Abstract
Ovarian cancer (OC) is one of the most prevalent and lethal malignancies affecting the female reproductive system, due to its tendency for metastasis and recurrence. This study identified the overexpression of LINC01320 (or long intergenic nonprotein coding RNA 1320) in tissues of ovarian cancer through the analysis of patient samples and online datasets. In vitro and in vivo experiments demonstrate that silencing of LINC01320 expression led to inhibition of proliferation and metastasis of OC cells. RNA pull-down followed by liquid chromatography tandem mass spectrometry (RNA pull-down-LC-MS/MS) revealed that LINC01320 interacted with purine-rich element binding protein B (PURB), a transcriptional repressor. Furthermore, the RNA-seq analysis identified damage-specific DNA binding protein 2 (DDB2) as a major common target of LINC01320 and PURB. Mechanistically, LINC01320 could recruit PURB to the promoter region of DDB2 to repress DDB2 transcription; thus, promoting the expression of NEDD4L and impeding the TGF-β/SMAD signaling pathway, and ultimately facilitating the progression of OC. Finally, rescue experiments confirmed the involvement of the DDB2/NEDD4L/TGF-β axis in LINC01320-mediated OC progression. In conclusion, this study unveils for the first time the pivotal function of the LINC01320/PURB/DDB2/NEDD4L/TGF-β axis and explores its prospective clinical implications in OC.
Similar content being viewed by others
Introduction
Ovarian cancer (OC) is a gynecological malignancy associated with high morbidity and mortality rates, featuring a five-year survival rate of less than 40%, owing to its dormant onset, absence of early diagnostic biomarkers, high invasiveness, and propensity for distant metastases1,2,3. Therefore, the exploration of valuable diagnostic and therapeutic biomarkers is of paramount importance for patients with OC.
Long non-coding RNAs (lncRNAs), functional RNA molecules exceeding 200 nucleotides in length without protein-coding capability4,5, play a pivotal role in numerous biological processes and are especially significant in the initiation and progression of various cancers6,7, including OC3,8,9. These RNAs influence gene regulation via epigenetic, transcriptional, post-transcriptional, and translational domains through their interactions with other RNAs10 and proteins11. LncRNAs are integral to the regulation of cancer cell proliferation, apoptosis, and migration, highlighting their potential as diagnostic markers or therapeutic targets in cancer treatment12. For instance, FEZF1-AS1 facilitates OC advancement via modulation of the miR-130a-5p/SOX4 axis13, whereas NRSN2-AS1 promotes OC progression by targeting the PTK2/β-catenin pathway3. Additionally, LINC00857 emerges as a regulator of OC progression by modulating the Hippo signaling pathway9.
LINC01320 (long intergenic non-protein coding RNA 1320) is chromosomally positioned at 2p22.3 in humans14,15. This lncRNA manifests marked upregulation within the lncRNA expression profiles of endometrial14, pancreatic16, and gastric cancers17. Meng et al. elucidated that LINC01320 impedes pancreatic cancer cell proliferation and migration through a targeted modulation of miR-324-3p16. Furthermore, Hu et al. unveiled the capacity of LINC01320 to expedite tumor progression in gastric cancer via the miR-495-5p/RAB19 axis17. Nevertheless, the functional implications of LINC01320 in the context of OC remain elusive to date.
This study evidenced a pronounced upregulation of LINC01320 in OC tissues, illustrating a direct positive correlation with the progression of OC. Employing RNA pull-down coupled with liquid chromatography tandem mass spectrometry (RNA pull-down-LC-MS/MS), we determined that LINC01320 engages in a direct interaction with purine rich element binding protein B (PURB), consequentially attenuating the transcriptional activity of damage-specific DNA binding protein 2 (DDB2). Antecedent investigations have elucidated that DDB2 orchestrates the modulation of TGF signal transduction in OC cells by downregulating the expression of the E3 ligase NEDD4L18. Thus, the unveiling of the LINC01320/PURB/DDB2/NEDD4L/TGF-β signal transduction pathway not only improves our understanding of the intricate role of LINC01320 within the OC environment, but also signals the arrival of new targets for both the prognosis and therapeutic intervention of OC.
Materials and methods
Bioinformatics analysis
Using UCSC Xena integrated computing19, transcript-level expression values were obtained, including 420 OC tissue samples from TCGA and 88 normal ovarian control samples from GTEx. The gene annotation was based on Gencode Release 23 (GRCh38.p3)20. Differential expression between the groups was compared using the limma-voom method.
Sample collection
OC tissues and the corresponding adjacent tissues were collected from 19 patients undergoing OC treatment from January 2021 and January 2023 at Suzhou Municipal Hospital. Before tissue collection, none of the patients received radiotherapy, chemotherapy, or molecular targeted therapy. All patients underwent a postoperative pathological examination to confirm the lesion as ovarian cancer. The collected tissue samples were washed with sterile PBS before being snap frozen in liquid nitrogen and stored in RNA Keeper Tissue Stabilizer (Vazyme, Nanjing, China), as previously described3,21. This study was approved by the Research Ethics Committee of Suzhou Municipal Hospital and written informed consent was obtained from all participants following the guidelines of the Declaration of Helsinki.
RNA extraction and real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) assays
As described in previous reports22,23, total RNA in tissues and cells was isolated employing TRIzol reagent (Vazyme), followed by reverse transcription into cDNA using the HiScript III RT SuperMix for the qPCR kit (Vazyme), adhering to the manufacturer’s instructions. The quantification of mRNA expression was conducted on an Applied Biosystems 7500 System utilizing the AceQ qPCR SYBR Green Master Mix kit (Vazyme). The relative expression of target genes was calculated based on the 2–ΔΔCT method normalized to 18sRNA24. The primers used in this study are listed in Table S1 and were synthesized by Sangon (Shanghai, China).
