Abstract
As one of the most common malignancies, colorectal cancer (CRC) usually starts with a benign lesion and accumulates DNA damage as it progresses to full-fledged cancer. Glycyrrhizin (GL) has been found to alleviate tumor growth and inflammation, while the role of GL influences DNA damage response (DDR) in colorectal cancer remains unclear. GL exposure significantly reduced cell colony formation and viability with a concomitant increase in DNA fragmentation in CRC, meanwhile GL induced apoptosis by activating caspase-3. Moreover, GL induced cell cycle arrest in CRC cells at S phase, which was associated with decreased cyclin D1 in vitro. GL treatment significantly ameliorated tumor growth and promoted DDR in vivo. Mechanism analysis revealed that GL significantly downregulated the NHEJ pathway via inhibiting HMGB1. Finally, the expression of HMGB1 was abnormal regulated in CRC tissue than in adjacent normal tissues and associated with TNM stage and overall survival. Our findings indicate that HMGB1 may be a novel therapeutic target in CRC, a result that GL may serve as a promising drug for CRC treatment.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) has barely any specific early symptoms, resulting in late diagnosis, has incompletely characterized genetic factors and environmental risk factors, and ranks as the second leading cause of cancer-related death worldwide1,2. Surgical resection remains the preferred radical treatment for CRC patients, especially patients with localized and locoregional tumors without resectable distant metastases3. Once distant metastasis occurs or after surgery, adjuvant chemoradiotherapy is still the mainstay. However, patients, even after undergoing radical surgery or adjuvant chemoradiotherapy, ultimately develop recurrence or metastasis, and subsequently, the 5-year survival rate has been hovering at approximately 10%4, demonstrating that current treatments for CRC still need further study. Therefore, there is an urgent need to understand the molecular mechanism of CRC and identify novel therapeutic targets for CRC treatment.
The DNA damage response (DDR) is a key factor in maintaining genome integrity and stability5, consisting of multiple kinase-dependent signaling pathways, and determines the severity of damage, such as cell cycle arrest, damage repair and apoptosis6. Colorectal cancer usually begins with benign lesions and accumulation of DNA damage as it develops into a more metastatic cancer and invasive form7,8. High mobility group protein 1 (HMGB1) is a highly conserved nonhistone chromosome protein in the nucleus, and plays an important role in DNA arrangement, replication, damage repair and transcription via binding to DNA to stabilize nucleosomes9. Additionally, HMGB1 plays a distinct role in the pathogenesis of several cancers by influencing cell apoptosis, autophagy and the tumor microenvironment10,11,12,13. Recent study has shown that HMGB1 promotes non-homologous end-joining (NHEJ), the major pathway of double-strand break (DSB) repair via interaction with Ku70, and inhibition of HMGB1 exceeds resistance to radiation and chemotherapy in nasopharyngeal carcinoma14. Nevertheless, the role of HMGB1 influences DNA damage in colorectal cancer remains unclear.
As an inhibitor of HMGB1, glycyrrhizin (GL) derived and identified from the traditional Chinese medicine licorice has attracted increasing attention15. Several studies have revealed that GL has antioxidative stress, anti-inflammatory, anticancer, and antibacterial effects16. It has reported that GL could reverse the cancer-promoting effects of IGF2BP3 overexpression in bladder cancer17. Moreover, GL has been shown to reduce the expression of HPV E6 and E7 and induce apoptosis in cervical cancer cells in numerous preclinical studies18. Herein, we demonstrated that GL could abolish cell progression in CRC cells both in vivo and in vitro. Importantly, GL augmented DNA damage by regulating NHEJ pathway via inhibiting HMGB1 in CRC cells. Moreover, overexpression of HMGB1 was negative correlated with TNM stage and overall survival in CRC tissue samples. Overall, our data suggest that GL is critical for HMGB1-dependent DNA damage in CRC cells and that targeting this molecule may be beneficial for CRC treatment.
Results
GL treatment inhibits the proliferation of CRC cells in vitro
To determine whether GL has an impact on the proliferation of CRC cells in vitro, we incubated HT29 and HCT116 cells with different concentrations of GL for 24 h, and the viability of human CRC cells was assessed. The CCK-8 assay results showed that GL inhibited cell viability in a dose-dependent manner (Fig. 1a). The IC50 values (causing 50% inhibition of viability) of GL in CRC cells were 267.3 µM (HCT116) and 258.3 µM (HT29). Therefore, 300 µM was selected for follow-up experiments (Fig. 1b). Afterward, the colony formation assay indicated that the number of colonies in CRC cells was decreased significantly after GL treatment (Fig. 1c). To investigate whether GL was related to cell apoptosis and cycle progression, we analyzed the apoptosis and cycle distribution of CRC cells with or without GL treatment by flow cytometry. As shown in Fig. 1d, Supplementary Fig. 1a, the apoptotic populations in CRC cells increased distinctly after GL treatment. In addition, GL treatment significantly enhanced the activity (Fig. 1e) and the protein level of caspase-3 (Fig. 1f, Supplementary Fig. 1b). The flow cytometry-based cell cycle assay results showed that the percentage of CRC cells in S phase was much higher than that of the control cells after GL treatment (Fig. 1g, Supplementary Fig. 1c). Moreover, the protein levels of cyclin D1 and CDK2, two cell cycle-associated genes, were examined in CRC cells after GL treatment by Western blotting. GL treatment induced the expression of cyclin D1, while GL had no effect on CDK2 expression (Fig. 1h, Supplementary Fig. 1d). These results demonstrated that GL inhibited cell proliferation in CRC.
