Abstract
India is accelerating efforts to eliminate kala-azar by aligning its National Kala-Azar Elimination Program with the World Health Organization’s (WHO) roadmap for Neglected Tropical Diseases (NTDs) 2021–2030. Elimination relies on comprehensive vector surveillance and integrated vector management. This study aimed to conduct nationwide entomological surveillance to detect Leishmania donovani in phlebotomine sand flies. A cross-sectional survey was conducted from January 2022 to December 2023 in five different biogeographical zones in India. Mechanical aspirator, light traps were used for sampling. The collected sand flies were identified to species level. Molecular xenomonitoring was conducted using kDNA qPCR, and parasite characterization targeting ITS1 gene sequencing and RFLP. Sand fly species was confirmed by DNA barcode. Molecular xenomonitoring revealed that Phlebotomus argentipes from Bihar, West Bengal, and Kerala exhibited high levels of L. donovani parasitic DNA. In Rajasthan, P. sergenti and P. papatasi and in Himachal Pradesh, P. longiductus, P. major, and P. bruneyi were positive. The high levels of L. donovani parasitic DNA detected in various Phlebotomus species, along with its presence in other sand fly species beyond the established vectors, underscore the urgent need for the National Kala-Azar Elimination Program to prioritize comprehensive and rigorous vector surveillance. Strengthening these efforts is crucial for achieving the program’s goal of eliminating the disease.
Similar content being viewed by others
Introduction
Leishmaniasis, a neglected tropical disease (NTD), is caused by over twenty species of parasites belonging to the genus Leishmania (Kinetoplastida: Trypanosomatidae). It is transmitted to humans through the bites of infected sand flies from the subfamily Phlebotominae (Diptera: Psychodidae)1. Leishmaniasis occurs in three main forms, such as visceral, the deadliest if untreated; cutaneous, the most common, characterized by skin ulcers, and mucocutaneous, which affects the mouth, throat, and nose. Visceral leishmaniasis (VL), also known as kala-azar, is the second most lethal parasitic disease after malaria2. According to the World Health Organization (WHO), leishmaniasis affects over 98 countries, including the Mediterranean, Central Asia, and East Africa. An estimated 350 million people are at risk, with over 90,000 new cases reported annually, though only 25–45% are officially reported to WHO3.
Countries such as India, Bangladesh, Sudan, Ethiopia, and Brazil account for about 95% of the global VL burden4, with India alone responsible for nearly 18% of the world’s kala-azar cases. In India, VL is primarily recorded in 54 districts across four states, Bihar (33 out of 38 districts), West Bengal (11 out of 23 districts), Uttar Pradesh (6 out of 75 districts) and Jharkhand (4 out of 24 districts)5. According to the National Centre of Vector Borne Disease Control (NCVBDC), India, 595 cases and 4 deaths due to VL and 314 cases of Post kala-azar dermal leishmaniasis (PKDL) were reported in 20236. Sporadic cases of VL are also reported in other states, including Kerala, Himachal Pradesh, Assam, Jammu & Kashmir, Puducherry, Sikkim, Gujarat, and Tamil Nadu5,7,8. Despite significant progress in reducing VL cases by 98% since 19925, the emergence of new endemic foci, like Kerala and Himachal Pradesh, poses challenges to the ongoing disease elimination target8,9. To address this, India’s National Kala-Azar Elimination Program (NKAEP) aligns with the WHO’s new NTD roadmap for 2021–20305,10. Integrated vector management (IVM) and vector surveillance are major arms of NKAEP, providing essential tools for controlling and eventually eliminating leishmaniasis by the targeted year.
Leishmania parasites have a digenetic life cycle, alternating between female sand fly vectors and mammalian hosts. In the old world, vectors belong to the Phlebotomus genera, while in the new world, they belong to Lutzomyia11. Of around 1000 sand fly species, about 10% are proven or suspected leishmaniasis vectors globally12. Whereas in India, 69 sand fly species have been documented, with 13 proven or suspected to transmit anthroponotic or zoonotic leishmaniasis13.
Phlebotomus argentipes is the primary vector for Leishmania donovani, causing VL in India13. Phlebotomus papatasi may be involved in transmitting L. tropica, responsible for CL in regions like Rajasthan14. While L. donovani typically causes VL, recent studies show it also causes atypical cutaneous leishmaniasis (ACL) in Kerala’s tribal populations8. Phlebotomus longiductus is suspected to transmit ACL in Himachal Pradesh7. Many indigenous cases of ACL caused by MON-37 strain of L. donovani have been reported in both regions during last decade7,8. Further, P. salehi has also been shown to transmit L. major experimentally1315.
Epidemiological surveillance of leishmaniasis typically relies on serological assays to detect infections in humans and reservoir hosts, while vector incrimination involves dissecting freshly caught or cryopreserved sand flies to isolate and characterize Leishmania strains from their midguts using iso-enzymatic methods16,17,18. However, PCR has become an essential tool for molecular xenomonitoring of leishmaniasis, enabling the identification of sand fly species involved in disease transmission19,20,21. Recently, PCR-based molecular techniques with high specificity and sensitivity have proven valuable for detecting and identifying Leishmania species in sand fly vectors22,23. Given that IVM and vector surveillance are key components of the NKAEP and the advancements in molecular detection of the parasite, the present study undertook a cross-sectional entomological survey focusing on molecular surveillance of leishmaniasis in five distinct bio-geographical zones of India. These zones were selected based on prior case reports and disease endemicity. The study aimed to conduct molecular xenomonitoring in Phlebotomus spp. to detect L. donovani, the parasite responsible for kala-azar in India.