Cell culture and treatments
Human epithelial OC cells (OVCAR3, SKOV3, A2780, and HO8910) and normal ovarian surface epithelial cell line (IOSE80) were purchased separately from Zhong Qiao Xin Zhou Biotechnology Company and the Chinese Academy of Cell Collection, both located in Shanghai, China. OVCAR3 cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 (Gibco, USA) medium supplemented with 20% fetal bovine serum (FBS) and 1% penicillin/streptomycin, whereas SKOV3, A2780, HO8910, and IOSE80 cells required 10% FBS in addition to 1% penicillin/streptomycin in RPMI-1640 medium. The cells were cultured under standard conditions at 37 °C with 5% CO2 in a humidified incubator.
OC cells were seeded in a 6-well plate and then transfected after 70% cell confluence. Lipofectamine 2000 (Invitrogen, USA) was utilized to transfect OC cells with small interference RNAs (siRNAs) (GenePharma, Suzhou, China) specifically targeting the genes LINC01320, PURB, and DDB2, in addition to a negative control siRNA (si-NC). Similarly, overexpression plasmids (GenePharma) pcDNA3.1-NC, pcDNA3.1-PURB and pGL6 were introduced into OC cells using X-treme GENE HP DNA transfection reagent (Mannheim, Germany). After transfection for 48 h, cells were harvested for subsequent experiments. The nucleotide sequences of these siRNAs can be found in Table S2.
Cell proliferation assays
OC cell proliferation was tested using a CCK-8 kit (Beyotime Biotechnology, Shanghai, China) and a colony formation assay. Transfected OC cells were inoculated into 96-well plates (1500 cells per well). CCK-8 reagent (Beyotime) was added and incubated for 4 h in each well. Absorbance readings were taken at an optical density (OD) of 450 nm utilizing a Bio-Rad Model 680 microplate reader (Richmond, CA, USA), and cellular proliferation rates were calculated based on the OD values.
For the 14-day cloning formation assay, OC cells were seeded in 6-well plates at a density of 600 to 800 cells per well. The culture medium was refreshed every 5 days. Clones were washed with PBS, fixed with methanol, and stained with 0.1% crystal violet (Beyotime). The clones were dried, counted, and photographed using a bright field microscope (Cossim, Beijing, China).
Cell migration assays
Transwell migration assays were performed using migration chambers with 8 μm pore size polycarbonate membranes (Corning, NY, USA) in 24-well plates. In brief, 5 × 104 OC cells in 300 µL of serum-free medium were seeded evenly in the upper chamber and 700 µL complete culture medium was added to the lower compartment as attractant. After a 48-hour incubation period, nonmigrated cells were gently removed using cotton swabs. The migrated cells adhering to the lower membrane surface were then fixed with methanol and stained with 0.1% crystal violet. Five random visual fields per well were selected for imaging and cell counting under a microscope (Axioskop 2 Plus; Carl Zeiss, Oberkochen, Germany).
Xenograft in mice
The in vivo xenograft experiments were authorized by the Animal Ethics and Welfare Committee of Nanjing Medical University and conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and ARRIVE guidelines. Female BALB/c nude mice aged 4 weeks (Vital River Laboratory, Beijing, China) were housed in a pathogen-free environment at a temperature of 24 ± 2 °C, with 50% humidity and a 12-h light-dark cycle, with ad libitum access to water and food.
Si-LINC01320-1 sequences were cloned into the LV-2 N/Puro lentiviral interfering vector (GenePharma). For stable transfection, lentiviral LV-2 N/Puro and packaging vectors were co-transfected into HEK-293T cells (ATCC, USA). The viruses were collected from the supernatant and used to infect OVCAR3 cells, which were subsequently selected for puromycin resistance for 2 weeks. The knockdown efficiency of LINC01320 was verified by RT-qPCR. To establish a xenograft tumor model25, mice (n = 5) were subcutaneously injected bilaterally with 1 × 107 cells stably transfected with sh-LINC01320 or sh-NC. The injection site was disinfected with 75% alcohol before injection. After injection, the site was pressed with a sterile cotton swab. Tumor growth was monitored every 3 days and tumor volume was calculated using the formula V = 0.5×D×d2 (V, volume; D, longitudinal diameter; d, latitudinal diameter). On day 15 post-injection, the mice were euthanized under carbon dioxide exposure followed by cervical dislocation, and tumors were collected and weighed.
To establish the lung metastasis model3, mice (n = 4) were injected through the tail vein with 1 × 107 transfected cells. Six weeks later, the mice were euthanized, the lungs were extracted, photographed, and embedded in formalin.
Immunofluorescence
Immunofluorescence assays were conducted as described previously26,27. The tissue-containing slides were incubated overnight with the anti-Ki-67 antibody (Table S3) at 4 °C, followed by incubation with Alexa Fluor-conjugated secondary antibodies (Thermo Scientific, Waltham, USA) at 37 °C for 1 h, and subsequently examined using a laser scanning confocal microscope (LSM-810, Carl Zeiss).
Hematoxylin and eosin (H&E) staining
Lung tissues were subjected to H&E staining, which involved sequential steps of deparaffinization, rehydration, hematoxylin and eosin staining, dehydration, xylene hyalinization, and resin mounting. Finally, the histopathological features were examined using a Zeiss Axioskop 2 Plus microscope.
RNA pull-down and LC-MS/MS analysis
The full-length LINC01320 transcript sequences were cloned into pEASY-T1 Cloning Vector (Transgen, Beijing, China). LINC01320 was transcribed and labeled using T7 RNA polymerase (Ambio Life, Shanghai, China) and a biotin RNA labeling mix (Ambio Life). The resultant biotinylated RNA was then attached to streptavidin magnetic beads (Thermo Scientific). Cell lysate was mixed with the RNA-bound beads and incubated, followed by the elution of the RNA-protein complexes. Finally, proteins that interact with LINC01320 were identified by LC-MS/MS, as described previously28,29,30.