Effect of GL mediated cytotoxicity on human colorectal cancer cells. a Percent cell viability at 24 h post-GL treatment for a range of 100–400 µM was assessed by CCK8 assays of HCT116 and HT29 cells. b CCK8 data at 24 h were used to calculate the IC50 values using the nonlinear regression function of GraphPad Prism. GL concentrations are reported in µM on a log (10) scale, DMSO: 0.2% DMSO (vehicle) negative control. c Cell colony formation was measured with or without GL treatment. d Apoptosis was measured using Annexin V/7-AAD double staining in CRC cells 24 h after GL at 300 µM. e Caspase-3 activity was utilized to examine the apoptosis ability of CRC cells post-GL at 300 µM. Data from the three experiments are presented as the mean ± SD. f The protein levels of Caspase-3 were measured via Western blotting in CRC cells after GL treatment. α-Tubulin was used as a loading control. g The effect of GL at 300 µM on the cell cycle distribution in CRC cells. h The protein levels of CDK2 and cyclin D1 were measured via Western blotting in CRC cells after GL treatment. α-Tubulin was used as a loading control. *P < 0.05, **P < 0.01.
GL treatment accelerates the DDR via inhibiting NHEJ pathway in CRC cells
DNA damage response mediated by DDR sensors is a phosphorylation-driven signaling event, initiated by damage sensing, following the conversion of damage signals to DDR mediators and downstream effectors by DDR transducers and finally exercising an appropriate response based on the severity of DNA damage7. We thus detected whether GL modulated DNA double strand breaks (DSBs) in CRC cells after GL treatment through γH2A.x staining. γH2A.x foci formation was assessed in CRC cells at different time periods after treatment with 300 µΜ GL. GL treatment resulted in a dramatically higher number of foci beginning at 0.25 h until 2 h, and the foci decreased after 2 h (Fig. 2a). The Western blotting results showed that the protein level of phospho-γH2A.x was increased after treatment with GL (Supplementary Fig. 2a). Next, we showed substantially blocked DNA repair after GL treatment in colorectal cancer cells by calculating the dimension of tail moments in the comet assay (Fig. 2b). Then, we used immunofluorescence staining to observe whether GL treatment augmented the production of 8-OHdG, the pivotal factor in the initiation and promotion of carcinogenesis caused by DNA damage. After 300 µM GL treatment, 8-OHdG was induced compared with that in the control group (Fig. 2c, Supplementary Fig. 2b). These results demonstrated that GL promoted DNA damage response.
Effect of GL on DNA damage in human colorectal cancer cells. a γH2A.x was measured using immunofluorescence in CRC cells 24 h after GL at 300 µM. The number of γ-H2A.x per cell at the indicated time points post-GL (scale bar: 50 μm). The foci were counted in 300 CRC cells. Data from the three experiments are presented as the mean ± SEM. b The DNA tail was measured using a comet assay in CRC cells 24 h after GL at 300 µM (scale bar: 20 μm). The DNA tail per cell was counted in 100 CRC cells. Data from the three experiments are presented as the mean ± SEM. c The 8-OHdG level was measured using immunofluorescence in CRC cells at 24 h post-GL at 300 µM (scale bar: 50 μm). d The mRNA expression of ATM, RAD51, Ku70, Ku80, XRCC4 was measured by qRT‒PCR in CRC cells at 24 h post-GL at 300 µM. e Western blotting was used to detect Ku70 and LIG4 protein expression levels in CRC cells at 24 h post-GL at 300 µM. α-Tubulin was used as a loading control. *P < 0.05, **P < 0.01.
DSBs were generally repaired by NHEJ and HR (homologous recombination)19. To investigate whether GL regulates DNA damage repair pathway in CRC, we analyzed the expression of a spectrum of key HR and NHEJ-related genes, including ataxia telangiectasia mutated (ATM), RAD51, Ku70, Ku80 and XRCC4 in CRC cells after GL treatment with real-time quantitative PCR (RT-qPCR). As shown in Fig. 2d, the expression level of Ku70, Ku80 and XRCC4, the key mediator in NHEJ pathway, was the most decreased in both HCT116 and HT29 cells after GL treatment. Additionally, Western blotting analysis revealed that the protein level of Ku70 and LIG4 was significantly reduced in CRC cells with GL treatment (Fig. 2e, Supplementary Fig. 2c). Thus, our results suggest that GL treatment accelerates the DDR via inhibiting NHEJ pathway.
GL treatment eliminated CRC progression based on the DDR in vivo
Given that GL exerts a key effect on CRC cell proliferation and the DNA damage response in vitro, we further assessed the effect of GL treatment in CRC in vivo. Balb/c mice bearing CT26 xenografts were treated with 15 mg/kg GL (Fig. 3a). Compared with the control treatment, GL treatment obviously suppressed the tumor growth of CT26 xenografts, while GL treatment had no significant effect on tumor weight (Fig. 3b, c). Then, we examined the proliferation marker Ki67 by immunohistochemistry staining in xenograft tumor tissues. As shown in Fig. 3d, GL treatment significantly reduced the number of Ki-67-positive cells in xenografts. The TUNEL assay was used to determine whether GL treatment induced apoptotic cells in xenograft tissues. Figure 3e shows that GL treatment increased the number of apoptotic cells in xenograft tissues compared with that of the control group. Furthermore, we used immunohistochemistry staining to determine the effect of GL on γH2A.x foci formation. The results showed that GL treatment obviously increased γH2A.x foci formation (Fig. 3f). These data suggest that GL could inhibit the proliferation on account of DNA damage response in colorectal cancer cells.
