Abstract
MicroRNAs(miRNAs) are promising biomarkers for early esophageal squamous cell carcinoma (ESCC) detection and prognostic prediction. This study aimed to explore the potential biomarkers and molecular pathogenesis in the early diagnosis of ESCC. Firstly, 48 differentially expressed miRNAs (DEMs) and 1319 differentially expressed genes (DEGs) were identified between 94 ESCC tissues and 13 normal esophageal tissues in TCGA. From miRNA–mRNA regulatory network, there are 6558 target genes of the 48 DEMs, where 400 target genes are also among 1319 DEGs. Then, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment indicate that the 400 DEGs significantly enriched in cell cycle, proteoglycans in cancer, p53 signaling pathway, protein digestion and absorption, transcriptional dysregulation in cancer, and oocyte meiosis. And there are 66 DEGs among these six biological pathways, which we called GO-DEGs. From miRNA–mRNA regulatory network, 32 DEMs regulated the 66 GO-DEGs, where 22 DEMs were verified by different types of experiments in ESCC tissues, cells, or serum from the literature. For the other novel 10 DEMs, single-factor Cox regression analysis show that only hsa-miR-34b-3p showed no significant correlation with the overall survival of ESCC patients. Finally, we obtained the novel 9 ESCC-related DEMs, where three are down-regulated, and six are up-regulated. We analyzed the expression trends of target genes for five miRNAs and identified three significantly different miRNAs (hsa-miR-205-3p, hsa-miR-452-3p, and hsa-miR-6499-3p) confirmed by qPCR. Moreover, the stage-specific miRNAs were also suggested. These three qPCR validated miRNAs are also specific to the early stages of ESCC: hsa-miR-452-3p is specific to Stage I, II and III; hsa-miR-205-3p is specific in Stage II and III; and hsa-miR-6499-3p is Stage II specific. They might be the potential biomarkers for ESCC stage diagnosis. This study identified three novel miRNA markers potentially related to the diagnosis of ESCC and participated in the occurrence and development of ESCC through cell cycle, proteoglycans in cancer, p53 signaling pathway, protein digestion and absorption, transcriptional dysregulation in cancer, and signaling pathway for oocyte meiosis.
Similar content being viewed by others
Introduction
Esophageal cancer is one of the most common gastrointestinal malignant tumors, the eighth incidence rate, the sixth mortality rate of all cancers in the world1, and the fourth leading cause of cancer-related deaths in China2. Esophageal cancer mainly has two pathological subtypes: esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC). And in China, about 90% of esophageal cancer are ESCC3, and most ESCC pathological stage, around 90% were at the severe stage4. The prognosis of ESCC in China is very poor, and 5-year survival rate is only 15–25%5. The ___location of ESCC were mainly in the upper and middle esophagus. The cause factors include smoking, alcohol abuse, dietary habits, and genetic factors6. However, the specific pathogenesis of ESCC remains unclear.
Studies have identified several miRNAs upregulated in ESCC patients, including miR-10a, miR-18a, miR-19b, miR-21, miR-22, miR-25, miR-31, miR-93, miR-129, miR-1246, miR-1322, miR-451, and miR-365. Conversely, miR-155, miR-203, miR-205, miR-375, miR-377, miR-486, and miR-718 are downregulated in these patients7. Recent findings indicate miRNA profiles are crucial for ESCC prognosis. High levels of plasma miR-21 and miR-16 correlate with poor survival, and low miR-375 levels predict an especially poor outcome8. A six-miRNA signature outperforms traditional tumour, node and metastasis (TNM) staging in accuracy, while miR-129, miR-103, and miR-107 also relate to worse survival rates9. Conversely, miR-377 and miR-15a levels are inversely related to survival, suggesting their potential as new prognostic markers, along with the rising importance of miR-367 serum levels. These results highlight the significant role of miRNA dysregulation in diagnosis and treatment of ESCC10.
Previous studies show that miRNA may participate in the occurrence, development, invasion, and metastasis of ESCC, thus, miRNAs are promising diagnosis and prognosis biomarkers of ESCC11,12. MiRNA is small non-coding RNA regulating gene expression through post-transcriptional regulation, and roles similarly as oncogene or tumor suppressor gene13. A meta-analysis14 of 11 studies revealed that miRNA could be used as a biomarker for early detection of ESCC. Most previous studies on ESCC miRNA markers used The Cancer Genome Atlas (TCGA) database, while few searched miRNACancerMap database and PubMed15,16,17.
In this study, we used the transcriptomic data of both miRNA and mRNA, clinical survival information of ESCC from TCGA to explore novel key miRNA markers participating in progression of ESCC, as well as their molecular pathogenic mechanism in ESCC.
Results
Differentially expressed miRNAs (DEM) and genes (DEG) between ESCC and normal
In total, 94 ESCC and 13 normal samples with both miRNA and mRNA transcriptome data from TCGA were included in the study. Compared to the normal esophageal tissue samples, 1319 differentially expressed genes (DEGs) with FDR < 0.05 and 4-FoldChange were obtained, as shown in the volcano plot of Fig. 1A, where 845 were upregulated and 474 were downregulated in ESCC samples. And 48 differentially expressed miRNAs (DEMs) with FDR < 0.05 and 4-FoldChange were identified as shown in the volcano plot of Fig. 1B, where 28 were upregulated and 20 were downregulated in ESCC samples. The heatmaps of these 1319 DEGs and 48 DEMs are shown in Fig. 1C and D, respectively.