Western blotting
Western blotting was performed according to the previous protocol22,23. Briefly, total protein was extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime) containing 1% protease inhibitor phenylmethylsulfonyl fluoride (PMSF, Beyotime). The proteins were denatured at 100 °C for 10 min and subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (GenScript, China), followed by transfer to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 5% nonfat milk and incubated with specific primary and horseradish peroxidase-conjugated secondary antibodies. An enhanced chemiluminescent substrate (Thermo Scientific) was used for the detection of immunocomplexes. Detailed information on the antibodies can be found in Table S3.
RNA immunoprecipitation (RIP) assays
RIP assays were performed using the RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) as previously described3. In summary, OC cells were lysed in RIP lysis buffer and then incubated with anti-PURB antibody (Table S3) overnight at 4 °C. Subsequently, the protein-RNA complexes were captured and purified. RT-qPCR was conducted to detect the interaction between PURB and LINC01320 using primers in Table S1.
RNA-seq
RNA-seq was conducted according to the standard procedure as reported previously31. Briefly, total RNA was isolated using TRIzol reagent (Vazyme) and quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). Poly (A) RNA was purified from 1 µg total RNA using Dynabeads Oligo (dT)25-61005 (Thermo Scientific). Poly(A) RNA was fragmented into small pieces and reverse-transcribed to create the cDNA library by SuperScript™ II Reverse Transcriptase (Invitrogen). Finally, we performed the 2 × 150 bp paired-end sequencing (PE150) on an Illumina Novaseq™ 6000 (LC-Bio Technology, Hangzhou, China). Differentially expressed transcripts (DET) were defined with a fold change > 2 and a false discovery rate (FDR) < 0.05.
Luciferase reporter gene assay
The wild-type (WT) or mutant (MUT) DDB2 promoter regions were subcloned into the pGL6-Basic vector (Beyotime). The pGL6-promoter-WT or pGL6-promoter-Mut was co-transfected with either the NC or PURB expression plasmids into OC cell lines. The Renilla luciferase vector was used as an internal control. Forty-eight hours post-transfection, cells were collected and luciferase activity was measured using the Dual Luciferase Reporter Assay System (Beyotime), as previously reported3,32.
Chromatin immunoprecipitation (ChIP)-qPCR assay
Chromatin immunoprecipitation (ChIP) assays were performed using the EZ-ChIP kit (Millipore, Billerica, MA, USA). In brief, OVCAR3 and SKOV3 cells were harvested, washed, and cross-linked with 1% formaldehyde. The cross-linked chromatin was sonicated (Branson Sonicator 250) to generate fragments of approximately 500 bp. Subsequently, the cell lysate was incubated with the anti-PURB antibody (Table S3) overnight at 4 °C. Immunoprecipitated DNA was analyzed by quantitative real-time PCR using primers in Table S1.
Statistical analysis
All data derived from at least three independent experiments and were subject to statistical analysis and are presented as the mean ± standard deviation (SD). Unpaired student t-tests were used to evaluate differences between two groups, except for special mention in figure legends, while one-way analysis of variance (ANOVA) was used to assess the significance of differences between groups of more than two, using GraphPad Prism 7.0 (La Jolla, CA, USA). P < 0.05 was considered statistically significant.
Results
Silencing LINC01320 inhibited the growth and metastasis of OC cellsin vitro and in vivo
We initially evaluated the expression of LINC01320 in OC tissues. TCGA and GTEx databases revealed that the expression of LINC01320 was higher in 420 OC tissues compared to 88 benign ovarian tissues (Figure S1A). Subsequently, RT-qPCR assays were performed using 19 paired OC tissues and their adjacent nontumor tissues, which confirmed elevated levels of LINC01320 in tissues from OC patients (Figure S1B). Next, we detected expression of LINC01320 in human OC cell lines (SKOV3, OVCAR3, A2780, and HO8910) and the normal ovarian surface epithelial cell line (IOSE80). The results showed that LINC01320 was upregulated in the SKOV3 and OVCAR3 cell lines (Figure S1C). Therefore, we used SKOV3 and OVCAR3 cell lines for the further study.
To determine the in vitro biological effects of LINC01320 on OC cells, siRNA-mediated silencing of LINC01320 was conducted in SKOV3 and OVCAR3 cells, with knockdown efficiency validated by RT-qPCR (Fig. 1A). We found that silencing of LINC01320 markedly reduced cell viability, colony-forming, and migratory capacity in both OC cell lines (Fig. 1B–G).
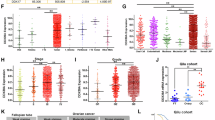
Silencing LINC01320 inhibits the growth and metastasis of OC cells both in vitro and in vivo. (A) Relative expression of LINC01320 in OC cells (SKOV3 and OVCAR3) transfected with si-LINC01320 or si-NC (negative control) tested by RT-qPCR, n = 3 per group. (B, C) The viability of OC cells was detected by CCK8 assays, n = 6 per group. (D, E) The proliferation ability of si-LINC01320-transfected OC cells was determined with a colony formation assay, n = 3 per group. (F, G) Transwell assays were performed to investigate the migratory ability of OC cells, n = 3 per group. Scale bar: 100 μm. (H–L) A total of 5 nude mice were injected subcutaneously with stably transfected OVCAR3 cells with sh-NC or sh-LINC01320. Tumors were collected and weighted (H, I). Scale bar: 1 cm. Paired Student’s t test for (I), p = 0.0358. Tumor volumes were calculated every 3 days (J). The expression of Ki-67 in tumors was evaluated with immunofluorescence (K, L). Scale bar: 20 μm. (M–O) OVCAR3 cells transfected with sh-NC or sh-LINC01320 were injected into the tails of nude mice (n = 4 per group). In both sh-NC and sh-LINC01320 mice, entire lungs were obtained (M) and nodules on the lung surfaces were counted (N). Lung sections were stained with H&E (O). For (M), scale bar: 2 mm; for (O), scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
To further elucidate the oncogenic role of LINC01320 in vivo, tumorigenesis experiments were performed by subcutaneous injection of OVCAR3 cells stably transfected with sh-LINC01320 or sh-NC in nude mice. Measurement of tumor volume and weight 15 days post-implantation revealed that xenograft tumors in the sh-LINC01320 group were smaller in size, slower in growth rate, and displayed a reduced proportion of Ki-67 positive cells (Fig. 1H–L). To assess the effects of LINC01320 on OC metastasis in vivo, OVCAR3 cells stably transfected with sh-LINC01320 or sh-NC were injected into the tail veins of nude mice. As a result, silencing of LINC01320 resulted in a decreased number of lung metastatic nodules compared with the control group (Fig. 1M–O). Taken together, the above results indicated that LINC01320 is required for the progression of OC both in vitro and in vivo.