GL treatment abrogates CRC cell proliferation in vivo. a Schematic overview of the CT26 syngeneic mouse tumor models and GL treatment. CT26 cells were inoculated under the skin of Balb/c mice. At Day 6 after translation, 15 mg/kg GL was intraperitoneally injected into Balb/c mice 6 times at 2-day intervals. The tumor size was measured at 2-day intervals. b Each group comprised 5 female Balb/c mice. Representative images of tumors formed by CT26 cells with or without GL treatment. c The tumor growth curves and weight of tumors formed by the indicated CT26 cells with or without GL treatment. The data are presented as the mean ± SEM (n = 5 mice per group). d Ki67 IHC staining in tumor tissues of the CT26 xenograft model with the indicated treatments (scale bar, 50 μm). Number of Ki67+ cells per visual field post-GL were quantified in 10 visual field per group. e TUNEL staining in tumor tissues of the CT26 xenograft model with the indicated treatments (scale bar, 50 μm). Number of TUNEL+ cells per visual field post-GL were quantified in 10 visual field per group. f γH2A.x IHC staining in the tumor tissues of the CT26 xenograft model with the indicated treatments (scale bar, 50 μm). Number of γH2A.x + cells per visual field post-GL were quantified in 10 visual field per group. ns, no significance, *P < 0.05, **P < 0.01.
GL treatment inhibits the proliferation of CRC cells via HMGB1
As an inhibitor of HMGB1, we further studied whether GL affects the biological characteristics of colorectal cancer cells via inhibiting the expression of HMGB1. Firstly, we measured the protein level of HMGB1 in CRC cells after GL treatment. As shown in Supplemental Fig. 3a, the HMGB1 protein level was reduced after GL treatment in CRC cells. Therefore, we overexpressed HMGB1 in colorectal cancer cells by lentivirus infection. qPCR and Western blotting further confirmed that both the mRNA and protein levels of HMGB1 were significantly increased (Fig. 4a). Meanwhile, we investigated the expression of HMGB1 in HMGB1-overexpression CRC cells after GL treatment. Western blotting confirmed that GL treatment decreased the expression of HMGB1, while overexpression HMGB1 could reverse the alleviate effect induced by GL (Supplemental Fig. 3b). Next, 300 µM GL was administered to the control and HMGB1-overexpressing CRC cells. The results of cell viability and colony formation assays showed that HMGB1 overexpression abolished the effect of GL in colorectal cancer cells (Fig. 4b, c). In addition, overexpression of HMGB1 significantly decreased the apoptotic population of colorectal cancer cells after treatment with GL (Fig. 4d, Supplementary Fig. 3c). Furthermore, the overexpression of HMGB1 decreased the activity and the protein level of Caspase-3 after GL treatment (Fig. 4e, f). Moreover, the cell cycle results showed that HMGB1 overexpression decreased the percentage of CRC cells in S phase with GL treatment (Fig. 4g, Supplementary Fig. 3d). As shown in the Fig. 4h, HMGB1 overexpression significantly reversed the upregulation of cyclin D1 protein level induced by GL, while CDK2 has no significantly change. In summary, these findings revealed that GL alleviates the progression of CRC via HMGB1.
GL treatment alleviates the proliferation of colorectal cancer cells via HMGB1. a The mRNA expression of HMGB1 was measured by qRT‒PCR in CRC cells. Western blotting was used to detect HMGB1 protein expression levels in CRC cells. α-Tubulin was used as a loading control. b Cell viability was measured using CCK-8 assays in HMGB1-overexpressing CRC cells 24 h after GL at 300 µM. c Colony formation assay of HMGB1-overexpressing CRC cells with or without GL treatment. d Apoptosis was measured using Annexin V/7-AAD double staining in HMGB1-overexpressing CRC cells 24 h after GL at 300 µM. e Caspase-3 activity was utilized to examine the apoptosis ability of CRC cells. f Western blotting was used to detect Caspase 3 protein expression levels in HMGB1-overexpression CRC cells post-GL at 300 µM. α-Tubulin was used as a loading control. g Cell cycle distribution was measured in HMGB1-overexpressing CRC cells 24 h after GL treatment at 300 µM. h Western blotting was used to detect CDK2 and cyclin D1 protein expression levels in HMGB1-overexpression CRC cells post-GL at 300 µM. α-Tubulin was used as a loading control. Data from the three experiments are presented as the mean ± SD. *P < 0.05, **P < 0.01.
GL treatment enhances the DDR via regulating NHEJ pathway by inhibiting HMGB1 in CRC cells
A previous study suggested that HMGB1 knockdown could increase the radiosensitivity by regulating DNA repair in esophageal squamous cell carcinoma20. Thus, we assumed that the inhibition effect of GL in CRC via regulating HMGB1. To further test the hypothesis, we detected γH2A.x foci formation at different time periods when GL was applied. As shown in Fig. 5a, overexpression of HMGB1 decreased γH2A.x foci formation in CRC cells after GL treatment. HMGB1 overexpression significantly increased the protein level of phospho-γH2A.x after GL treatment (Fig. 5b, Supplemental Fig. 4a). Additionally, HMGB1 overexpression significantly decreased the DNA tail percentage in CRC cells with GL treatment (Fig. 5c). Then, immunofluorescence staining of 8-OHdG showed that HMGB1 overexpression significantly decreased the production of 8-OHdG in CRC cells after GL treatment (Fig. 5d, Supplementary Fig. 4b). Meanwhile, HMGB1 overexpression abolished the GL-induced decrease in the protein level of Ku70 and LIG4 in CRC cells (Fig. 5e, Supplementary Fig. 4c). These results indicate that GL promotes the DNA damage response via regulating NHEJ pathway in an HMGB1-dependent manner.
GL treatment enhances the DDR via regulating NHEJ by HMGB1 in colorectal cancer cells. a γH2A.x was measured using immunofluorescence in HMGB1-overexpressing CRC cells at 24 h post-GL at 300 µM (scale bar: 50 μm). The number of γ-H2A.x per cell at the indicated time points post-GL. The foci were counted in 300 CRC cells. Data from the three experiments are presented as the mean ± SEM. b Western blotting was used to detect phosphor-γH2A.x protein expression levels in HMGB1-overexpression CRC cells at 24 h post-GL at 300 µM. α-Tubulin was used as a loading control. c The DNA tail was measured using immunofluorescence in HMGB1-overexpressing CRC cells at 24 h post-GL at 300 µM (scale bar: 20 μm). The DNA tail per cell was counted in 100 CRC cells. Data from the three experiments are presented as the mean ± SEM. d The 8-OHdG level was measured using immunofluorescence in HMGB1-overexpressing CRC cells at 24 h post-GL at 300 µM (scale bar: 50 μm). e Western blotting was used to detect Ku70 and LIG4 protein expression levels in HMGB1-overexpression CRC cells at 24 h post-GL at 300 µM. α-Tubulin was used as a loading control. *P < 0.05, **P < 0.01.
