Abstract
This study was performed to investigate the diagnostic value of radiomics models constructed by fat suppressed T2-weighted imaging (T2WI-FS) and contrast-enhanced T1-weighted imaging (CET1) based on magnetic resonance imaging (MRI) for differentiation of osteosarcoma (OS) and chondrosarcoma (CS). In this retrospective cohort study, we included all inpatients with pathologically confirmed OS or CS from Second Xiangya Hospital of Central South University (Hunan, China) as of October 2020. Demographic and imaging variables were extracted from electronic medical records and compared between OS and CS group. Totals of 530 radiomics features were extracted from CET1 and T2WI-FS sequences based on MRI. The least absolute shrinkage and selection operator (LASSO) method was used for screening and dimensionality reduction of the radiomics model. Multivariate logistic regression analysis was performed to construct the radiomics model, and receiver operating characteristic curve (ROC) was generated to evaluate the diagnostic accuracy of the radiomics model. The training cohort and validation cohort included 87 and 29 patients, respectively. 8 CET1 features and 15 T2WI-FS features were screened based on the radiomics features. In the training group, the area under the receiver-operator characteristic curve (AUC) value for CET1 and T2WI-FS sequences in the radiomics model was 0.894 (95% CI 0.817–0.970) and 0.970 (95% CI 0.940–0.999), respectively. In the validation group, the AUC value for CET1 and T2WI-FS sequences in the radiomics model was 0.821 (95% CI 0.642–1.000) and 0.899 (95% CI 0.785–1.000), respectively. In this study, we developed a radiomics model based on T2WI-FS and CET1 sequences to differentiate between OS and CS. This model exhibits good performance and can help clinicians make decisions and optimize the use of healthcare resources.
Similar content being viewed by others
Introduction
Both osteosarcoma (OS) and chondrosarcoma (CS) are common primary malignant bone tumors1,2,3,4,5,6,7, but their treatment methods vary greatly due to the varying sensitivity of radiotherapy/chemotherapy8,9. Therefore, early differential diagnosis is of great significance. At present, bone tissue biopsy is still the gold standard for diagnosis, but biopsy has invasiveness, sampling errors, and potential risks of tumor spread10. Therefore, it is necessary to find a non-invasive and highly accurate method for early differentiation between OS and CS. Magnetic resonance imaging (MRI) is the preferred detection method for diagnosing, staging, and monitoring bone and soft tissue lesions, especially fat suppressed T2-weighted imaging (T2WI-FS) and contrast-enhanced T1-weighted imaging (CET1), which have high sensitivity in describing lesion morphology and surrounding structures involved11. However, there are still great difficulties in the differential diagnosis between OS and CS by the above sequences alone12. The concept of radiomics was first reported in 2012, which converts images into mineable data with high fidelity and throughput for extraction and application in clinical decision support systems13,14,15. It has been widely applied in tumor staging, differential diagnosis, and efficacy prediction16,17,18. However, little study of MRI-based radiomics to differentiate OS from CS has been reported19,20. In this study, we aim to construct a radiomics model based on the CET1 and T2WI-FS sequences of MRI, and explore its value in distinguishing between OS and CS.
Methods
Study design and consent
This is a retrospective cohort study (Level III evidence) in which subjects received a image examination. This study was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University (No. K0340) and was conducted in accordance with the Declaration of Helsinki. All data used in the present study were obtained from the electronic medical records of our hospital and treated with strict confidentiality. The requirement of obtaining specific informed consent for this retrospective study was waived. All patients provided written informed consent for receiving this image examination. Due to the retrospective nature of the study, Jiangdong Ni (member of Institutional Review Board) waived the need of obtaining informed consent.
Patients
The general information of patients who met the following inclusion criteria and exclusion criteria in the Second Xiangya Hospital of Central South University from January 2014 to October 2020 was retrospectively analyzed, including patient gender, age, tumor localization and histopathological results. Inclusion criteria: (1) preoperative MRI examination, including at least CET1 and T2WI-FS sequences; (2) pathologically confirmed OS or CS cases by needle biopsy; (3) preoperative MRI and biopsy interval not exceed 3 months. Exclusion criteria: (1) patients with other tumors than OS or CS; (2) patients received surgery, radiotherapy, chemotherapy or molecular targeted therapy before MRI; (3) occult lesions which were poorly showed on the images, and difficult to delineate the region of interest (ROI) and extract the radiomics characteristics; (4) images or related sequences were lost.