The target genes of 48 DEMs
There are 6,559 target genes of 48 DEMs according to miRTarBase database (https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php).
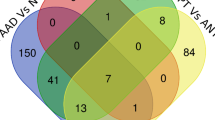
The comparison of these 6559 target genes with 1319 DEGs was shown in Fig. 2, where Venn diagram in Fig. 2A shows that 400 DEGs are also the target genes regulated by the 48 DEMs, i.e., DEM regulated DEGs. The heatmap of 400 DEM-regulated DEGs in Fig. 2B shows that they well separate the ESCC and normal samples.
GO and KEGG enrichment
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes(KEGG) enrichment analysis of 1319 DEGs, 6559 target genes of the 48 DEMs, and 400 DEM-regulated DEGs were shown in Fig. 3.
GO and KEGG enrichment of DEGs and DEMs. A Dot plot of top GO terms enriched from 6559 target gene of 48 DEMs; B Dot plot of top GO terms enriched from 1319 DEGs; C Dot plot of top GO terms enriched from 400 DEM-regulated DEGs; D Dot plot of top KEGG pathways enriched from 6559 target gene of 48 DEMs; E Dot plot of top KEGG pathways enriched from 1319 DEGs; F Dot plot of top KEGG pathways enriched from 400 DEM-regulated DEGs; G The occurrence heatmap shows that only 66 DEM-regulated DEGs participate in the top six KEGG pathways.
From GO enrichment of 6559 DEMs target genes in Fig. 3A, the top enriched biological processes include cell growth, mitotic cell cycle phase transition, gland development, regulation of apoptotic signaling pathway, and regulation of binding; the top cell components include cell-substrate junction, focal adhesion, spindle, chromosomal region, and cell leading edge; and the top molecular function include DNA-binding transcription factor binding, DNA-binding transcription activator activity, RNA polymerase II-specific, RNA polymerase II-specific DNA-binding transcription factor binding cadherin binding.
From GO enrichment of 1319 DEGs in Fig. 3B, the top enriched biological processes include epidermis development, organelle fission, nuclear division, skin development, and mitotic nuclear division. The top enriched cell components include the collagen-containing extracellular matrix, chromosome, chromosomal region, and basal part of the cell. The top enriched molecular functions include glycosaminoglycan binding, DNA-binding transcription activator activity, RNA polymerase II-specific, glycosaminoglycan binding, extracellular matrix structural constituent, and heparin binding.
From GO enrichment of 400 DEM-regulated DEGs in Fig. 3C, the top enriched biological processes include organelle fission, nuclear division, chromosome segregation, mitotic nuclear division, nuclear chromosome segregation, mitotic cell cycle phase transition, sister chromatid segregation, mitotic sister chromatid segregation, microtubule cytoskeleton organization involved in mitosis, and regulation of chromosome segregation; the top cell components include the spindle, chromosomal region, condensed chromosome, chromosome centromeric region, condensed chromosome centromeric region, kinetochore, mitotic spindle, midbody, kinesin complex, and outer kinetochore; and the top molecular functions include DNA-binding transcription activator activity RNA polymerase II-specific, DNA-binding transcription activator activity, tubulin binding, microtubule binding, catalytic activity acting on DNA, microtubule motor activity, cytoskeletal motor activity, DNA secondary structure binding, extracellular matrix structural constituent conferring tensile strength, and single-stranded DNA helicase activity.
From KEGG enrichment of 6559 DEMs target genes in Fig. 3D, the top enriched KEGG pathways include Human papillomavirus infection, Proteoglycans in cancer, Cellular senescence, and Cell cycle.
From KEGG enrichment of 1319 DEGs in Fig. 3E, the top enriched KEGG pathways include Cell Cycle, Oocyte meiosis, Hippo signaling pathway, Melanogenesis, IL-17 signaling pathway, and p53 signaling pathway.
Furthermore, KEGG enrichment of 400 DEM-regulated DEGs in Fig. 3F shows that the top enriched KEGG pathways include cell cycle, proteoglycan in cancer, p53 signaling pathway, protein digestion and absorption, transcriptional dysregulation in cancer, and oocyte meiosis.
Both GO and KEGG enrichment indicates that these six pathways were the main mechanisms of both DEM target genes and regulated DEGs, which related to ESCC. We thus filtered the DEM-regulated DEGs in these six ESCC-related pathways, and there are 66 DEM-regulated DEGs among them as shown in their occurrence heatmap of Fig. 3G.
The heatmaps of these 66 DEM-regulated DEGs in the six ESCC-related pathways are shown in Fig. 4A–F, respectively. They show that these 66 DEMs regulated DEGs well separate ESCC and normal samples. We thus looked for the DEMs regulating these 66 DEGs. From the miRNA–mRNA regulatory network in miRTarBase database, there are 32 DEMs in total regulating these 66 DEGs, called as ESCC-related DEMs.
