-1, is an important membrane skeleton protein belonging to the protein 4.1 family. As a candidate tumor suppressor gene, EPB41L3 inhibits cell overgrowth by inducing cell apoptosis and arresting the cell cycle28. It was found that the expression of EPB41L3 in various malignant tumors such as CC was significantly down-regulated compared with that in normal tissues, and the aberrant methylation of its promoter played an important role in the regulation of the expression of EPB41L3 genes29. Boers et al.30 showed that the sensitivity and specificity of EPB41L3 promoter methylation for the screening of CIN3 cervical precancerous lesions reached 79% and 88%, respectively, with a high clinical value for the diagnosis of cervical lesions in HPV-positive patients. In our study, methylation levels of EPB41L3 gene promoter had a higher sensitivity and specificity for the prediction of HSIL + CC, at 86.3% and 95.3%, respectively. The sensitivity and specificity of further differentiating HSIL from CC were 54.9% and 81.8%, respectively.
PAX1 has been revealed to be a key tumor suppressor gene that regulates cell differentiation and maturation. In cervical cells, once the PAX1 gene promoter is methylated, it will cause the gene to be silenced or inactivated, thus losing the function of inhibiting tumor growth and resulting in CC, which has been widely recognized and studied internationally31. In 2010, Lai’s research team32 found that PAX1 methylation assay has a sensitivity of 78% and a specificity of 91% for CIN3 + cervical precancer. In 2014, Kan’s study33 confirmed that methylation detection of PAX1 has an 86% sensitivity and 85% specificity for CIN3 + cervical precancer. In another study, Yang L et al.34 found that in non-HPV16/18 hrHPV-positive populations, PAX1 methylation detection had a sensitivity and specificity of 86.2% and 75.5%, respectively, for CIN2 cervical precancerous lesions, while CIN3 cervical precancerous lesions had a sensitivity and specificity of 90% and 69.3%, respectively, which were significantly better than cytology detection. According to the latest research, hypermethylation of PAX1 was positively associated with high HPV viral load, especially HPV16/18, and PAX1 methylation had a specificity of 86.6% for detecting CIN2+35. From our experimental data, the methylation detection of gene PAX1 also had high sensitivity and specificity for the diagnosis of HSIL + CC, which is 88.0% and 97.7%, respectively. The sensitivity and specificity of further differentiating HSIL from CC were 81.4% and 58.7%, respectively.
Although the methylation status of these three genes has been assessed separately in several cervical cancer early screening studies, and there are several approved kits on the market, the innovation of our research is that for the first time, these three genes have been detected and validated simultaneously in a larger sample. We would be interested in developing a three-gene assay to significantly improve the sensitivity and specificity of the test. There are currently no kits on the market that contain all three genes. In addition, the accuracy of disease prediction by methylation assay also depends on the genomic CpG loci selected in each study. Through initial testing, we identified the optimal genetic detection site and designed our own probe and primer sequences to improve the sensitivity and specificity of the assay.
Meanwhile, there were also some limitations in this study. First of all, previous studies may have biased results due to small sample sizes, and in order to obtain more accurate conclusions, we studied the correlation between methylation levels of these three genes and CC in a larger sample population. However, in subsequent studies, further sample collection for validation is needed, due to the fact that the variability of sensitivity and specificity depending on the characteristics of the study population. Second, due to the fact that there was no difference in methylation degree of the three genes in the control cases and the LSIL cases, other screening indicators need to be further explored and confirmed for cervical lesions below HSIL. Last but not least, on the basis that both HSIL and CC cases had high methylation levels of FAM19A4, EPB41L3 and PAX1 and high rates of hrHPV16/18 infection, further studies are needed to investigate the relationship between methylation alterations and hrHPV16/18 viral load. Moreover, further basic researches are needed to explore the mechanism of these tumor suppressor genes with abnormal methylation status in the pathogenesis of CC, so as to provide more effective and precise therapies and interventions.
Conclusion
In all, this exploratory study aimed to explore the possible role of methylation detection in CC screening. We firstly provided evidence that the methylation levels of FAM19A4, EPB41L3 and PAX1 increased as cervical lesions worsened, thus promising to be new biomarkers for monitoring disease progression. In particular, if possible, the combinationed performance of tri-gene methylation assay and logistic regression model will have the greatest predictive value. Our experimental results revealed that methylation detection of these three genes has extremely high sensitivity and specificity for HSIL and above, including CC. however, if further differentiation between HSIL and CC is required, additional ancillary evidence may be necessary to ensure that the diagnosis is not flawed.