Data collection of MRI
Preoperative examination was performed using a SIEMENS 3.0T MR scanner (Magnetom Skyra, Siemens Healthcare, Erlangen, Germany), at least including CET1 and T2-FS sequences. In this study, CET1 and T2WI-FS sequences were selected for radiomics feature extraction and analysis. Dimeglumine gadopentetate injection (Magnevist®, Bayer HealthCare AG, Daxing District, Beijing) was intravenously injected with weight-based dosing regimen (0.1 mmol/L/kg body weight) and injection rate of 2.5 mL/s. All MR data were retrieved from our hospital’s Picture Archiving and Communication System (PACS) and saved in DICOM format for further analysis.
Preprocessing of MRI images
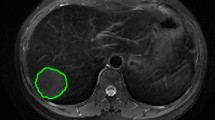
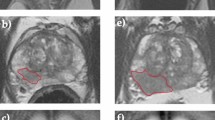
All images were transferred to ITK-SNAP software (version 3.8.0, http://www.itksnap.org/) for delineation of tumor area. We draw the region of interest for each layer of MRI and finally generate three-dimensional segmentation of the region of interest. All lesions were manually segmented by a radiologist with more than 5 years of experience in MRI diagnosis. The accuracy of the segmentation was confirmed by an other radiologist with more than 10 years of experience in MRI diagnosis. Two radiologists modified the specific segmentation of image when there was disagreement after discussion. ROI was delineated on one image of each sequence, including the entire tumor, peritumoral edema, and periosteal reaction. The selected slice or image showed the maximum diameter of the lesion.
Extraction of radiomics features
Pyradiomics (https://pyradiomics.readthedocs.io/en/latest/), a commonly used Python open-source software package for radiomics studies, is applied to extract radiomics features from medical images (CT, PET, MRI). Five categories of radiomics features defined by Pyradiomics include first-order gray level features, shape features, gray level co-occurrence matrix (GLCM), gray level run length matrix (GLRLM), and wavelet transform features21.
Selection of features
The least absolute shrinkage and selection operator (LASSO) method was used for the screening of the optimal features in the training set. The regularization parameter (λ) in the feature selection was adjusted using the minimum criterion, with 10 cross-validations used. The selected features with non-zero coefficients were used to construct the radiomics model. The radiology score (Rad-Score) of each patient was calculated with the weighting coefficient of logistic regression model.
Statistical analysis
R software (R Foundation for Statistical Computing, Vienna, Austria. URL: http://www.R-project.org), and SPSS 22.0 software were applied for statistical analysis, in which LASSO Logistic regression analysis and cross-validation were performed with the “glmnet” software package, with P < 0.05 as statistically significant.
Results
Extraction/selection of radiomics feature
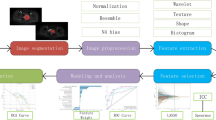
Figure 1 showed the study workflow and the basic procedure was shown in Fig. 2. 116 patients were finally enrolled in this study, including 52 females and 64 males, aged 5–82 (28.9 ± 19.8) years. Those cases included 79 OS cases and 37 CS cases, and were randomly assigned to the training group (n = 87) and the validation group (n = 29) in a ratio of 3:1. The training group was used for establishing the diagnostic model, and the validation group for verifying the diagnostic efficacy of model in differentiating OS from CS. The clinical characteristics of the patients in this study were shown in Table 1.
Basic procedures for the study. (1) Screen patients suitable for this study from all OS and CS patients in our hospital and obtain CET1 and T2WI-FS sequence data; (2) experienced radiologist outlines the tumor on CET1 and T2WI-FSMR images; (3) extract 530 initial radiomics features from the tumor area delineated by the original MR image; (4) select radiomics features by least absolute shrinkage and selection operator (LASSO) logistic regression; (5) use ROC analysis to evaluate the established radiomics model.