The heatmaps of the DEM-regulated DEGs in six ESCC-related pathways. A Heatmap of DEM-regulated DEGs in cell cycle pathway; B Heatmap of DEM-regulated DEGs in proteoglycans in cancer pathway; C Heatmap of DEM-regulated DEGs in p53 signaling pathway; D Heatmap of DEM-regulated DEGs in protein digestion and absorption pathway; E Heatmap of DEM-regulated DEGs in transcriptional misregulation in cancer pathway; F Heatmap of DEM-regulated DEGs in oocyte meiosis pathway.
Comparison with ESCC-related DEMs reported in the literature
Among the 32 ESCC-related DEMs identified from our analysis in “GO and KEGG enrichment” section, 22 DEMs (hsa-miR-133a-3p, hsa-miR-196a-5p, hsa-miR-205-5p, hsa-miR-129-5p, hsa-miR-196b-5p, hsa-miR-30a-5p, hsa-miR-30a-3p, hsa-miR-148a-3p, hsa-miR-149-5p, hsa-miR-139-5p, hsa-miR-224-5p, hsa-miR-204-5p, hsa-miR-455-3p, hsa-miR-338-3p, hsa-miR-153-5p, hsa-miR-192-5p, hsa-miR-1-3p, hsa-miR-29c-3p, hsa-miR-375, hsa-miR-338-5p, hsa-miR-194-5p, hsa-miR-675-3p) were previously reported in miRCancerMap and PubMed, as verified by different experiments in ESCC tissues, cell lines, or serum, as shown in Table 1.
Univariate Cox regression analysis shows that hsa-miR-34b-3p has no significant correlation with the overall survival of ESCC patients. The other nine ESCC-related DEMs were significantly related to ESCC survival, and have been reported in the literature, thus might be novel miRNA markers related to ESCC. Among them, six upregulated (hsa-miR-944, hsa-miR-205-3p, hsa-miR-4652-5p, hsa-miR-452-3p, hsa-miR-6499-3p, hsa-miR-767-5p) and three downregulated (hsa-miR-215-5p, hsa-miR-194-3p, hsa-miR-29b-2-5p) in ESCC. The differential expression details of these nine miRNA markers, along with their regulated target genes and the ESCC pathways they participate in, were shown in Table 2.
miRNA–mRNA regulatory network
The regulatory network of the 32 DEMs and the 66 DEM-regulated DEGs drawn by Cytoscape 3.7.2 was shown in Fig. 5. The heatmap of the 9 novel ESCC-related DEMs was shown in Fig. 6A. They well separated the ESCC from normal. The regulatory network of the 9 novel and their regulated DEGs was shown in Fig. 6B. The target DEGs of six up-regulated miRNAs and three down-regulated miRNAs were shown in Table 2. The Sankey diagram of the novel 9 DEMs, their regulated DEGs, and the ESCC pathways these DEGs participate in was shown in Fig. 6C.
The regulatory network of the 32 DEMs and the 66 DEM-regulated DEGs was drawn by Cytoscape 3.7.2, where red squares represent DEMs which previously reported related to ESCC in literature; red triangles represent DEMs which were novel reported related to ESCC in our study, and yellow circles represent DEM-regulated DEGs in the cell cycle pathway. Light blue represents DEM-regulated DEGs in proteoglycans in the cancer pathway, and dark blue represents DEM-regulated DEGs in Transcriptional misregulation in the cancer pathway. Orange represents common DEM-regulated DEGs in the protein digestion and absorption pathway, purple represents DEM-regulated DEGs in the p53 signaling pathway, and green represents DEM-regulated DEGs in the Oocyte meiosis pathway.
The 9 ESCC-related DEMs target genes and their GO and KEGG enrichment results
There are 2280 target genes of 9 ESCC-related DEMs according to the miRTarBase database. They were top enriched in biological processes such as organelle fission, chromosome segregation, mitotic cell cycle phase transition, nuclear division, and regulation of cell cycle phase transition; cell components such as spindle, chromosomal region, condensed chromosome, and kinetochore; and molecular functions such as in single-stranded DNA binding, tubulin binding, ubiquitin-like protein ligase binding, and ubiquitin protein ligase binding; and KEGG pathways such as Cell Cycle, p53 signaling pathway pathway, which are consistent with our previous analysis, as shown in Fig. 7.
GO and KEGG enrichment of 2280 target genes of 9 ESCC-related DEMs. A Dot plot of top GO terms enriched from 2280 target genes of 9 ESCC-related DEMs; B Dot plot of top KEGG pathways enriched from 2280 target genes of 9 ESCC-related DEMs; C Chord plot of top KEGG pathways enriched from 2280 target genes of 9 ESCC-related DEMs.
Identification of novel miRNA markers related to ESCC
To evaluate the expression patterns of novel miRNA biomarkers in ESCC, we employed qRT-PCR to measure the expression levels of nine miRNAs in the ESCC cell line KYSE-30 and in the normal human esophageal epithelial cell line Het-1 A. The qRT-PCR results in Fig. 8 showed that the expression levels of five miRNAs—hsa-miR-944, hsa-miR-205-3p, hsa-miR-452-3p, hsa-miR-6499-3p, and hsa-miR-767-5p—were significantly elevated in the KYSE-30 cell line compared to the Het-1 A cell line. These findings align with the expression trends observed in the TCGA database.