Data availability
Data are available from the corresponding author on request.
Abbreviations
- LSIL:
-
Low-grade squamous intraepithelial lesion
- HSIL:
-
High-grade squamous intraepithelial lesion
- CC:
-
Cervical cancer
- hrHPV:
-
High-risk human papillomavirus
- HPV:
-
human papillomavirus
- ROC:
-
The receiver-operating characteristic curve
- AUC:
-
Area under the curve
- CI:
-
Confidence interval
- MSP:
-
Methylation-specific PCR
- ANOVA:
-
One-way analysis of variance
- VAR:
-
Value at risk
- SCC:
-
Squamous cell carcinoma
- AC:
-
Adenocarcinoma
- TCT:
-
Thinprep cytologic test
References
Singh, D. et al. Global estimates of incidence and mortality of cervical cancer in 2020: a baseline analysis of the WHO Global Cervical Cancer Elimination Initiative. Lancet Glob. Health. 11, E197–E206 (2023).
Sankaranarayanan, R., Prabhu, P. R. & Pawlita, M. Immunogenicity and HPV infection after one, two, and three doses of quadrivalent HPV vaccine in girls in India: a multicentre prospective cohort study. Lancet Oncol. 17 (1), 67–77 (2016).
MacLaughlin, K. L. et al. Trends over time in pap and Pap-HPV cotesting for cervical cancer screening. J. Womens Health. 28 (2), 244–249 (2019).
Nkwabong, E. et al. Pap smear accuracy for the diagnosis of cervical precancerous lesions. Trop. Doct. 49 (1), 34–39 (2019).
Lechner, M. et al. Cancer epigenome. Adv. Genet. 70, 247–276 (2010).
Kristensen, L. S. & Hansen, L. L. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin. Chem. 55 (8), 1471–1483 (2009).
Wentzensen, N. et al. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-thescience. Gynecol. Oncol. 112 (2), 293–299 (2009).
Yu-Ligh, L. et al. Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clin. Epigenetics. 8 (0), 0 (2016).
Qiaowen, B. The clinical significance of FAM19A4 methylation in high-risk HPV-positive cervical samples for the detection of cervical (pre)cancer in Chinese women. BMC Cancer 18 (1), 0 (2018).
Ying, S. Exploring the diagnostic potential of EPB41L3 methylation in Cervical Cancer and Precancerous lesions: a systematic review and Meta-analysis. Gynecol. Obstet. Invest. 89 (1), 0 (2023).
Xiang, L. et al. BRIF-Seq: Bisulfite-Converted randomly Integrated fragments sequencing at the single-cell level. Mol. Plant. 12 (3), 0 (2019).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin. 68 (6), 394–424 (2018).
Denny, L. et al. Interventions to close the divide for women with breast and cervical cancer between low-income and middle-income countries and high-income countries. Lancet. 389, 861–870 (2017).
Jee, B. et al. Immunology of HPV-mediated cervical cancer: current understanding. Int. Rev. Immunol. 40, 359–378 (2021).
Koliopoulos, G. et al. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst. Rev. 8 (8), CD008587 (2017).
Kremer, W. W. et al. The use of host cell DNA methylation analysis in the detection and management of women with advanced cervical intraepithelial neoplasia: a review. BJOG. 128 (3), 0 (2020).
de Waard, J. et al. Identification of a methylation panel as an alternative triage to detect CIN3 + in hrHPV-positive self-samples from the population-based cervical cancer screening programme. Clin. Epigenetics. 15 (1), 103 (2023).
Reuter, C. et al. Consistency of the S5 DNA methylation classifier in formalin-fixed biopsies versus corresponding exfoliated cells for the detection of pre-cancerous cervical lesions. Cancer Med. 10 (8), 2668–2679 (2021).
Zhang, J. et al. DNA methyltransferases in cancer: biology, paradox, aberrations, and targeted therapy. Cancers (Basel). 12 (8), 2123 (2020).
Feng, C. et al. The progress of methylation regulation in gene expression of cervical cancer. Int. J. Genomics. 2018, 8260652 (2018).
Hernandez-Lopez, R. et al. Methylation estimates the risk of precancer in HPV-infected women with discrepant results between cytology and HPV16/18 genotyping. Clin. Epigenetics. 11 (1), 140 (2019).