Totals of 530 initial features were extracted from the MR raw data and were divided into first-order features (n = 18), shape features (n = 8), GLCM features (n = 24), GLRLM features (n = 16), and wavelet transform features (n = 464). All sequence parameters were shown in Table 2. In this study, a total of 530 radiomics features were extracted in CET1WI images, including 18 first order features, 8 shape features, 16 GLRLM features, 24 GLCM features, and 464 wavelet transform features. Some features are reported in Table 3. Intra-class correlation coefficients (ICCs) ranged from 0.66 to 0.99 (average = 0.90) for all extracted features. The twenty-three selected radiomics features showed good inter-reader agreement, with ICCs ranging from 0.90 to 0.99.
LASSO regression is the most widely used shrinkage method, especially in reducing overfitting and enhancing prediction accuracy. Previous studies have directly used LASSO for feature shrinkage and dimensionality reduction to screen out features able to used in the construction of logistic regression models22,23,24,25,26,27,28,29. Hence, the LASSO method was performed to analyze the extracted features in this study. We selected the 15 features which differentiated OS from CS significantly from the T2WI-FS images and the 8 most significant features from the CET1 images using the LASSO method (Fig. 3). The selected features were shown in Table 4. The distribution of CET1 and T2-FS sequence-based radiomics features was shown by violin diagram (Figs. 4, 5).
Radiomics features were selected from CET1 and T2WI-FS using the LASSO binary logistic regression model. (a,c) is the tuning parameter (λ) in the LASSO model selected by ten cross-validations based on the minimum criterion. The lower x axis represents log (λ), the upper x axis represents the number of elements, and the y axis represents the binomial deviation. The dashed vertical line indicates the deviation value for each model with a given λ. The vertical dashed black line indicates the optimal value of λ. When the optimal λ is selected as 0.0008 in (a), log (λ) value is −3.13, and when the optimal λ is selected as 0.0031 in (c), log value (λ) is −2.506352. (b,d) Shows the LASSO coefficient profiles of 530 radiomics features. (b) Shows 15 selected features with non-zero coefficients in the T2WI-FS sequence and (d) shows 8 selected features with non-zero coefficients in the CET1 sequence.
Differentiation performance
Table 5 present the sensitivity and specificity of the training group and validation group in distinguishing OS from CS using T2WI-FS and CET1 data. In the training group, the specificity of the radiomics model constructed based on CET1 was 0.897 and the sensitivity was 0.81, and the specificity of the radiomics model constructed based on T2-FS was 0.862 and the sensitivity was 0.966. Radiomics models based on CET1 and T2WI-FS images in the training group obtained AUC values of 0.894 (95% CI 0.817–0.970) and 0.970 (95% CI 0.940–0.999), respectively, by ROC curve analysis. In the validation group, the specificity of the radiomics model constructed based on CET1 was 0.75 and the sensitivity was 0.81, and the specificity of the radiomics model constructed based on T2-FS was 0.875 and the sensitivity was 0.857. Radiomics models based on CET1 and T2WI-FS images in the validation group obtained AUC values of 0.821 (95% CI 0.642-1.000) and 0.899 (95% CI 0.785-1.000), respectively (Fig. 6).
ROC curves for the training and validation groups of the radiomics model. ROC curves for the T2WI-FS and CET1 based radiomics models for OS and CS in the training (a) and validation (b) groups. The AUC of CET1 and T2WI-FS in the training group was 0.894 (95% CI 0.817–0.970) and 0.970 (95% CI 0.940–0.999), respectively; and the AUC in the validation group was 0.821 (95% CI 0.642–1.000) and 0.899 (95% CI 0.785–1.000), respectively.
Discussion
In this study, we developed a radiomics model based on T2WI-FS and CET1 sequences to differentiate between OS and CS. This radiomics model performed satisfactorily, with AUC-based accuracies of 0.97 and 0.89 in the training and validation cohorts, respectively. Meantime, the specificity (75% and 87.5%) and sensitivity (81% and 85.7%) of CET1 and T2-FS were high in the verification group, suggesting that the diagnostic model has good diagnostic efficacy.