Given that miRNAs act as negative regulators of mRNA expression, we excluded miRNAs whose expression patterns were congruent with those of their target mRNAs. Consequently, through qRT-PCR analysis coupled with the consideration of miRNA–mRNA negative regulatory relationships, we identified three differentially expressed miRNAs with statistical significance that were also validated by PCR: namely, hsa-miR-205-3p, hsa-miR-452-3p, and hsa-miR-6499-3p (Table 3).
To substantiate the robustness of these three differentially expressed miRNAs and to affirm their consistent differential expression in esophageal squamous cell carcinoma, we extended our qRT-PCR validation on another ESCC cell line, KYSE-180, alongside the Het-1 A, the normal human esophageal epithelial cell line. The validation results as shown in Fig. 9 demonstrated that the expression levels of these three miRNAs were consistent with the discovery results from TCGA database and the validation results observed in the ESCC cell line KYSE-30.
These validation results confirm the results from our bioinformatics analyses, indicating the potential utilization of these novel miRNA markers in the diagnosis and therapeutics of esophageal squamous cell carcinoma. These three miRNAs are now at the forefront of our ongoing research.
The stage specificity of the novel miRNA markers related to ESCC
We also performed the differential analysis of miRNAs on patients at four ESCC pathologic stages to show the stage specificity of the novel miRNA markers. The results are shown in Supplementary Table 1, among the 9 novel DEMs, hsa-miR-944, hsa-miR-29b-2-5p were common to all of the four stages in ESCC; hsa-miR-452-3p (qPCR validated) and hsa-miR-215-5p is common to Stage I, II and III; hsa-miR-205-3p (qPCR validated), hsa-miR-4652-5p and hsa-miR-194-3p were specific in Stage II and III; hsa-miR-6499-3p (qPCR validated) and hsa-miR-767-5p were Stage II specific. The three qPCR validated miRNAs were also specific to the first three stages of ESCC. They might be the potential biomarkers for ESCC stage diagnosis.
Materials and methods
Data download
The bioinformatics analysis flow chart of this study is shown in Fig. 10. The gene expression profile data, miRNA expression profile data and Clinical data of ESCC were downloaded from the TCGA database website (https://portal.gdc.cancer.gov/) on 11 May 2023. The gene expression profile data include 95 tumor samples and 13 normal samples. The miRNA expression profile data include 96 tumor samples and 13 normal samples. By removing duplicate samples and recurrent samples, 13 normal and 94 ESCC samples have both miRNA and mRNA expression data were included in this study. Among the 94 ESCC samples, 6 patients were in the stage I (Stage IA or Stage IB), 55 patients were in Stage II (Stage II, Stage IIA or Stage IIB), 26 patients were in Stage III (Stage III, Stage IIIA, Stage IIIB, or Stage IIIC), 4 patients in Stage IV (Stage IVA or Stage IV), and the other two have no information about the stage.
The bioinformatics analysis flow chart of this study. (Note: TCGA stands for The Cancer Genome Atlas, also known as the Cancer Genome Atlas Project; GO refers to Gene Ontology, the gene ontology; KEGG stands for Kyoto Encyclopedia of Genes and Genomes, the encyclopedia of genes and genomes in Kyoto; DEMs refer to differentially expressed miRNAs, differentially expressed microRNAs; DEG refers to differentially expressed gene; qRT-PCR stands for Quantitative Real-time PCR, quantitative real-time polymerase chain reaction).
Data preprocessing and differential expression analysis
The miRNAs and mRNAs expression data were normalized by using the “GDCRNATools” package in R 4.2.2. The miRNAs and mRNAs with no expression on more than 50% samples were excluded in the following analysis.
The differentially expression analysis of miRNAs and mRNAs between tumor and adjacent normal tissues were conducted using the “limma” package. The screening thresholds are FDR < 0.05 and logFC > 2 (where logFC > 0 indicates upregulated, and logFC < 0 indicates downregulated in tumor samples). The miRNAs and mRNAs satisfying the screening thresholds are the differentially expressed miRNAs (DEMs) and differentially expressed genes (DEGs), respectively. The visualization of differentially expression analysis results was shown in VolcanoPlot using the “gdcVolcanoPlot” package in R 4.2.2. The heatmaps of DEMs and DEGs expression patterns were drawn using the “ComplexHeatmap” package in R 4.2.2, where the clustering distance used Euclidean distance of normalized expression data.
Prediction of DEM target genes
The miRNA–mRNA interactions were collected from miRTarBase database (https://mirtarbase.cuhk.edu.cn). The target genes of the DEMs were then filtered based on these miRNA–mRNA interactions. The intersection of the target genes of DEMs and DEGs, i.e., the DEM-regulated DEGs, were analyzed by “VennDiagram” package in R 4.2.2.
GO and KEGG analysis
The functional annotations of Gene ontology (GO) included biological process (BP), cell component (CC), and molecular function (MF). GO and KEGG41,42,43 pathway enrichment analysis of the target genes of DEMs, DEGs and DEM-regulated DEGs, were performed on “clusterProfiler” package in R 4.2.2. The statistically significant enriched functional annotations and pathways were those with enrichment adjusted p-value, i.e., q-value < 0.05. The heatmap of the top enriched KEGG pathway with DEGs expression was drawn by “enrichplot” package in R 4.2.2.