LINC01320 interacted with PURB, and PURB knockdown phenocopies the effects of LINC01320 loss in OC cells
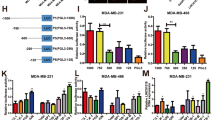
To further investigate the potential molecular mechanisms of LINC01320 in OC, we used the RNA pull-down-LC-MS/MS assay to identify the potential protein partners of LINC01320 (Fig. 2A). Venn diagrams generated from three independent RNA pull-down experiments revealed 25 overlapping proteins that interacted with LINC01320 (Fig. 2B and Table S4), and the top 10 proteins ranked by intensity-based absolute quantification (iBAQ) are illustrated in (Fig. 2C). PURB attracted our attention, not only because it is among the top 10 proteins but also because PURB always interacts with lincRNAs to regulate the progression of various tumor types, such as breast cancer33 and non-small cell lung cancer34. Subsequently, RNA pull-down and RIP-qPCR experiments were used to verify the interaction between LINC01320 and PURB in OC cells (Fig. 2D, E). Furthermore, the impact of PURB on cell behavior was assessed in vitro by transfecting OC cells with si-PURB (Fig. 2F). The results demonstrated that silencing PURB significantly suppressed cell viability, colony formation, and migratory capacity of OC cells (Fig. 2G–L), which phenocopied the effects of loss of LINC01320 (Fig. 1B–G).
LINC01320 interacts with PURB, and PURB is required for OC cell proliferation and migration. (A) Flow chart of the RNA pull-down assays. (B) Venn diagram of proteins identified by MS from three independent RNA pull-down assays. (C) Top 10 interactors ranked by iBAQ algorithm. (D) Western blotting analysis of the expression levels of PURB in LINC01320 pull-down products. The antisense strand of LINC01320 was served as the negative control (NC). (E) RIP assays for LINC01320 binding to PURB in OC cells, with rabbit anti-IgG as the negative control, n = 3 per group. (F) qRT-PCR analysis of PURB expression in OC cells transfected with si-NC or si-PURB, n = 3 per group. (G, H) CCK-8 assays of cell viability in OC cells transfected with si-NC or si-PURB, n = 6 per group. (I, J) Colony-formation assays in OC cells transfected with si-NC or si-PURB, n = 3 per group. (K, L) Transwell assays of OC cells transfected with si-NC or si-PURB, n = 3 per group. Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
The RNA sequence revealed DDB2 as a potential downstream target of LINC01320/PURB
PURB is a transcriptional regulator that can bind to purine-rich tracts of single-stranded nucleic acids to repress gene transcription35,36,37. However, knockdown of PURB did not alter LINC01320 expression (Fig. 3A); PURB expression also remained unchanged after LINC01320 depletion (Fig. 3B, C). Taken together, these results suggested that LINC01320 might interact with PURB to form a co-regulatory axis, thus regulating the transcriptional activity of downstream genes. To verify this hypothesis, we employed unbiased RNA-seq to identify differentially expressed transcripts (DET) regulated by the knockdown of LINC01320 and PURB in OVCAR3 cells (Fig. 3D, E). Compared with the control group (si-NC), the LINC01320 knockdown group exhibited 3646 DETs, whereas the PURB knockdown group showed 3191 DETs. A Venn diagram indicated an overlap of 930 DETs at the intersection of these two gene sets, in which the expression trend showed a highly positive correlation (Fig. 3F, G and Supplementary Dataset 1). Gene Ontology (GO) analysis based on the 930 common DETs (corresponding to 838 differentially expressed genes [DEG]) indicated that biosynthesis of amino acids, degradation of amino acids, protein processing in the endoplasmic reticulum, ubiquitin mediated proteolysis, and metabolic pathways were the prominent pathways involved in “LINC01320/PURB” modulatory biological processes (Fig. 3H, I). Among these DEGs/pathways, DDB2 has been identified as a tumor suppressor associated with tumorigenicity of OC cells18,38. In addition, RNA-seq revealed a remarkable upregulation of DDB2 upon LINC01320/PURB knockdown, which was subsequently corroborated by RT-qPCR (Fig. 3J). Hence, we chose DDB2 for further study.
RNA-seq analysis reveals common targets regulated by LINC01320 and PURB. (A) Relative expression of LINC01320 in OC cells (SKOV3 and OVCAR3) transfected with si-PURB or si-NC (negative control) tested by RT-qPCR, n = 3 per group. (B, C) Western blotting was used to determine the PURB protein level in OC cells (SKOV3 and OVCAR3) transfected with si-LINC01320 or si-NC, tubulin was used as an internal control, n = 3 per group. (D) Scatter plots showing DETs between si-LINC01320- and si-NC-transfected OVCAR3 cells based on RNA-seq data. (E) Scatter plots showing DETs between si-PURB- and si-NC-transfected OVCAR3 cells based on RNA-seq data. (F) Venn plot showing the intersection between DETs in (D) and (E). (G) Correlation analysis of the common DETs in (F). (H, I) Gene Ontology analysis based on the 930 common DETs (corresponding to 838 differentially expressed genes). (J) RT-qPCR experiments were performed to detect DDB2 expression in the si-LINC01320 or si-PURB groups, n = 3 per group. *P < 0.05, **P < 0.01, ***P < 0.001, n.s., not significant.