OS and CS are often difficult be accurately differentiated in clinical practice based on clinical features and imaging data, and ultimately need to be confirmed by needle biopsy. In terms of the age of predilection, although OS mainly occurs in children and young people, and CS mainly occurs in the elderly, the highest incidence rate is between 50 and 60 years old, but it occasionally occurs in younger individuals, so it is impossible to absolutely distinguish OS from CS based on age30. In addition, although pathological biopsy has a sensitivity and specificity of 86% and 95% in diagnosing bone tumors, it is still difficult to distinguish cartilage lesions. Pathological biopsy highly relies on the biopsy site and fixed tumor components, making it difficult to distinguish some subtypes of OS and CS25,26,27. For example, the common site and imaging characteristics of dedifferentiated chondrosarcoma (DDCS) are similar to those of OS, involving the proximal femur and presenting as a lytic, destructive, and invasive lesion with elastic regions and cortical de struction, both of which are easily confused with each other. Chondroblastic osteosarcoma (COS) is prone to producing cartilage like matrix, with the most common being high-grade transparent cartilage, making it extremely difficult to distinguish from CS31. MRI is a routine diagnostic tool for various tumors, as it has good soft tissue resolution and can more clearly display bone marrow replacement and cortical destruction21. It provides good tissue contrast, significantly better than CT and X-ray examinations. However, considering age distribution, high incidence sites, and similar pathological tissue types, MRI is also difficult to accurately distinguish between OS and CS. The emergence of radiomics can precisely be used to solve the above-mentioned problems.
This study is the first attempt to use MRI radiomics to differentiate the diagnosis of OS and CS. Previous studies have explored the effectiveness of CT and Micro CT radiomics in distinguishing between OS and CS, but the results are not very satisfactory as they cannot distinguish the differentiation of tumor stroma in detail and determine the boundaries32,33. Pathological biopsy is prone to loss of information due to tissue processing, while MRI can preserve the initial information of the tumor to the greatest extent possible. Therefore, MRI radiomics is more suitable for clinical differential diagnosis and tumor classification through raw feature extraction34.
Previous studies have reported that CET1 and T2WI-FS are widely used in the differentiation of malignant tumors. Moreover, T2WI-FS can effectively display the edge of the lesion, while CET1 can display the degree of blood supply to the lesion. Both have unique advantages in distinguishing between necrosis and solid tumors35,36,37,38,39,40,41,42. Therefore, this study selected the above sequence to verify that both have high accuracy in distinguishing between OS and CS, and further found that T2WI-FS has better diagnostic value than CET1, with higher sensitivity and specificity.
Our study has several limitations. First, our study has a small sample size, which may affect the diagnostic performance of radiomics models. However, we believe that our findings have significant clinical implications and require multi-sample center research to improve diagnostic efficiency. Second, our study used LASSO regression for dimensionality reduction and selected the final features, without using other statistical methods for initial screening. However, there is still controversy in previous studies regarding whether to conduct initial screening, and LASSO regression is more suitable for analyzing a large number of radiomics features with a small sample size, so our results are reliable. Third, future studies should focus on other factors that may affect diagnostic performance of radiomics models for bone malignant tumors.
Conclusions
In this study, we developed a radiomics model based on T2WI-FS and CET1 sequences to differentiate between OS and CS. This model exhibits good performance and can help clinicians make decisions and optimize the use of healthcare resources.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CET1:
-
Contrast-enhanced T1-weighted imaging
- COS:
-
Chondroblastic osteosarcoma
- CS:
-
Chondrosarcoma
- DDCS:
-
Dedifferentiated chondrosarcoma
- GLRLM:
-
Gray level run length matrix
- LASSO:
-
The least absolute shrinkage and selection operator
- GLCM:
-
Gray level co-occurrence matrix
- MRI:
-
Magnetic resonance imaging
- OS:
-
Osteosarcoma
- PET:
-
Positron emission tomograp
- ROC:
-
Receiver operating characteristic curve
- T2WI-FS:
-
Fat suppressed T2-weighted imaging
References
Philip, T. et al. Osteosarcoma. Br. J. Cancer 84(Suppl 2), 78–80 (2001).
Hu, X. et al. Effect of cisplatin arterial infusion (CAI) on primary nonmetastatic pelvic osteosarcoma: a preliminary study. Cancer Manag. Res. 13, 1491–1503 (2021).
Lin, Z., Xie, X., Lu, S. & Liu, T. Noncoding RNAs in osteosarcoma: implications for drug resistance. Cancer Lett. 504, 91–103 (2021).
Liu, T., Ling, L., Zhang, Q., Liu, Y. & Guo, X. Evaluation of the efficacy of pasteurized autograft and intramedullary vascularized fibular transfer for osteosarcoma of the femoral diaphysis. Orthop. Surg. 11(5), 826–834 (2019).