Abnormal expression of HMGB1 was correlated with overall survival in CRC tissue samples
To investigate HMGB1 expression in human colorectal cancer tissues, paired human CRC (n = 80) as well as matched adjacent normal tissues (NAT) were analyzed. We found that HMGB1 expression was significantly higher in CRC tissues than in the matched adjacent normal tissues with IHC staining. Interesting, we observed HMGB1 staining displayed nuclear staining (Fig. 6a). Additionally, it was observed that the expression of HMGB1 increased with tumor stage. The level of HMGB1 was higher in advanced clinical stages (III and IV) than in early stages (I and II) (Fig. 6b, Supplemental Table 2). Kaplan–Meier analysis revealed that the overall survival of patients with low HMGB1 expression was significantly longer than that of patients with high HMGB1 expression (Fig. 6c). These results indicated that high HMGB1 expression is a potential indicator of a poor clinical outcome in patients with CRC.
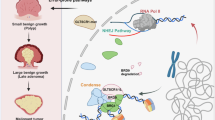
a One representative image of the IHC analysis of HMGB1 protein expression in CRC (n = 80) tissue sections at different stages (scale bar, 50 μm). b Individual staining intensities are represented by the heat map (up panel). HMGB1 protein expression based on the staining index in nonmalignant adjacent tissues (NAT) and CRC specimens and different clinical stages (down panel). Values are expressed as the mean ± SEM. c Kaplan-Meier curves of overall survival according to HMGB1 expression in patients with CRC. d Schematic illustration of plausible molecular mechanistic action of GL in CRC cells. GL is critical for HMGB1-dependent DNA damage response through regulating NHEJ pathway in CRC cells, and targeting this molecule may be beneficial for CRC treatment. **P < 0.01, *P < 0.05.
Discussion
Chemoresistance, radioresistance caused by cancer stem cells, DNA damage repair, and tumor microenvironment seriously affect the prognosis of patients with CRC15,21,22. In this study, we focused our research on the inhibition of DNA damage response of GL in CRC. Although previous study shown that GL induced apoptosis and autophagy and suppressed cell migration and invasion in CRC via regulating MMP-9 and MMP-2, the efficacy of GL on DNA damage remains poorly understood in CRC cells23,24. Here, we demonstrated the antiproliferative effect of GL in CRC cells. To address this, we attempted to systematically elucidate the antiproliferative effect of GL depending on the DDR in CRC cells. Originally, we found that the proliferation of CRC cells was significantly mitigated, as shown by a cell viability assay, in a concentration-dependent manner after GL treatment. Human cancer usually proliferates and progresses via escaping apoptosis; hence, phytocompounds could be applied as new therapeutics for cancer drug development due to their modulation of apoptotic pathways25. Our results showed that GL induced cell apoptosis in CRC cell lines. Moreover, we observed that GL arrested the progression of the cell cycle at S phase in CRC cells. Cell cycle regulatory gene alteration deregulates proliferation and cell cycle progression, resulting in diverse pathophysiological conditions26. During Western blot analysis, the protein level of cyclin D1 was increased after GL treatment, which could arrest cancer cells at S phase (Fig. 1). Taken together, these results indicate that GL may play a vital role in CRC treatment.
Previous studies have shown that GL could prevent the oxidative stress-mediated DNA damage response in UV-B-irradiated human primary dermal fibroblasts27. Upadhyay et al. also found that GL aided in repairing DNA damage and mitochondrial function by altering ROS levels and repairing the antioxidant status28. DNA damage induced by therapeutic drugs contributes to cell cycle arrest and cell apoptosis, whether it is endogenous or exogenous DNA damage, forming DSBs, which is always followed by the phosphorylation of the histone H2A.x. Therefore, GL-induced DNA damage was determined by γH2A.x foci formation, and our results showed that GL significantly increased the number of γH2A.x foci and the protein level of phospho-γH2A.x. Meanwhile, GL treatment increased the DNA tail moments in the comet assay. Moreover, 8-OHdG staining showed that GL treatment augmented the level of 8-OHdG (Fig. 2). GL treatment inhibited the growth of CRC tumors based on a reduction in Ki67 and enhanced apoptosis and DNA damage response in vivo (Fig. 3). These data showed the inhibition efficacy of GL via induced DNA damage response in CRC.
As the ligand of HMGB1, GL could bind with each of the two neighboring HMG boxes of the nonglycosylated protein29. HMGB1 is a multifunctional protein hinge on its ___location. In the nucleus, HMGB1 plays a key role in the process of DNA replication, transcription, chromatin remodeling, and V(D)J recombination, thus regulating DNA damage repair and the maintenance of genome stability as a DNA chaperone9. HMGB1 located in the nucleus has been presumed to replace linker histone 1, subsequently combining with DNA and disentangling chromatin. Furthermore, it has been assumed that HMGB1 empowers repair proteins by binding around the damage site30. Rohde et al. found that the expression of H2A.x and HMGB1 was abnormally expressed in subcutaneous adipose tissue and omental visceral adipose tissue, which was linked to DNA methylation and genetic variation31. Zhu et al.. researches showed that HMGB1 promoting NHEJ through interacting with Ku70 contributed to resistance to radiation and cisplatin14. Beside the effect in human cancer, HMGB1 signaling also phosphorylates Ku70 and impairs DNA damage repair in Alzheimer’s disease32. The present results showed that GL treatment could inhibit NHEJ pathway, while the overexpression of HMGB1 reversed GL-induced cell apoptosis and DNA damage response via promoting NHEJ pathway in CRC cells (Figs. 4 and 5). The nuclear staining of HMGB1 in CRC tissues also hinted that HMGB1 participated in DNA damage response (Fig. 6a). These data indicated that the DNA damage response caused by GL was in an HMGB1-dependent manner through regulating NHEJ pathway in CRC. Nevertheless, we have yet to illustrate the special mechanism by which HMGB1 influences the DNA damage repair. Hence, the manner in which HMGB1 regulates the DNA damage repair requires further detection in future research.