LINC01320 repressed the transcriptional activity of DDB2 by recruiting PURB to the promoter region of DDB2
To further elucidate the mechanism by which LINC01320/PURB regulated DDB2 transcription, we predicted the PURB binding site (PNR)33,37,38,39 in the promoter region of DDB2, and identified four putative PNRs at -400 bp of the DDB2 promoter (Fig. 4A). ChIP-qPCR confirmed that PURB occupied the DDB2 promoter fragment, which included the purine-rich region, in OC cells (Fig. 4B, C). qRT-PCR validated that PURB suppressed DDB2 expression in OC cells, which could be reversed by silencing LINC01320 (Fig. 4D). We cloned both full-length (-2,000 ~ + 100 bp) and mutant (lacking PNRs) DDB2 promoter sequences in the pGL6 luciferase reporter plasmid. Compared with the empty vector pGL6 (pGL6-EV), pGL6-promoter-WT and pGL6-promoter-Mut could effectively drive luciferase expression; however, the luciferase activity in the pGL6-promoter-Mut group was significantly higher than in the pGL6-promoter-WT group, suggesting that the PNR sequence was required for DDB2 transcriptional repression (Fig. 4E). Subsequently, the PURB overexpression plasmid was cotransfected with the reporter vectors into OC cells. The results demonstrated that PURB could repress DDB2 transcription, which could be reversed by silencing LINC01320 (Fig. 4F, G). However, in the pGL6-promoter-Mut group, the above results were not observed (Fig. 4F, G). The above results indicated that LINC01320 is required for the PURB-mediated transcriptional repression of DDB2.
LINC01320 recruits PURB to the DDB2 promoter. (A) Diagram showing the PNRs in the promoter region of DDB2. (B, C) ChIP-qPCR of PURB-associated DNA sequences in the PURB-binding region of the DDB2 promoter in (B) SKOV3 and (C) OVCAR3 cells. The GAPDH gene was used as a negative control, n = 3 per group. (D) After treatment of OC cells with OE-EV, OE-PURB, or OE-PURB + si-LINC01320 for 48 h, DDB2 expression levels were determined by qRT-PCR assays, n = 4 per group. (E) The dual luciferase reporter assay measured the luciferase activity of wild-type (WT) and mutant (Mut) DDB2 reporter genes, n = 6 per group. (F, G) After treatment of OC cells with OE-EV, OE-PURB, or OE-PURB + si-LINC01320 for 48 h, dual luciferase report analysis verified the luciferase activity of WT and Mut DDB2 reporter genes in SKOV3 (F) and OVCAR3 (G) cells, n = 6 per group. (H, I) After treatment of OC cells with si-NC or si-LINC01320 for 48 h, ChIP-qPCR of PURB-associated DNA sequences in the PURB binding region of the DDB2 promoter in SKOV3 (H) and OVCAR3 (I) cell lines transfected with si-LINC01320 or si-NC, n = 3 per group. *P < 0.05, **P < 0.01, ***P < 0.001, n.s., not significant.
Next, we investigated how LINC01320 affected the transcriptional repression effect of PURB on DDB2. Our results showed that silencing LINC01320 led to a significant decrease in the binding ability of PURB to the DDB2 promoter (Fig. 4H, I).
LINC01320 promoted the progression of OC by targeting the DDB2/NEDD4L/TGF-β axis
DDB2 can transcriptionally repress the expression of the E3 ligase NEDD4L18. In turn, NEDD4L negatively regulates the TGF-β signaling pathway by targeting phosphorylated Smad2 (p-Smad2) to proteasomal degradation40. Therefore, DDB2 exerts tumor-suppressive effects by increasing the enhancing the TGF-β pathway18. In line with the above findings, we observed that the expression of the NEDD4L transcript was significantly reduced when silencing LINC01320 and PURB in OC cells, while increased when silencing DDB2 (Fig. 5A). To investigate whether LINC01320 exerts its oncogenic role via the DDB2/NEDD4L/TGF-β signaling, we detected DDB2/NEDD4L/TGF-β levels in LINC01320-knockdown cells by Western blotting. As expected, suppression of LINC01320 resulted in increased expression of DDB2 and p-Smad2 and reduced the levels of NEDD4L, whereas total Smad expression remained unchanged (Fig. 5B-E). To provide further evidence for the involvement of the DDB2/NEDD4L/TGF-β axis in the LINC01320-mediated progression of OC, si-DDB2 and the small-molecule inhibitor of the TGF-β/Smad pathway, SB431542, were applied to LINC01320-silenced OC cell lines. Cell-based functional assays demonstrated that inhibition of DDB2 or the TGF-β pathway significantly restored the proliferation and migration capacity of LINC01320-knockdown OC cells (Fig. 5F-K). Furthermore, receiver operating characteristic (ROC) curve analyses further confirmed the prognostic value of the integrated LINC01320/PURB/DDB2 gene signature in OC (Figure S2).