Ouyang, Z. et al. Therapeutic effect of palbociclib in chondrosarcoma: implication of cyclin-dependent kinase 4 as a potential target. Cell. Commun. Signal. 17(1), 17 (2019).
Wang, S., Ma, F., Feng, Y., Liu, T. & He, S. Role of exosomal miR–21 in the tumor microenvironment and osteosarcoma tumorigenesis and progression (review). Int. J. Oncol. 56(5), 1055–1063 (2020).
Xiao, Z. et al. Targeting CD44 by CRISPR-Cas9 in multi-drug resistant osteosarcoma cells. Cell. Physiol. Biochem. 51(4), 1879–1893 (2018).
Mirabello, L., Troisi, R. J. & Savage, S. A. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int. J. Cancer 125(1), 229–234 (2009).
Gelderblom, H. et al. The clinical approach towards chondrosarcoma. Oncologist 13(3), 320–329 (2008).
Di Tommaso, L. et al. Role of liver biopsy in hepatocellular carcinoma. World J. Gastroenterol. 25(40), 6041–6052 (2019).
Dai, Y. et al. Differentiation of pelvic osteosarcoma and ewing sarcoma using radiomic analysis based on T2-weighted images and contrast-enhanced T1-weighted images. Biomed. Res. Int. 2020, 9078603 (2020).
Jobke, B. & Werner, M. Osteogenic tumors of bone. Der Radiol. 56(6), 489–506 (2016).
Kumar, V. et al. Radiomics: the process and the challenges. Magn. Reson. Imaging 30(9), 1234–1248 (2012).
Lambin, P. et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 14(12), 749–762 (2017).
Lambin, P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 48(4), 441–446 (2012).
Aerts, H. J. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014).
Huang, Y. Q. et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J. Clin. Oncol. 34(18), 2157–2164 (2016).
Trebeschi, S. et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 30(6), 998–1004 (2019).
Zwirner, K. et al. Radiogenomics in head and neck cancer: correlation of radiomic heterogeneity and somatic mutations in TP53, FAT1 and KMT2D. Strahlenther. Onkol. 195(9), 771–779 (2019).
Mu, W. et al. Staging of cervical cancer based on tumor heterogeneity characterized by texture features on (18)F-FDG PET images. Phys. Med. Biol. 60(13), 5123–5139 (2015).
Sopta, J., Dordević, A., Tulić, G. & Mijucić, V. Dedifferentiated chondrosarcoma: our clinico-pathological experience and dilemmas in 25 cases. J. Cancer Res. Clin. Oncol. 134(2), 147–152 (2008).
Alhamzawi, R. & Ali, H. T. M. The bayesian adaptive lasso regression. Math. Biosci. 303, 75–82 (2018).
Mao, B. et al. Preoperative classification of primary and metastatic liver cancer via machine learning-based ultrasound radiomics. Eur. Radiol. 1, 1 (2021).
Han, Y. et al. Distinguishing brain inflammation from grade II glioma in population without contrast enhancement: a radiomics analysis based on conventional MRI. Eur. J. Radiol. 134, 109467 (2021).
Jiang, Y. et al. Radiomic signature based on CT imaging to distinguish invasive adenocarcinoma from minimally invasive adenocarcinoma in pure ground-glass nodules with pleural contact. Cancer Imaging 21(1), 1 (2021).
Liu, L., Lu, F., Pang, P. & Shao, G. Can computed tomography-based radiomics potentially discriminate between anterior mediastinal cysts and type B1 and B2 thymomas? Biomed. Eng. Online 19(1), 89 (2020).
Yu, X. P. et al. MDCT-based radiomics features for the differentiation of serous borderline ovarian tumors and serous malignant ovarian tumors. Cancer Manag. Res. 13, 329–336 (2021).
Jiang, Y. et al. Radiomics signature on computed tomography imaging: association with lymph node metastasis in patients with gastric cancer. Front. Oncol. 9, 340 (2019).
Zheng, Y. M. et al. MRI-based radiomics nomogram for differentiation of benign and malignant lesions of the parotid gland. Eur. Radiol. (2020).