Additionally, HMGB1 is secreted into the cytoplasm or outside the cells after stimulation and interacts with multiple receptors, including RAGE and TLR33,34. As a damage-associated related pattern (DAMP), HMGB1 derived from dying cells could affect cancer migration via EMT and PI3K/pAKT activation by binding with TLR2 in pancreatic carcinoma35. HMGB1 derived from HIF-1α facilitated macrophage infiltration and successive chemoresistance development in CRC after chemotherapy treatment36. The present research showed that compared with normal adjacent tissues, HMGB1 was upregulated in CRC tissue samples and positive with TNM stage and poor prognosis in CRC patients (Fig. 6b, c). Given the key immunological and nonimmunological functions of HMGB1 in tumors, it will be valuable to intercept HMGB1 for tumor therapy.
Compared with conventional medicine, natural compounds derived from plant could be capitalized as the complementary or alternative medicine without too toxicity. In addition to its direct antitumor effect, GL can also be used as a drug carrier material. As the metabolite product of GL in the liver, glycyrrhetinic acid (GA) has been combined with polymers to deliver nanoformulations targeting hepatocellular carcinoma, including GA combined with docetaxel (DTX) and nanographene (NGO) and GA-modified hyaluronic acid combined with DTX37,38. However, on side effect caused by GL is pseudohyperaldosteronism due to long-term consumption39, and the patient was monitored for potassium and blood pressure. These studies have shown the development of GL as a new drug with clinical therapeutic effects for cancer.
In conclusion, we demonstrated that GL attenuated the progression of CRC via NHEJ through inhibiting HMGB1. Furthermore, GL treatment was associated with the DNA damage repair in an HMGB1-dependent manner (Fig. 6d). Additionally, abnormal expression of HMGB1 is positively correlated with TNM stage and prognosis in CRC tissues. Overall, our results illustrate that HMGB1 is a potential biomarker utilized to identify the prognosis of patients and that GL may accelerate the therapeutic regimen against CRC.
Materials and methods
Cell lines and cell culture
The HT29 (Research Resource Identifier CVCL_0320), HCT116 (CVCL_0291) and CT26 (CVCL_7254) cell lines were purchased from the Procell Life Science & Technology Co., Ltd (Wuhan, China) with the cell line authentication. These cells were cultured in DMEM (CellMax, Beijing, China) with 10% fetal bovine serum (FBS, CellMax), 100 U/ml penicillin and 100 mg/ml streptomycin (Solarbio, Beijing, China) in a humidified atmosphere at 37 °C and 5% CO2. The cells were verified by STR analysis before use. All cell lines were periodically tested to confirm that they remained mycoplasma-free.
Cell transfection and lentivirus infection
Lentivirus vectors carrying human HMGB1 cDNA were generated by GeneChem Co., Ltd. (Shanghai, China). The empty vector control lentivirus (LV-NC) was used as a control. For lentivirus infection, HCT116 and HT29 cells at 30% confluence in 6-well plates were transduced with lentiviral particles at an MOI of 10. After 48 h of infection with lentivirus, the infection efficiency was validated by counting GFP-expressing cells under a fluorescence microscope 72 h after infection. Transfection efficiency was determined by RT‒qPCR and Western blot assays.
RNA isolation and RT‒qPCR
Total RNA from cultured cells was prepared using RNAiso reagent (Vazyme, Nanjing, China) according to the manufacturer’s instructions. A total of 1 µg of RNA was reverse-transcribed using a cDNA Reverse Transcription Kit (TaKaRa) according to the manufacturer’s instructions. The acquired cDNA was analyzed in triplicate by real-time PCR on an ABI 7500 Real-Time PCR system (Applied Biosystems, MA, USA) using a SYBR PrimeScript RT‒qPCR Kit (TaKaRa). The PCR conditions were as follows: 95 °C for 30 s, followed by 40 cycles of amplification for 3 s at 95 °C and 30 s at 60 °C. Individual gene expression was normalized to that of GAPDH mRNA. The primer sequences for RT‒qPCR are provided in Supplemental Table 1.
Protein extraction and Western blotting analysis
HCT116 or HT29 cells in 6-well plates were lysed with RIPA lysis buffer (Solarbio) containing a protease inhibitor cocktail (Solarbio) according to the manufacturer’s instructions. Protein concentrations were measured with a Pierce BCA protein assay kit (Beyotime, Shanghai, China). Equal amounts of protein were separated by 12% SDS‒PAGE and transferred to a PVDF membrane (Merck Millipore, Darmstadt, Germany). The antibodies used for Western blotting analysis in this study were as follows: mouse anti-human HMGB1 (Proteintech, #66525-1-Ig), mouse anti-human cyclin D1 (Proteintech, #60186-1-Ig), mouse anti-human CDK2 (Proteintech, #60312-1-Ig), mouse anti-human caspase-3 (Proteintech, #66470-2-Ig), rabbit anti-human/mouse phospho-Histone H2A.x-S139 (Abclonal, #AP0099), rabbit anti-human Ku70 (Proteintech, #10723-1-AP), rabbit anti-human LIG4 (Proteintech, #12695-1-AP) and mouse anti-human/mouse α-Tubulin (Proteintech, #66031-1-Ig). The membranes were developed with Clarity Western ECL substrate (Vazyme) and visualized with a ChemiDocTM MP imaging system (Tacan). The band density was analyzed using Image J software.