LINC01320 promotes the progression of OC by targeting /DDB2/NEDD4L/TGF-β axis. (A) OC cells transfected with si-NC, si-LINC01320, si-PURB, or si-DDB2 were analyzed by RT-qPCR for NEDD4L expression, n = 3 per group. (B–E) Western blotting was used to determine the protein expression of DDB2, NEDD4L, SMAD2, and p-SMAD2 in SKOV3 (B and D) and OVCAR3 (C and E) cell lines transfected with si-LINC01320 or si-NC, tubulin was used as an internal control, n = 3 per group. (F–K) OC cells were transfected with si-NC, si-LINC01320, si-LINC01320 + si-DDB2, or si-LINC01320 + SB431542 (TGF-β inhibitor, 30 µM). CCK-8 assays were used to verify the viability of OC cells (F, G), n = 6 per group. Colony formation assays were performed to detect proliferation ability (H, I), n = 3 per group. Transwell assays were used to investigate changes in migratory ability (J, K), n = 3 per group. Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Recent investigations have elucidated that the oncogene LINC01320 is markedly overexpressed in both pancreatic16 and gastric cancers17, promoting cancer cell proliferation and migration through targeted modulation of the miR-324-3p and miR-495-5p/RAB19 axis, respectively. However, the role and molecular mechanisms of LINC01320 in OC remain to be elucidated. In this study, based on the analysis of patient samples and OC databases, LINC01320 was found to be highly expressed in OC tissues. Previous studies have shown that lncRNAs can interact with transcriptional regulatory/auxiliary factors, forming ribonucleoprotein (RNP) complexes that regulate the expression of downstream genes41. In the present study, through RNA pull-down-LC-MS/MS assays, PURB emerged as a molecule of significant interest. PURB, a known evolutionarily conserved modulator of transcription ubiquitously expressed across eukaryotes42,43,44, can bind specifically and with high affinity to purine-rich single-stranded DNA (ssDNA) or RNA45,46, and is associated with tumor progression, cellular senescence, and postnatal brain development47,48,49. Previous research has indicated that PURB is a key regulator of gene transcription and cellular physiology. For instance, linc-HOXA1 represses the expression of Hoxa1 in embryonic stem cells by recruiting PURB protein as a transcriptional auxiliary factor35. Similarly, mammary tumor-associated RNA 25 (MaTAR25), also known as LINC01271, modulates the expression of the downstream target gene Tensin1 (Tns1) by interacting with PURB, thus influencing the progression of breast cancer33. Consequently, in this study, PURB was selected as a candidate protein for interaction with LINC01320, which was further confirmed by RNA extraction and RIP-qPCR experiments. Furthermore, silencing PURB phenocopied the effects of LINC01320 knockdown in OC cells.
Given that PURB acts as a transcriptional repressor37, we hypothesized that LINC01320 modulates the expression of downstream genes in OC by recruiting PURB. Through RNA-seq, we identified DDB2 as a previously undiscovered downstream target gene regulated by LINC01320 and PURB in OC cells and observed that knockdown of either LINC01320 or PURB resulted in the upregulation of DDB2 expression. DDB2, a 48 kDa protein initially identified for its capacity to bind UV-damaged DNA as an initial damage recognition factor50,51, has been implicated in tumor progression, with attenuated expression in cisplatin-resistant OC cell lines52, colorectal53 and skin cancers54. Moreover, low DDB2 expression correlates with poor prognosis in OC38, a relationship that extends to breast cancer patients55. Through sequence analysis, we found that the promoter region of DDB2 is enriched with PNR. ChIP-qPCR and dual-luciferase reporter assays delineated the interaction between PURB and PNR within the DDB2 promoter. Subsequently, our study revealed that LINC01320 is required for PURB-DDB2 interactions and further transcriptional repression, since knockdown of LINC01320 abolished PURB-DDB2 interactions without altering PURB expression. Collectively, our findings suggest that LINC01320 is instrumental in recruiting PURB to the DDB2 promoter, thus mediating transcriptional repression of DDB2.
Previous studies have indicated that overexpression of DDB2 alleviates the progression of OC by downregulating NEDD4L, thus amplifying the antiproliferative effects of the TGF-β signaling pathway18. In this study, we observed a reduction in NEDD4L expression after silencing LINC01320 and PURB in OC cells, whereas silencing DDB2 led to its increase. Considering that NEDD4L plays a pivotal role in suppressing the TGF-β signaling pathway by targeting activated Smad2 (p-Smad2) for proteasomal degradation18,40. Our data suggest that inhibition of LINC01320 decreases NEDD4L levels while increasing p-Smad2, with the total Smad expression remaining relatively stable. Furthermore, rescue experiments further affirmed that LINC01320 promotes the proliferation and migration of OC by impeding the DDB2-mediated TGF-β signaling pathway in OC. In summary, this study provides further evidence of the involvement of the DDB2/NEDD4L/TGF-β axis in LINC01320-mediated OC progression.
Our study delineates a novel mechanism in which the LINC01320/PURB/DDB2/NEDD4L/TGF-β axis serves as the foundation for the progression of OC (Fig. 6). The interaction between LINC01320 and PURB mediates the transcriptional repression of DDB2, concurrently substantiating the participation of the DDB2/NEDD4L/TGF-β axis in LINC01320-mediated OC progression. Despite enriching and expanding understanding of the pathogenesis of OC, limitations in our study should be acknowledged, notably the few clinical samples examined. Consequently, discoveries concerning clinical features, such as TNM staging, require validation through larger-scale multicenter studies. Nonetheless, this study offers deeper insight into the role of LINC01320 in OC, facilitating the early diagnosis and treatment of ovarian cancer.
Data availability
All data pertinent to this study are accessible through the corresponding author, subject to a reasonable request.
References
Lheureux, S., Gourley, C., Vergote, I. & Oza, A. M. Epithelial ovarian cancer. Lancet (London England) 393, 1240–1253. https://doi.org/10.1016/s0140-6736(18)32552-2 (2019).
Eisenhauer, E. A. Real-world evidence in the treatment of ovarian cancer. Ann. Oncol. Official J. Eur. Soc. Med. Oncol. 28, viii61–viii65. https://doi.org/10.1093/annonc/mdx443 (2017).
Wu, Y. B. et al. Long non-coding RNA NRSN2-AS1 promotes ovarian cancer progression through targeting PTK2/β-catenin pathway. Cell Death Dis. 14 https://doi.org/10.1038/s41419-023-06214-z (2023).
Mercer, T. R., Dinger, M. E. & Mattick, J. S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. https://doi.org/10.1038/nrg2521 (2009).
Chandra Gupta, S. & Nandan Tripathi, Y. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int. J. Cancer 140, 1955–1967. https://doi.org/10.1002/ijc.30546 (2017).
Schwarzmueller, L., Bril, O., Vermeulen, L. & Léveillé, N. Emerging role and therapeutic potential of lncRNAs in Colorectal Cancer. Cancers 12 https://doi.org/10.3390/cancers12123843 (2020).
Schmitt, A. M. & Chang, H. Y. Long noncoding RNAs in Cancer pathways. Cancer cell. 29, 452–463. https://doi.org/10.1016/j.ccell.2016.03.010 (2016).