Comparison to miRNACancerMAP and PubMed
The miRNACancerMAP (https://cis.hku.hk/miRNACancerMAP) consists of miRNA expression, related pathways, disease prognosis, and regulatory target genes in various cancers reported in PubMed. Compared the miRNA markers related to ESCC from miRNACancerMAP with our DEMs, the novel ESCC-related miRNA markers were obtained.
Construction of miRNA–mRNA regulatory network
The miRNA–mRNA interactions between DEMs and DEM-regulated DEGs inferred from miRTarBase database was drawn on Cytoscape 3.7.2.
Quantitative real-time polymerase chain reaction (qPCR)
The mRNA levels of newly identified miRNAs were measured using qPCR, with each sample tested in triplicate for accuracy. Primer sequences are provided in Supplementary Table 2, designed for specific primer fragments to ensure reliable gene expression data.
Statistical analysis
Statistical analysis was conducted using R language and GraphPad prism 9. Images were processed with ImageJ, Adobe Photoshop, and Adobe Illustrator CS2 software. Comparisons between two groups were done with t test and multiple groups with one-way analysis of variance (ANOVA). The data are presented as mean ± standard error of the mean (SEM). Normally distributed variables were compared by Student’s t test, or one-way analysis of variance (ANOVA) followed by Bonferroni correction for multiple comparisons. Chi-square test was used to analyze the counting data. P < 0.05 was considered statistically significant.
Discussion and conclusion
ESCC is a prominent subtype of esophageal cancer in Asia. However, both early diagnosis and effective treatment are quite limited. The pathogenic mechanisms of ESCC remains unclear. The treatment approaches include surgical procedures, chemoradiotherapy, targeted drug therapy, and immunotherapy.
Recent studies demonstrated that miRNA roled as an essential marker for early diagnosis and potential target of cancer. MiRNAs are not very specific as potential drug targets as they negatively regulate many target genes. Thus, the integration analysis of miRNA and mRNA are very important to figure out the specific miRNA–mRNA regulations related to disease progression. TCGA is currently the biggest dataset on various cancers, it included miRNA, mRNA expression data, and the clinical traits. The integration analysis can help to make the target miRNAs more specifically as potential markers for ESCC, and also compensate the information loss from small sample set.
In this study, we jointly analyzed ESCC RNA-seq and miRNA-seq data in the TCGA database. From differential expression analysis, 48 DEMs, 1319 DEGs, and 400 DEM-regulated DEGs were identified. According to KEGG enrichment of 400 DEM-regulated DEGs, the top enriched KEGG pathways include cell cycle, proteoglycan in cancer, p53 signaling pathway, protein digestion and absorption, transcriptional dysregulation in cancer, and oocyte meiosis. We call these six KEGG pathways as ESCC-related pathways. And there are 66 DEM-regulated DEGs in these ESCC-related pathways, and 32 DEMs were their regulators. Among them, 22 DEMs were reported to relate ESCC in PubMed. Survival analysis on the remaining 10 DEMs confirmed their significant relatedness to ESCC except hsa-miR-34b-3p. The other 9 DEMs (hsa-miR-944, hsa-miR-205-3p, hsa-miR-4652-5p, hsa-miR-452-3p, hsa-miR-6499-3p, hsa-miR-767-5p, hsa-miR-215-5p, hsa-miR-194-3p, and hsa-miR-29b-2-5p) might be novel ESCC-related miRNA markers.
Previous studies have reported abnormal expression of hsa-miR-944 in various cancers, such as esophageal adenocarcinoma, gastric cancer, colorectal cancer, breast cancer, oral cancer, lung cancer, cervical cancer, nasopharyngeal cancer, and liver cancer44. It plays a vital role in tumor proliferation, invasion, migration, epithelial-mesenchymal transition, apoptosis, and drug resistance. Moreover, it has significant clinical application value in cancer diagnosis, treatment, and prognosis judgment. However, its expression in esophageal squamous cell carcinoma (ESCC) remains unexplored. Another novel marker, miR-205-3p is up-regulated in non-small cell lung cancer tissues and cell lines, and it regulates the expression of APBB2, promoting the progression of non-small cell lung cancer45. In addition, it is significantly associated with poor prognosis in breast cancer patients46. MiR-205-3p also exhibits a role in inhibiting cancer in ovarian cancer and gastric cancer47,48. Similarly, miR-4652-5p is involved in regulating cell adhesion through down-regulating RND1 expression, thereby driving the progression of lung squamous cell carcinoma49. It could be a potential molecular diagnostic biomarker for laryngeal squamous cell carcinoma and head and neck squamous cell carcinoma50,51. Moreover, miR-452-3p is significantly up-regulated in liver cancer tissues and cells, and it promotes the proliferation and migration of liver cancer cells by targeting CPEB352. Long-chain noncoding RNA ZFAS1 promotes the development of oral squamous cell carcinoma through the regulation of the miR-6499-3p/CCL5 axis53. Similarly, miR-767-5p acts as an oncogene, promoting the progression of hepatocellular carcinoma by regulating the expression of the downstream target gene PMP2254. Studies further highlight its significant relationship with the prognosis of liver cancer patients through weighted gene co-expression network analysis55. Additionally, miR-767-5p is one of the ten miRNAs in a prognostic model that accurately predicts tumor recurrence in hepatocellular carcinoma patients after surgical treatment56. Conversely, miR-215-5p has been identified as a tumor suppressor gene in several human cancers. For example, it inhibits tumor cell proliferation by targeting RUNX1 in multiple myeloma57, inhibits tumor progression in colorectal cancer by regulating the expression of epiregulin and HOXB958, and inhibits cancer cell invasion by down-regulating the expression of Sox9 in breast cancer59. In addition, miR-215-5p down-regulates RAD54B expression, promoting cancer cell apoptosis60. The molecular panel consisting of miR-215-5p, miR-190b-5p, and miR-527 can serve as a serum biomarker for prostate cancer61. Furthermore, miR-194-3p inhibits the proliferation, migration, invasion, and docetaxel resistance of colorectal cancer cells through KLK10 regulation62,63. Moreover, high expression of miR-194-3p is associated with early cancer staging64. In pancreatic ductal adenocarcinoma, miR-29b-2-5p inhibits cell proliferation by directly targeting Cbl-b65. This miRNA is regarded as one of the most significant hinge genes in digestive system cancers, including esophageal cancer, and may be related to cancer prognosis66.