Kelly, H. et al. Performance of DNA methylation assays for detection of high-grade cervical intraepithelial neoplasia (CIN2+): a systematic review and Meta-analysis. Br. J. Cancer. 121 (11), 954–965 (2019).
Tom, T. Y. et al. TAFA: a novel secreted family with conserved cysteine residues and restricted expression in the brain. Genomics. 83 (4), 727–734 (2004).
Wang, W. et al. FAM19A4 is a novel cytokine ligand of formyl peptide receptor 1 (FPR1) and is able to promote the migration and phagocytosis of macrophages. Cell. Mol. Immunol. 12 (5), 615–624 (2015).
De Strooper, L. M. A. et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev. Res. (Phila). 7 (12), 1251–1257 (2014).
Luttmer, R. et al. Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre)cancer in high-risk HPV-positive women of a gynecologic outpatient population (COMETH study). Int. J. Cancer. 138 (4), 992–1002 (2016).
Bu, Q. et al. The clinical significance of FAM19A4 methylation in high-risk HPV-positive cervical samples for the detection of cervical (pre)cancer in Chinese women. BMC Cancer. 18 (1), 1182 (2018).
Xiaofeng, Y. Pivotal roles of protein 4.1B/DAL-1, a FERM–___domain containing protein, in tumor progression (review). Int. J. Oncol. 55 (5), 0 (2019).
Kelly, H. A. et al. Associations of human gene EPB41L3 DNA methylation and cervical intraepithelial neoplasia in women living with HIV-1 in Africa. AIDS. 32 (15), 2227–2236 (2018).
Boers, A. et al. DNA methylation analysis in self-sampled brush material as a triage test in hrHPV-positive women. Br. J. Cancer. 111 (6), 1095–1101 (2014).
Li, X. et al. PAX1 hypomethylation as a prognostic biomarker for radioresistance of cervical cancer. Clin. Epigenetics. 15 (1), 123 (2023).
Lai, H. C. et al. Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer. 116, 4266–4274 (2010).
Kan, Y. Y. et al. PAX1 methylation as a potential biomarker for cervical cancer screening. Int. J. Gynecol. Cancer. 24, 928–934 (2014).
Yang, L. et al. Utilization of PAX1 methylation test for cervical cancer screening of non-HPV16/18 high-risk HPV infection in women. Future Oncol. 19, 1917–1927 (2023).
Li, M. et al. Association and Effectiveness of PAX1 methylation and HPV viral load for the detection of Cervical High-Grade squamous intraepithelial lesion. Pathogens. 12 (1), 63 (2022).
Acknowledgements
We would like to thank all the participants and their families who agreed on taking part into the study. We also thank all the researchers and clinical staff that helped collecting the data used in this research. In particular, we acknowledge Dr. Wu Kang2,3 for giving professional advice on this article.
Funding
This study was supported by the Science and Technology Bureau of Putuo District, Zhoushan City (2023YB003) and “Zhou Creates Future” Haina Plan Innovative Talent (Team) ([2023]1–08).
Author information
Authors and Affiliations
Contributions
Y.K, WF.L and T.Z made substantial contributions in conception and design; XB.C, RR.C, YJ.C, FQ.F, YF.Y, GQ.J, TT.L, H.W, JW.D, M.Z, YH.H, QZ.T and ZQ.S made contributions to acquisition of clinical data and samples; WW.S and T.Z performed experiments and data analysis; YQ.J and T.Z wrote the original draft of the manuscript under the supervision of Y.K and WF.L; XB.C took part in reviewing the article or revising it critically for important intellectual content. All authors read and approved the final manuscript, and agreed to submit to the current journal and to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All study protocols comply with the Declaration of Helsinki. All participants provided written and informed consent before being included in the research. The protocol for the study has been approved by the Institutional Ethics Committee of Obstetrics and Gynecology Hospital Affiliated to Fudan University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
41598_2024_75047_MOESM2_ESM.tif
Supplementary Material 2. Fig. 1. Methylation levels of three genes in cervical squamous cell carcinoma and adenocarcinoma (p > 0.05).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, X., Chai, R., Zhang, T. et al. A novel methylation-detection panel for HPV associated high-grade squamous intraepithelial lesion and cervical cancer screening. Sci Rep 14, 25556 (2024). https://doi.org/10.1038/s41598-024-75047-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-75047-3