Li, J. et al. Long noncoding RNA-JPX predicts the poor prognosis of ovarian cancer patients and promotes tumor cell proliferation, invasion and migration by the PI3K/Akt/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 22, 8135–8144. https://doi.org/10.26355/eurrev_201812_16505 (2018).
Lin, X., Feng, D., Li, P. & Lv, Y. LncRNA LINC00857 regulates the progression and glycolysis in ovarian cancer by modulating the Hippo signaling pathway. Cancer Med. 9, 8122–8132. https://doi.org/10.1002/cam4.3322 (2020).
Liang, H. et al. LncRNA PTAR promotes EMT and invasion-metastasis in serous ovarian cancer by competitively binding mir-101-3p to regulate ZEB1 expression. Mol. Cancer 17 https://doi.org/10.1186/s12943-018-0870-5 (2018).
Wang, C. et al. LncRNA SPOCD1-AS from ovarian cancer extracellular vesicles remodels mesothelial cells to promote peritoneal metastasis via interacting with G3BP1. J. Exp. Clin. cancer Res. CR 40, 101. https://doi.org/10.1186/s13046-021-01899-6 (2021).
Wu, W., Guo, L., Liang, Z., Liu, Y. & Yao, Z. Lnc-SNHG16/miR-128 axis modulates malignant phenotype through WNT/beta-catenin pathway in cervical cancer cells. J. Cancer 11, 2201–2212. https://doi.org/10.7150/jca.40319 (2020).
Sun, Z., Gao, S., Xuan, L. & Liu, X. Long non-coding RNA FEZF1-AS1 induced progression of ovarian cancer via regulating miR-130a-5p/SOX4 axis. J. Cell. Mol. Med. 24, 4275–4285. https://doi.org/10.1111/jcmm.15088 (2020).
Suhorutshenko, M. et al. Endometrial receptivity revisited: endometrial transcriptome adjusted for tissue cellular heterogeneity. Hum. Reprod. (Oxford England) 33, 2074–2086. https://doi.org/10.1093/humrep/dey301 (2018).
Sun, C., Jiang, H., Sun, Z., Gui, Y. & Xia, H. Identification of long non-coding RNAs biomarkers for early diagnosis of myocardial infarction from the dysregulated coding-non-coding co-expression network. Oncotarget 7, 73541–73551. https://doi.org/10.18632/oncotarget.11999 (2016).
Meng, H., Guo, K. & Zhang, Y. Effects of lncRNA LINC01320 on Proliferation and Migration of Pancreatic Cancer cells through targeted regulation of miR-324-3p. J. Healthc. Eng. 2021 (4125432). https://doi.org/10.1155/2021/4125432 (2021).
Hu, N. & Ji, H. N6-methyladenosine (m6A)-mediated up-regulation of long noncoding RNA LINC01320 promotes the proliferation, migration, and invasion of gastric cancer via miR495-5p/RAB19 axis. Bioengineered 12, 4081–4091. https://doi.org/10.1080/21655979.2021.1953210 (2021).
Zhao, R. et al. DDB2 modulates TGF-β signal transduction in human ovarian cancer cells by downregulating NEDD4L. Nucleic Acids Res. 43 7838–7849. https://doi.org/10.1093/nar/gkv667 (2015).
Goldman, M. J. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678. https://doi.org/10.1038/s41587-020-0546-8 (2020).
Frankish, A. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773. https://doi.org/10.1093/nar/gky955 (2019).
Liu, J. Y. et al. BMI-1 promotes breast cancer proliferation and metastasis through different mechanisms in different subtypes. Cancer Sci. 114, 449–462. https://doi.org/10.1111/cas.15623 (2023).
Yu, X. et al. E3 ubiquitin ligase RNF187 promotes growth of spermatogonia via lysine 48-linked polyubiquitination-mediated degradation of KRT36/KRT84. FASEB J. Official Publ. Feder. Am. Soc. Exp. Biol. 37, e23217. https://doi.org/10.1096/fj.202301120R (2023).
Xu, B. Y. et al. RNF187 governs the maintenance of mouse GC-2 cell development by facilitating histone H3 ubiquitination at K57/80. Asian J. Androl. https://doi.org/10.4103/aja202368 (2023).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. https://doi.org/10.1093/nar/29.9.e45 (2001).
Wang, Q. et al. BMI1 promotes osteosarcoma proliferation and metastasis by repressing the transcription of SIK1. Cancer Cell Int. 22 https://doi.org/10.1186/s12935-022-02552-8 (2022).
Yu, J. et al. Srlp is crucial for the self-renewal and differentiation of germline stem cells via RpL6 signals in Drosophila testes. Cell Death Dis. 10, 294. https://doi.org/10.1038/s41419-019-1527-z (2019).
Zhou, H. et al. The plasminogen receptor directs maintenance of spermatogonial stem cells by targeting BMI1. Mol. Biol. Rep. 49, 4469–4478. https://doi.org/10.1007/s11033-022-07289-1 (2022).
Wu, L., Li, S., Xu, J., Shen, C. & Qian, Q. AGAP2-AS1/BRD7/c-Myc signaling axis promotes skin cutaneous melanoma progression. Am. J. Transl. Res. 15, 350–362 (2023).
Liu, Y. et al. INTS7-ABCD3 Interaction stimulates the proliferation and osteoblastic differentiation of mouse bone marrow mesenchymal stem cells by suppressing oxidative stress. Front. Physiol. 12, 758607. https://doi.org/10.3389/fphys.2021.758607 (2021).
Zhang, K. et al. BMI1 promotes spermatogonia proliferation through epigenetic repression of Ptprm. Biochem. Biophys. Res. Commun. 583, 169–177. https://doi.org/10.1016/j.bbrc.2021.10.074 (2021).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. https://doi.org/10.1186/gb-2010-11-2-r14 (2010).
Yu, J. et al. BMI1 drives steroidogenesis through Epigenetically repressing the p38 MAPK pathway. Front. cell. Dev. Biol. 9, 665089. https://doi.org/10.3389/fcell.2021.665089 (2021).