In summary, these nine ESCC-related novel miRNAs (hsa-miR-944, hsa-miR-205-3p, hsa-miR-4652-5p, hsa-miR-452-3p, hsa-miR-6499-3p, hsa-miR-767-5p, hsa-miR-215-5p, hsa-miR-194-3p, and hsa-miR-29b-2-5p) involved in cancer inhibition or carcinogenic effects, and might be promising potential biomarkers for ESCC diagnosis and treatment targets. They participate in the occurrence and development of ESCC through various pathways, including the cell cycle, proteoglycans in cancer, p53 signaling pathway, protein digestion and absorption, transcriptional dysregulation in cancer, and the signaling pathway of oocyte meiosis. These results require further validation in clinical samples. We also performed the qPCR on two ESCC cell lines, and confirmed three novel miRNA markers, hsa-miR-205-3p, hsa-miR-452-3p, hsa-miR-6499-3p, might be the ESCC biomarkers. Interestingly, these three markers were also specific to the first three stages of ESCC, indicating they might be the potential diagnosis biomarkers of ESCC stage.
Data availability
The ESCC RNA-Seq, miRNA-Seq data, clinical data were obtained from The Cancer Genome Atlas(TCGA) (https://portal.gdc.cancer.gov/projects/TCGA-ESCA; dbGaP Study Accession number is phs000178). This study complies with its data use and publication rules.
Abbreviations
- ESCC:
-
Esophageal squamous cell carcinoma
- DEMs:
-
Differential expression of miRNAs
- DEGs:
-
Differentially expressed RNAs
- BP:
-
Biological process
- CC:
-
Cell component
- MF:
-
Molecular function
References
Siegel, R. L. et al. Cancer statistics, 2022. CA Cancer J. Clin. 72 (1), 7–33 (2022).
Chen, W. et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 66 (2), 115–132 (2016).
Lu, T. et al. Systematic profiling of ferroptosis gene signatures predicts prognostic factors in esophageal squamous cell carcinoma. Mol. Ther. Oncolytics. 21, 134–143 (2021).
Yang, Z. et al. Annual cost of illness of stomach and esophageal cancer patients in urban and rural areas in China: a multi-center study. Chin. J. Cancer Res. 30 (4), 439–448 (2018).
Safiri, S. et al. The global, regional, and national burden of stomach cancer in 195 countries, 1990–2017: a systematic analysis for the Global Burden of Disease study 2017. Lancet Gastroenterol. Hepatol. 5(1), 42–54 (2020).
Abnet, C. C., Arnold, M. & Wei, W. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 154 (2), 360–373 (2018).
Fujihara, S. et al. MicroRNA expression profiles in superficial esophageal squamous cell carcinoma before endoscopic submucosal dissection: a pilot study. Int. J. Mol. Sci. 22, 4789 (2021).
Guo, Y. et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 68, 26–33 (2008).
Zheng, D. et al. Identification of serum microRNAs as novel biomarkers in esophageal squamous cell carcinoma using feature selection algorithms. Front. Oncol. 8, 674 (2019).
Mohammadi, E. et al. MicroRNAs in esophageal squamous cell carcinoma: application in prognosis, diagnosis, and drug delivery. Pathol. Res. Pract. 240, 154196 (2022).
Bertoli, G., Cava, C., Castiglioni, I. & MicroRNAs New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 5 (10), 1122–1143 (2015).
Cheng, G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv. Drug Deliv. Rev. 81, 75–93 (2015).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136(2), 215–233 (2009).
Bahn, J. H. et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 61(1), 221–230 (2015).
Koga, N. et al. Clinical significance of signal regulatory protein alpha (SIRPalpha) expression in esophageal squamous cell carcinoma. Cancer Sci. 112 (8), 3018–3028 (2021).
Shao, M. et al. Identification of key genes and pathways associated with esophageal squamous cell carcinoma development based on weighted gene correlation network analysis. J. Cancer. 11 (6), 1393–1402 (2020).
Luo, H. S. & Wu, D. H. Identification of miR-375 as a potential prognostic biomarker for esophageal squamous cell cancer: a bioinformatics analysis based on TCGA and meta-analysis. Pathol. Res. Pract. 215 (3), 512–518 (2019).