CCK-8 assay
Cells (5*103 cells/well) were seeded in 96-well plates and then treated with different concentrations of GL (0, 100, 200, 300 and 400 µM, Selleck, Shanghai, China) for 24 h. Then, the cells were stained with 10 µl of sterile CCK-8 (Dojindo Laboratory, Mashikimachi, Japan) for 4 h at 37 °C. Finally, OD values were measured at 450 nm to indicate cell viability.
Cell colony formation
Cells (800 cells/well) were seeded in 12-well plates and then treated with 300 µM GL. After 2 weeks, the colonies were fixed with methanol for 30 min and then stained with 1% crystal violet (Solarbio) for 30 min. Finally, cell colonies with more than 50 cells were counted.
Flow cytometry
Cells were harvested and washed with cold PBS, and the cell cycle distribution and apoptosis rate were analyzed by flow cytometry (Beckman Coulter, CA, USA). Briefly, CRC cells were plated into 6-well plates at 2*105 cells per well and then treated with or without GL at 300 µM. For analysis of the cell cycle distribution, cells were collected after 24 h, washed twice with PBS, fixed in 70% ethanol containing 0.5% FBS and stored at -20 °C for at least 24 h. Samples were washed twice with PBS, treated with cell cycle reagent (Solarbio) and analyzed by flow cytometry using ModFit LT 5.0. For apoptosis analysis, the cells were washed with cold PBS and stained with Annexin V-PE and 7-AAD (Elabscience, Hubei, China) according to the manufacturer’s instructions. Annexin-V+/7-AAD− cells and Annexin-V+/7-AAD+ cells were considered apoptotic cells.
Caspase 3 activity assay
Cells were harvested after stimulation with 300 µM GL for 24 h, and then, caspase 3 activity was detected using a Caspase 3 Activity Assay Kit (Beyotime) according to the manufacturer’s instructions.
Immunofluorescence
Cells were washed with PBS, fixed with 4% paraformaldehyde and blocked with 5% BSA/PBS for 30 min at 37 °C. Then, primary antibodies, rabbit anti-human/mouse phospho-Histone H2A.x-S139 (Abclonal, 1:500), and mouse anti-human 8-OHdG (Santa Cruz, sc66036, 1:500), were incubated with cells overnight at 4 °C, after which a CoraLite549-conjugated mouse (Proteintech, SA00013-3, 1:1000) and Dylight594-conjugated rabbit (Abbkine, A23320, 1:1000) secondary antibody was incubated with the cells for 1 h at 37 ℃. The cells were counterstained with DAPI to visualize the nuclei and examined using a confocal microscope (Leica, Wetzlar, German).
Comet assay
Cells were harvested after stimulation with 300 µM GL for 24 h and then subjected to the comet assay using a DNA Damage Detection Kit (KeyGen, Jiangsu, China) according to the manufacturer’s instructions. Briefly, cell suspensions (10 µl) were embedded in 0.7% low-melting-point agarose (final concentration of 1%, 1*105 cells/slide) and incubated at 4℃ for 10 min. Afterward, the slides were lysed in lysis buffer for 2 h at 4 °C. After lysis, the slides were washed once with PBS, denatured in chilled alkaline electrophoresis solution for 40 min at room temperature and then electrophoresed under the same conditions for 30 min (25 V/cm). After electrophoresis, the microscopic slides were neutralized three times in 0.4 M Tris-HCl buffer (pH = 7.5). The slides were stained with propidium iodide, covered with a cover slip and analyzed. The slides were observed at 40× magnification using a fluorescence microscope (Olympus) and analyzed with CASP software. One hundred cells per sample were analyzed. The mean of two medians from each slide was used as a statistical unit. Hedgehog cells were excluded from the analysis. The software provides a variety of parameters for each cell (tail moment, percent of DNA in the tail and tail length). We used the percent of DNA in the tail for damage assessment.
Mouse xenograft tumor
Female Balb/c mice (6–8 weeks, 18–20 g) were purchased from PengYue Experiment Animal Breeding Co. and maintained under specific pathogen-free conditions. All experiments conducted on animals were approved by the Institutional Ethical Committee of Binzhou Medical University. For generation of tumors, mice were injected subcutaneously with 5*106 CT26 cells suspended in 100 µl of sterile PBS in the flank of each mouse at Day 0. The mice were randomly divided into the control and GL (15 mg/kg15) groups (5 mice per group). Tumor size was measured with a caliper every third day. The length and width measurements were obtained with calipers, and the tumor size was calculated. On Day 18, the animals were euthanized by cervical dislocation, and the tumors were excised and weighed. Tumor size (mean ± SEM; mm2) was calculated according to the following equation: tumor size (mm2) = S (mm) × L (mm), where S and L are the smallest and largest perpendicular tumor diameters, respectively40.
TUNEL assay
For the apoptosis assay, the xenografted tumor tissues of Balb/c mice were examined using a One-step TUNEL In Situ Apoptosis Kit (Elabscience) according to the manufacturer’s instructions. Briefly, sections from paraffin-embedded xenograft tumor tissues were dewaxed and rehydrated and then incubated with TUNEL reaction mixture at 37 °C for 1 h in a chamber with a humidified atmosphere. Nuclei were stained with DAPI. The numbers of TUNEL-positive cells and total cells were analyzed using a confocal microscope.
Tissue samples
Between January 2015 and September 2020, paraffin blocks of 80 CRC tissue samples and the corresponding normal adjacent tissue samples were obtained from the Yantaishan Hospital (Yantai, China), which were identified as CRC with hematoxylin and eosin (H&E) staining. All patients have signed the informed consent, and ethical committee was approved by Binzhou Medical University (Ethics No.: 2022 − 121). The clinicopathological features was recorded in Supplemental Table 2.