Chang, K. C. et al. MaTAR25 lncRNA regulates the Tensin1 gene to impact breast cancer progression. Nat. Commun. 11, 6438. https://doi.org/10.1038/s41467-020-20207-y (2020).
Pan, J. et al. Luteolin inhibits viability, migration, angiogenesis and invasion of non-small cell lung cancer vascular endothelial cells via miR-133a-3p/purine rich element binding protein B-mediated MAPK and PI3K/Akt signaling pathways. Tissue cell. 75, 101740. https://doi.org/10.1016/j.tice.2022.101740 (2022).
Maamar, H., Cabili, M. N., Rinn, J. & Raj, A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 27, 1260–1271. https://doi.org/10.1101/gad.217018.113 (2013).
Pandey, P. R. et al. circSamd4 represses myogenic transcriptional activity of PUR proteins. Nucleic Acids Res. 48, 3789–3805. https://doi.org/10.1093/nar/gkaa035 (2020).
Wang, J. et al. The novel long noncoding RNA Lnc19959.2 modulates triglyceride metabolism-associated genes through the interaction with Purb and hnRNPA2B1. Mol. Metab. 37, 100996. https://doi.org/10.1016/j.molmet.2020.100996 (2020).
Han, C. et al. DDB2 suppresses tumorigenicity by limiting the cancer stem cell population in ovarian cancer. Mol. cancer Res. MCR. 12, 784–794. https://doi.org/10.1158/1541-7786.Mcr-13-0638 (2014).
Ramsey, J. E. & Kelm, R. J. Mechanism of strand-specific smooth muscle alpha-actin enhancer interaction by purine-rich element binding protein B (purbeta). Biochemistry 48, 6348–6360. https://doi.org/10.1021/bi900708j (2009).
Gao, S. et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol. Cell 36, 457–468. https://doi.org/10.1016/j.molcel.2009.09.043 (2009).
Balas, M. M. & Johnson, A. M. Exploring the mechanisms behind long noncoding RNAs and cancer. Non-coding RNA Res. 3, 108–117. https://doi.org/10.1016/j.ncrna.2018.03.001 (2018).
Daniel, D. C. & Johnson, E. M. PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions. Gene 643, 133–143. https://doi.org/10.1016/j.gene.2017.12.004 (2018).
Liu, H. & Johnson, E. M. Distinct proteins encoded by alternative transcripts of the PURG gene, located contrapodal to WRN on chromosome 8, determined by differential termination/polyadenylation. Nucleic Acids Res. 30, 2417–2426. https://doi.org/10.1093/nar/30.11.2417 (2002).
Kelm, R. J., Lamba, G. S., Levis, J. E. & Holmes, C. E. Characterization of purine-rich element binding protein B as a novel biomarker in acute myelogenous leukemia prognostication. J. Cell. Biochem. 119, 2073–2083. https://doi.org/10.1002/jcb.26369 (2018).
Kelm, R. J., Cogan, J. G., Elder, P. K., Strauch, A. R. & Getz, M. J. Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle alpha-actin promoter. J. Biol. Chem. 274, 14238–14245. https://doi.org/10.1074/jbc.274.20.14238 (1999).
Kelm, R. J., Wang, S. X., Polikandriotis, J. A. & Strauch, A. R. Structure/function analysis of mouse purbeta, a single-stranded DNA-binding repressor of vascular smooth muscle alpha-actin gene transcription. J. Biol. Chem. 278, 38749–38757. https://doi.org/10.1074/jbc.M306163200 (2003).
Johnson, E. M., Daniel, D. C. & Gordon, J. The pur protein family: genetic and structural features in development and disease. J. Cell. Physiol. 228, 930–937. https://doi.org/10.1002/jcp.24237 (2013).
Khalili, K. et al. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol. Cell. Biol. 23, 6857–6875. https://doi.org/10.1128/mcb.23.19.6857-6875.2003 (2003).
Mulnix, R. E. et al. hnRNP C1/C2 and pur-beta proteins mediate induction of senescence by oligonucleotides homologous to the telomere overhang. OncoTargets Ther. 7, 23–32. https://doi.org/10.2147/ott.S54575 (2013).
Dualan, R. et al. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics 29, 62–69. https://doi.org/10.1006/geno.1995.1215 (1995).
Tang, J. & Chu, G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair. 1, 601–616. https://doi.org/10.1016/s1568-7864(02)00052-6 (2002).
Barakat, B. M. et al. Overexpression of DDB2 enhances the sensitivity of human ovarian cancer cells to cisplatin by augmenting cellular apoptosis. Int. J. Cancer 127, 977–988. https://doi.org/10.1002/ijc.25112 (2010).
Roy, N. et al. DDB2 suppresses epithelial-to-mesenchymal transition in colon cancer. Cancer Res. 73, 3771–3782. https://doi.org/10.1158/0008-5472.Can-12-4069 (2013).
Stoyanova, T. et al. p21 cooperates with DDB2 protein in suppression of ultraviolet ray-induced skin malignancies. J. Biol. Chem. 287, 3019–3028. https://doi.org/10.1074/jbc.M111.295816 (2012).
Ennen, M. et al. DDB2: a novel regulator of NF-κB and breast tumor invasion. Cancer Res. 73, 5040–5052. https://doi.org/10.1158/0008-5472.Can-12-3655 (2013).
Acknowledgements
This work was supported by the Suzhou Gu Su Health Talent Research Project (GSWS2023056).
Author information
Authors and Affiliations
Contributions
C.S., X.C., and T.Z conceived and designed the research. G.W., B.X., X.Y., M.L., T.W., W.G., H.H., and B.J. conducted the experiments. G.W., B.X., X.Y., and Y.W. analyzed the data. G.W., B.X., and C.S. wrote the manuscript. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, G., Xu, B., Yu, X. et al. LINC01320 facilitates cell proliferation and migration of ovarian cancer via regulating PURB/DDB2/NEDD4L/TGF-β axis. Sci Rep 14, 26233 (2024). https://doi.org/10.1038/s41598-024-78255-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-78255-z