Yin, Y. et al. miR-133a-3p suppresses cell proliferation, migration, and invasion and promotes apoptosis in esophageal squamous cell carcinoma. J. Cell. Physiol. 234 (8), 12757–12770 (2019).
Sang, C. et al. Identification and validation of hub microRNAs dysregulated in esophageal squamous cell carcinoma. Aging (Albany NY). 12 (10), 9807–9824 (2020).
Ishibashi, O. et al. MiR-141-3p is upregulated in esophageal squamous cell carcinoma and targets pleckstrin homology ___domain leucine-rich repeat protein phosphatase-2, a negative regulator of the PI3K/AKT pathway. Biochem. Biophys. Res. Commun. 501 (2), 507–513 (2018).
Wang, H. et al. Long non-coding RNA XIST promotes the progression of esophageal squamous cell carcinoma through sponging mir-129-5p and upregulating CCND1 expression. Cell. Cycle. 20 (1), 39–53 (2021).
Chen, Z. et al. ADAMTS9-AS2 regulates PPP1R12B by adsorbing miR-196b-5p and affects cell cycle-related signaling pathways inhibiting the malignant process of esophageal cancer. Cell. Cycle. 21 (16), 1710–1725 (2022).
Yan, Q. et al. Long non-coding RNA OIP5-AS1 inhibits the proliferation and migration of esophageal squamous carcinoma cells by targeting FOXD1/miR-30a-5p axis and the effect of micro- and nano-particles on targeting transfection system. J. Biomed. Nanotechnol. 17(7), 1380–1391 (2021).
Qi, B. et al. Down-regulation of miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell proliferation by activating the wnt signaling pathway. World J. Gastroenterol. 23 (45), 7965–7977 (2017).
Tang, Y. et al. LncRNA TUG1 contributes to ESCC progression via regulating miR-148a-3p/MCL-1/Wnt/beta-catenin axis in vitro. Thorac. Cancer. 11 (1), 82–94 (2020).
Xu, Z. et al. Circular RNA hsa_circ_0000654 promotes esophageal squamous cell carcinoma progression by regulating the miR-149-5p/IL-6/STAT3 pathway. IUBMB Life. 72 (3), 426–439 (2020).
Zheng, L. J. et al. The suppressive effects of microRNA-139-5p on proliferation and invasion of esophageal squamous cell carcinoma. Zhonghua Yi Xue Za Zhi 101(13), 956–965 (2021).
Cheng, J. et al. Circ_0007624 suppresses the development of esophageal squamous cell carcinoma via targeting miR-224-5p/CPEB3 to inactivate the EGFR/PI3K/AKT signaling. Cell. Signal. 99, 110448 (2022).
Luo, H. et al. Mir-204-5p inhibits cell proliferation and induces cell apoptosis in esophageal squamous cell carcinoma by regulating Nestin. Int. J. Med. Sci. 19 (3), 472–483 (2022).
Yang, H. et al. MiR-455-3p acts as a prognostic marker and inhibits the proliferation and invasion of esophageal squamous cell carcinoma by targeting FAM83F. Eur. Rev. Med. Pharmacol. Sci. 21 (14), 3200–3206 (2017).
Yang, M. et al. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of esophageal squamous cell carcinoma. Oncol. Rep. 29 (1), 169–176 (2013).
Zuo, J. et al. MicroRNA-153 inhibits tumor progression in esophageal squamous cell carcinoma by targeting SNAI1. Tumour Biol. (2016).
Huang, Z. et al. A novel serum microRNA signature to screen esophageal squamous cell carcinoma. Cancer Med. 6 (1), 109–119 (2017).
Zhang, D. et al. LINC01518 knockdown inhibits tumorigenicity by suppression of PIK3CA/Akt pathway in oesophageal squamous cell carcinoma. Artif. Cells Nanomed. Biotechnol. 47 (1), 4284–4292 (2019).
Wang, H. et al. MiR-29c-3p suppresses the migration, invasion and cell cycle in esophageal carcinoma via CCNA2/p53 axis. Front. Bioeng. Biotechnol. 8, 75 (2020).
Wu, K. et al. miR-375 suppresses the growth and metastasis of esophageal squamous cell carcinoma by targeting PRDX1. J. Gastrointest. Oncol. 13 (5), 2154–2168 (2022).
Wu, W. et al. miR-375 inhibits the proliferation, migration and invasion of esophageal squamous cell carcinoma by targeting XPR1. Curr. Gene Ther. 21(4), 290–298 (2021).
Lin, W. C. et al. Mir-338-5p inhibits cell proliferation, colony formation, migration and cisplatin resistance in esophageal squamous cancer cells by targeting FERMT2. Carcinogenesis. 40 (7), 883–892 (2019).
Qu, F. et al. Circular RNA circ_0006168 enhances taxol resistance in esophageal squamous cell carcinoma by regulating miR-194-5p/JMJD1C axis. Cancer Cell. Int. 21 (1), 273 (2021).
Xiao, Q. et al. MicroRNA-675-3p promotes esophageal squamous cell cancer cell migration and invasion. Mol. Med. Rep. 18(4), 3631–3640 (2018).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28 (11), 1947–1951 (2019).
Kanehisa, M. et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51 (D1), D587–D592 (2023).