Immunohistochemistry
Sections from paraffin-embedded tissues were incubated with a rabbit anti-human/mouse Ki67 antibody (Proteintech, 1:3000), a rabbit anti-human/mouse phospho-Histone H2A.x-S139 antibody (Abclonal, 1:150) and mouse anti-human HMGB1 (Proteintech, 1:1000) overnight at 4 °C. This step was followed by staining (30 min at room temperature) with the corresponding HRP-labeled rabbit anti-goat secondary antibody or goat anti-mouse/rabbit secondary antibody (ZSGB-BIO, Beijing, China). Next, the sections were visualized by staining with 3,3’-diaminobenzidine (ZSGB-BIO) and counterstaining with hematoxylin (Solarbio). Images of tissue sections were captured using a Leica optical microscope.
All sections were reviewed blindly by two experienced pathologists (Dr. Sheng and Dr. Liu). The scoring criteria for HMGB1 immunostaining was adopted the semiquantitative immunoreactive score (IRS) system based on clinical data40. Briefly, category A (intensity of immunostaining) was scored using the following criteria: negative (0); weak (1); moderate (2); strong (3). Category B (percentage of immunoreactive cells) was scored using the following criteria: 0–25% (1); 26–50% (2); 51–75% (3); and 76–100% (4). Final scores were calculated by multiplying the scores of categories A and B in the same section and the scores ranged from 0 to 12. Cells were considered positive for HMGB1 if the product of IHC and percentage scores was >7.35.
Statistical analysis
All statistical analyses were performed using GraphPad 9.0 statistical software packages. Statistically significant differences between groups were determined using Student’s t test. A P value of < 0.05 was considered statistically significant in all cases.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33 (2022).
Siegel, R. L., Wagle, N. S., Cercek, A., Smith, R. A. & Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 73, 233–254 (2023).
Vodicka, P., Vodenkova, S., Buchler, T. & Vodickova, L. DNA repair capacity and response to treatment of colon cancer. Pharmacogenomics. 20, 1225–1233 (2019).
Yang, I. P. et al. MicroRNAs as predictive biomarkers in patients with colorectal cancer receiving chemotherapy or chemoradiotherapy: a narrative literature review. Cancers. 15, 1358 (2023).
Deng, S. et al. Targeting the DNA damage response and DNA repair pathways to enhance radiosensitivity in colorectal cancer. Cancers (Basel). 14, 4874 (2022).
Mauri, G., Arena, S., Siena, S., Bardelli, A. & Sartore-Bianchi, A. The DNA damage response pathway as a land of therapeutic opportunities for colorectal cancer. Ann. Oncol. 31, 1135–1147 (2020).
Mirza-Aghazadeh-Attari, M. et al. DNA damage response and repair in colorectal cancer: defects, regulation and therapeutic implications. DNA Repair. (Amst). 69, 34–52 (2018).
Tomasini, P. P. et al. Analyzing the opportunities to target DNA double-strand breaks repair and replicative stress responses to improve Therapeutic Index of Colorectal Cancer. Cancers (Basel). 13, 3130 (2021).
Mandke, P. & Vasquez, K. M. Interactions of high mobility group box protein 1 (HMGB1) with nucleic acids: implications in DNA repair and immune responses. DNA Repair.(Amst). 83, 102701 (2019).
Cheng, K. J. et al. HMGB1: an overview of its versatile roles in the pathogenesis of colorectal cancer. Cell. Oncol. (Dordr). 43, 177–193 (2020).
Ma, H. et al. High mobility group box 1 promotes radioresistance in esophageal squamous cell carcinoma cell lines by modulating autophagy. Cell. Death Dis. 10, 136 (2019).
Min, H. J. et al. Cytoplasmic HMGB1 and HMGB1-Beclin1 complex are increased in radioresistant oral squamous cell carcinoma. Br. J. Oral Maxillofac. Surg. 57, 219–225 (2019).
Li, B. et al. Tumor-derived exosomal HMGB1 promotes esophageal squamous cell carcinoma progression through inducing PD1(+) TAM expansion. Oncogenesis. 8, 17 (2019).
Zhu, X. et al. Inhibition of HMGB1 overcomes resistance to radiation and chemotherapy in nasopharyngeal carcinoma. OncoTargets Ther. 13, 4189–4199 (2020).
Liu, X. et al. Section 62 promotes stemness and chemoresistance of human colorectal cancer through activating Wnt/β-catenin pathway. J. Experimental Clin. cancer Research: CR. 40, 132 (2021).
Jain, R. et al. Oncopreventive and oncotherapeutic potential of licorice triterpenoid compound glycyrrhizin and its derivatives: molecular insights. Pharmacol. Res. 178, 106138 (2022).
Lv, L. et al. IGF2BP3 prevent HMGB1 mRNA decay in bladder cancer and development. Cell. Mol. Biol. Lett. 29, 39 (2024).
Bravo, V., Serrano, M., Duque, A., Ferragud, J. & Coronado, P. J. Glycyrrhizinic acid as an antiviral and anticancer agent in the treatment of human papillomavirus. J. Personalized Med. 13, 1639 (2023).
Nickoloff, J. A. et al. Roles of homologous recombination in response to ionizing radiation-induced DNA damage. Int. J. Radiat. Biol. 99, 903–914 (2021).
Han, G. et al. HMGB1 knockdown increases the radiosensitivity of esophageal squamous cell carcinoma by regulating the expression of molecules involved in DNA repair. Oncol. Lett. 22, 503 (2021).
Liu, T. et al. SENP5 promotes homologous recombination-mediated DNA damage repair in colorectal cancer cells through H2AZ deSUMOylation. J. Experimental Clin. cancer Research: CR. 42, 234 (2023).