VandenBussche, C. J., Sathiyamoorthy, S., Wakely, P. E. & Ali, S. Z. Chondroblastic osteosarcoma: cytomorphologic characteristics and differential diagnosis on FNA. Cancer Cytopathol. 124(7), 493–500 (2016).
Mamachan, P., Dang, V., Bharadwaj, N. S., DeSilva, N. & Kant, P. Chondroblastic osteosarcoma—A case report and review of literature. Clin. Case Rep. 8(11), 2097–2102 (2020).
Tossato Pdos, S., Pereira, A. C. & Cavalcanti, M. G. Osteosarcoma and chondrosarcoma–radiographic differentiation with computerized tomography. Braz. Oral Res. 16(1), 69–76 (2002).
Langheinrich, A. C. et al. Diagnostic value of ex-vivo three-dimensional micro-computed tomography imaging of primary nonhematopoietic human bone tumors: osteosarcoma versus chondrosarcoma. Acta Radiol. 49(8), 940–948 (2008).
Ni, M. et al. Prediction of the clinicopathological subtypes of breast cancer using a fisher discriminant analysis model based on radiomic features of diffusion-weighted MRI. BMC Cancer 20(1), 1073 (2020).
Jung, H. N. et al. Discrepant lesion size estimated on T1- and fat-suppressed T2-weighted MRI: diagnostic value for differentiation between inflammatory pseudotumor and carcinoma of the nasopharynx. Diagn. Interv. Radiol. 23(3), 199–205 (2017).
Yin, P. et al. A triple-classification radiomics model for the differentiation of primary chordoma, giant cell tumor, and metastatic tumor of sacrum based on T2-weighted and contrast-enhanced T1-weighted MRI. J. Magn. Reson. Imaging 49(3), 752–759 (2019).
Gerges, L., Popiolek, D. & Rosenkrantz, A. B. Explorative investigation of whole-lesion histogram MRI Metrics for differentiating uterine leiomyomas and leiomyosarcomas. Am. J. Roentgenol. 210(5), 1172–1177 (2018).
Li, L. et al. Radiomic analysis of multiparametric magnetic resonance imaging for differentiating skull base chordoma and chondrosarcoma. Eur. J. Radiol. 118, 81–87 (2019).
Sarioglu, F. C. et al. MRI-based texture analysis for differentiating pediatric craniofacial rhabdomyosarcoma from infantile hemangioma. Eur. Radiol. 30(10), 5227–5236 (2020).
Wengert, G. J. et al. Differentiation of intrahepatic cholangiocellular carcinoma from hepatocellular carcinoma in the cirrhotic liver using contrast-enhanced MR imaging. Acad. Radiol. 24(12), 1491–1500 (2017).
Rahman, T. Y., Mahanta, L. B., Chakraborty, C., Das, A. K. & Sarma, J. D. Textural pattern classification for oral squamous cell carcinoma. J. Microsc. 269(1), 85–93 (2018).
Vamvakas, A. et al. Imaging biomarker analysis of advanced multiparametric MRI for glioma grading. Phys. Med. 60, 188–198 (2019).
Acknowledgements
We would like to thank all patients for their devotion to taking unidentified risks in this clinical trial. We would like to thank radiologists Ju Chao and Longtao Yang from the Second Xiangya Hospital of Central South University for their professional guidance on magnetic resonance imaging (MRI) film reading.
Author information
Authors and Affiliations
Contributions
Zhi Gao and Zhongshang Dai are co-first authors. Deye Song and Yongfang Jiang are co-corresponding authors. Zhi Gao and Zhongshang Dai wrote the main manuscript text. Zhengxiao Ouyang prepared Figs. 1 and 2. Dianqing Li prepared Figs. 3 and 4. Sihuai Tang prepared Figs. 5 and 6. Penglin Li prepared Tables 1, 2 and 3. Xudong Liu prepared Tables 4 and 5. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Consent for publication
This study was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University (No. K0340) and was conducted in accordance with the Declaration of Helsinki. The requirement of obtaining specific informed consent from the patients for the publication of any accompanying images.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gao, Z., Dai, Z., Ouyang, Z. et al. Radiomics analysis in differentiating osteosarcoma and chondrosarcoma based on T2-weighted imaging and contrast-enhanced T1-weighted imaging. Sci Rep 14, 26594 (2024). https://doi.org/10.1038/s41598-024-78245-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-78245-1