Shen, J. et al. Novel insights into miR-944 in cancer. Cancers (Basel). 14 (17) (2022).
Xu, L. B. et al. miR-205-3p promotes lung cancer progression by targeting APBB2. Mol. Med. Rep. 24 (2) (2021).
Qiu, C. et al. Mir-205-3p promotes proliferation and reduces apoptosis of breast cancer MCF-7 cells and is associated with poor prognosis of breast cancer patients. J. Clin. Lab. Anal. 33 (8), e22966 (2019).
Qiao, B. et al. Mir-205-3p functions as a tumor suppressor in ovarian carcinoma. Reprod. Sci. 27(1), 380–388 (2020).
Ma, X. et al. Oncosuppressive role of microRNA-205-3p in gastric cancer through inhibition of proliferation and induction of senescence: oncosuppressive role of microRNA-205 in gastric cancer. Transl. Oncol. 14(11), 101199 (2021).
Zhou, Y. et al. MiR-4652-5p targets RND1 to regulate cell adhesion and promote lung squamous cell carcinoma progression. Appl. Biochem. Biotechnol. 194(7), 3031–3043 (2022).
Luo, K. et al. Identification of critical miRNAs as novel diagnostic markers for laryngeal squamous cell carcinoma. Dis. Mark. 2022, 6858411 (2022).
Wu, C. et al. Two miRNA prognostic signatures of head and neck squamous cell carcinoma: a bioinformatic analysis based on the TCGA dataset. Cancer Med. 9 (8), 2631–2642 (2020).
Tang, H. et al. Mir-452-3p: a potential tumor promoter that targets the CPEB3/EGFR axis in human hepatocellular carcinoma. Technol. Cancer Res. Treat. 16(6), 1136–1149 (2017).
Qiu, X., Li, C. & Chen, H. Long noncoding RNA ZFAS1 promotes progression of oral squamous cell carcinoma through targeting miR-6499-3p/CCL5 axis. In Vivo 35(6), 3211–3220 (2021).
Zhang, L. et al. Functional analysis of mir-767-5p during the progression of hepatocellular carcinoma and the clinical relevance of its dysregulation. Histochem. Cell. Biol. 154 (2), 231–243 (2020).
Shen, B., Li, K. & Zhang, Y. Identification of modules and novel prognostic biomarkers in liver cancer through integrated bioinformatics analysis. FEBS Open Bio 10(11), 2388–2403 (2020).
Zhang, X. et al. Identification of a novel miRNA-based recurrence and prognosis prediction biomarker for hepatocellular carcinoma. BMC Bioinform. 23 (1), 479 (2022).
Liu, S. et al. Mir-215-5p is an anticancer gene in multiple myeloma by targeting RUNX1 and deactivating the PI3K/AKT/mTOR pathway. J. Cell. Biochem. 121 (2), 1475–1490 (2020).
Vychytilova-Faltejskova, P. et al. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis. 6 (11), 399 (2017).
Gao, J. B., Zhu, M. N. & Zhu, X. L. miRNA-215-5p suppresses the aggressiveness of breast cancer cells by targeting Sox9. FEBS Open Bio 9(11), 1957–1967 (2019).
Wang, M. et al. Integrated study of miR-215 promoting breast cancer cell apoptosis by targeting RAD54B. J. Cell. Mol. Med. 25 (7), 3327–3338 (2021).
Khan, M. M., Serajuddin, M. & Bharadwaj, M. Potential plasma microRNAs signature miR-190b-5p, mir-215-5p and miR-527 as non-invasive biomarkers for prostate cancer. Biomarkers. 28 (2), 227–237 (2023).
Liu, T. & Fang, Y. MiR-194-3p modulates the progression of colorectal cancer by targeting KLK10. Histol. Histopathol. 37(3), 301–309 (2022).
Zhao, J. et al. Mir-194-3p represses the docetaxel resistance in colon cancer by targeting KLK10. Pathol. Res. Pract. 236, 153962 (2022).
Wang, Y. et al. Establishing a three-miRNA signature as a prognostic model for colorectal cancer through bioinformatics analysis. Aging (Albany NY). 13 (15), 19894–19907 (2021).
Li, C. et al. MicroRNA-29b-2-5p inhibits cell proliferation by directly targeting Cbl-b in pancreatic ductal adenocarcinoma. BMC Cancer. 18 (1), 681 (2018).
Chen, Z. et al. RNA-associated co-expression network identifies novel biomarkers for digestive system cancer. Front. Genet. 12, 659788 (2021).
Funding
This work was supported by Grants from the Natural Science Foundation project of Hubei Province(2021CFB158), the Education Research Project of Hubei Province (No. D20222106), and the National Natural Science Foundation of China (No. 32060150).
Author information
Authors and Affiliations
Contributions
WL and JG supervised the study. PY, XG, and MX conduct the bioinformatics analysis and qPCR validation. PY wrote the first draft of the manuscript. WL, XG, MX, LQ, ZQ, JS, ZX, HX, RY, LZ, XL and JG revised the manuscript. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yuan, P., Gao, X., Xu, M. et al. Novel miRNA markers and their mechanism of esophageal squamous cell carcinoma (ESCC) based on TCGA. Sci Rep 14, 27261 (2024). https://doi.org/10.1038/s41598-024-76321-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-76321-0