Li, M. et al. PREX2 contributes to radiation resistance by inhibiting radiotherapy-induced tumor immunogenicity via cGAS/STING/IFNs pathway in colorectal cancer. BMC Med. 22, 154 (2024).
Ahmad, A., Tiwari, R. K., Saeed, M., Ahmad, I. & Ansari, I. A. Glycyrrhizin mediates downregulation of Notch pathway resulting in initiation of apoptosis and disruption in the cell cycle progression in cervical cancer cells. Nutr. Cancer. 74, 622–639 (2022).
Zhihang, Z., Meng, X., Xu, S. & Xu, G. Naturally occurring glycyrrhizin triterpene exerts anticancer effects on colorectal cancer cells via induction of apoptosis and autophagy and suppression of cell migration and invasion by targeting MMP-9 and MMP-2 expression. J. BUON. 25, 188–193 (2020).
Kapinova, A. et al. Dietary phytochemicals in breast cancer research: anticancer effects and potential utility for effective chemoprevention. Environ. Health Prev. Med. 23, 36 (2018).
Su, W. C. et al. DPY30 promotes proliferation and cell cycle progression of colorectal cancer cells via mediating H3K4 trimethylation. Int. J. Med. Sci. 20, 901–917 (2023).
Umar, S. A. et al. Glycyrrhizic acid prevents oxidative stress mediated DNA damage response through modulation of autophagy in Ultraviolet-B-Irradiated human primary dermal fibroblasts. Cell. Physiol. Biochem. 53, 242–257 (2019).
Upadhyay, S., Mantha, A. K. & Dhiman, M. Glycyrrhiza glabra (licorice) root extract attenuates doxorubicin-induced cardiotoxicity via alleviating oxidative stress and stabilising the cardiac health in H9c2 cardiomyocytes. J. Ethnopharmacol. 258, 112690 (2020).
Vergoten, G. & Bailly, C. N-glycosylation of high mobility Group Box 1 protein (HMGB1) modulates the interaction with glycyrrhizin: a molecular modeling study. Comput. Biol. Chem. 88, 107312 (2020).
Kozlova, A. L., Valieva, M. E., Maluchenko, N. V. & Studitsky, V. M. HMGB proteins as DNA chaperones that modulate chromatin activity. Mol. Biol. 52, 637–647 (2018).
Rohde, K. et al. Role of the DNA repair genes H2AX and HMGB1 in human fat distribution and lipid profiles. BMJ Open. Diabetes Res. Care. 8, e000831 (2020).
Tanaka, H. et al. HMGB1 signaling phosphorylates Ku70 and impairs DNA damage repair in Alzheimer’s disease pathology. Commun. Biol. 4, 1175 (2021).
Qian, F., Xiao, J., Gai, L. & Zhu, J. HMGB1-RAGE signaling facilitates ras-dependent Yap1 expression to drive colorectal cancer stemness and development. Mol. Carcinog. 58, 500–510 (2019).
Zhang, L. et al. Dedifferentiation process driven by radiotherapy-induced HMGB1/TLR2/YAP/HIF-1alpha signaling enhances pancreatic cancer stemness. Cell. Death Dis. 10, 724 (2019).
Chen, X. et al. Radiotherapy-induced cell death activates paracrine HMGB1-TLR2 signaling and accelerates pancreatic carcinoma metastasis. J. Exp. Clin. Cancer Res. 37, 77 (2018).
Zheng, H. et al. HIF1alpha promotes tumor chemoresistance via recruiting GDF15-producing TAMs in colorectal cancer. Exp. Cell. Res. 398, 112394 (2021).
Zhu, X. L. et al. Preparation and characterization of glycyrrhetinic acid-modified nano graphene oxide drug delivery system. Zhongguo Zhong Yao Za Zhi. 44, 4621–4626 (2019). (Chinese).
Xue, H. et al. Preparation of docetaxel-loaded, glycyrrhetinic acid-modified nanoparticles and their liver-targeting and antitumor activity. Exp. Ther. Med. 22, 1144 (2021).
Palermo, M., Armanini, D. & Delitala, G. Grapefruit juice inhibits 11beta-hydroxysteroid dehydrogenase in vivo, in man. Clin. Endocrinol. (Oxf). 59, 143–144 (2003).
Ma, Y. et al. B7-H3 regulates KIF15-activated ERK1/2 pathway and contributes to radioresistance in colorectal cancer. Cell. Death Dis. 11, 824 (2020).
Acknowledgements
We would like to thank Springer Nature (https://authorservices.springernature.cn/) for English language editing service.
Funding
This study was supported by a grant from the National Natural Science Foundation of China (82203543); Project ZR2022QH267 and ZR2023LZL008 supported by Shandong Province Natural Science Foundation; Initial project of Binzhou Medical University (50012304458); Medical and Health Science and Technology Project of Shandong Province (202309030619) and the Shandong Provincial College Youth Innovation and Talent Cultivation Plan.
Author information
Authors and Affiliations
Contributions
Yuhui Han and Yanchao Ma performed the experiments, analyzed the data and wrote the manuscript. Xiuxin Liu and Xinyu Jia generated the animal model. Wenjiong Sheng and Haide Liu helped with the tumor analysis. Honghui Li and Changyuan Wang conducted some cell experiments. Tao Hu and Bin Wang proposed and supervised the project. Yanchao Ma designed and performed the experiments. All authors approved the final version of manuscript including the authorship list.
Corresponding author
Ethics declarations
Ethics approval
All animal experiments were performed in accordance with the ARRIVE guidelines and approved by the Animal Experimental Ethics Committee of the Binzhou Medical University.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Han, Y., Sheng, W., Liu, X. et al. Glycyrrhizin ameliorates colorectal cancer progression by regulating NHEJ pathway through inhibiting HMGB1-induced DNA damage response. Sci Rep 14, 24948 (2024). https://doi.org/10.1038/s41598-024-76155-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-76155-w