Results
Sand fly diversity and distribution
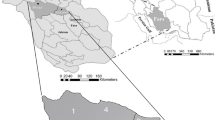
Between January 2022 and December 2023, a total of 6,278 sand fly specimens were collected across various biogeographical zones in India as part of a cross-sectional survey (Fig. 1 [QGIS version 3.36.0-Maidenhead software]). The collection sites were primarily cattle sheds, which yielded more samples compared to human dwellings (10:1). All Phlebotomus spp. samples were mainly collected from cattle shed, whereas Phlebotomus, Sergentomyia, and Grassomyia sp. samples were collected from human dwellings. Eastern zone i.e., Rajasthan was an exception where both Phlebotomus and Sergentomyia spp. samples were found from cattle shed collection with majority being Sergentomyia spp. In the eastern zone, comprising Bihar and West Bengal, the sand fly specimens collected included three different genera: Phlebotomus, Sergentomyia, and Grassomyia. Eight species were identified, with Phlebotomus argentipes, P. papatasi, Sergentomyia babu, S. bailyi, S. insularis, S. punjabaensis, and Grassomyia indica. Among these, P. argentipes was the predominant species. In the northern zone, particularly in Himachal Pradesh, the sand fly specimens were from two genera: Phlebotomus and Sergentomyia. The identified species included P. longiductus, P. major, P. bruneyi, P. colabaensis, P. argentipes, S. babu, S. arboris, and S. kauli. Phlebotomus longiductus was the predominant species, followed by P. bruneyi and P. major. In the western zone, represented by Rajasthan, the sand fly specimens belonged to three genera: Phlebotomus, Sergentomyia, and Grassomyia, totalling thirteen different species. The predominant species recorded during this survey were S. christophersi, followed by S. punjabaensis, S. babu, P. sergenti, S. baghdadis, P. papatasi, P. argentipes, S. shorttii, S. bailyi, S. cyledi, S. eadithae, P. colabaensis, and G. indica. In Kerala, the southern zone, the sand fly specimens belonged to two genera: Phlebotomus and Sergentomyia, with a total of fourteen different species. P. argentipes was the most common, followed by S. babu, S. monticola, S. jerighatiansis, S. zeylanica, P. colabaensis, S. himalayensis, S. dhandai, S. bailyi, S. shorttii, S. insularis, S. baghdadis, S. linearis, and S. hospitii. In the central zone, which included study sites in Madhya Pradesh, sand fly specimens belonged to three genera: Phlebotomus, Sergentomyia, and Grassomyia. Six species were recorded, with P. argentipes as the predominant species, followed by S. babu, S. bailyi, S. punjabaensis, G. indica, and P. colabaensis (Fig. 2 [Microsoft Office 2019 version]).
Molecular xenomonitoring and characterization ofLeishmania parasite
Real-time PCR targeting the kDNA of the Leishmania parasite revealed that different Phlebotomus spp. detected with Leishmania parasitic DNA. In the eastern and southern zones, particularly in Bihar, West Bengal, and Kerala, P. argentipes was identified with Leishmania DNA. In the western zone, P. papatasi and P. sergenti from Rajasthan were detected with the kDNA gene of the Leishmania parasite. In the northern zone, comprising Himachal Pradesh, P. longiductus, P. bruneyi, and P. major were identified as sand fly species with the presence of Leishmania parasitic DNA. Notably, none of the sand fly specimens from Madhya Pradesh contained kDNA gene of the Leishmania parasite (Supplementary Fig. 1).
The cycle quantification (Cq) values for the sand fly specimens found positive for Leishmania kDNA ranged from 15.72 to 39.19 across various biogeographical zones of India (Supplementary Fig. 2). Phlebotomus longiductus from Himachal Pradesh showed the highest levels of Leishmania parasitic DNA among pooled sand flies, with a rate of 61.11%, while P. argentipes from Bihar exhibited the highest individual parasitic DNA levels at 57.14% among the sand fly specimens (Table 1).
The species of the Leishmania parasite was confirmed through ITS1 gene amplification and subsequent RFLP analysis using the HaeIII enzyme. The characteristic banding pattern of 180, 90, and ~ 50 base pairs indicated the presence of L. donovani (Supplementary Fig. 3). This identification was further supported by the ITS1 gene sequence data generated from the positive sand fly specimens, with accession numbers PP481746 to PP481750, PP062796, and PP062797. Phylogenetic analysis of these sequences reinforced the confirmation of the Leishmania species and linked its lineage with other strains of L. donovani from India and Sri Lanka (Fig. 3 [MEGA 7.0.26]).
Phylogenetic analysis of ITS1 gene of Leishmania donovani detected and amplified from sandfly samples detected with parasitic DNA from different bio-geographical zones of India using Kimura 2-parameter model with bootstrap analysis of 1000 replications and maximum likelihood statistical method aligned L. donovani sequence from other countries and L. tropica and L. major as an outgroup.
Molecular confirmation of sand fly species detected with Leishmania parasitic DNA
The molecular identification of the sand flies from various biogeographical zones in India was confirmed through COI barcoding. The sand fly species confirmed to be detected with the Leishmania parasite DNA includes P. argentipes, P. papatasi, P. sergenti, P. longiductus, P. bruneyi, and P. major. The COI sequences generated for these sand fly specimens were submitted to GenBank with accession numbers PP495525 to PP495531 and PP501171. To further validate the species identification, these sequences were aligned with existing sequences of the same species in GenBank, and a phylogenetic tree was constructed. This phylogenetic analysis supported the species confirmation, reinforcing the findings of the study and ensuring accurate identification of the sand fly species carrying the Leishmania parasite (Fig. 4 [MEGA 7.0.26]).
Phylogenetic analysis of COI gene of sand fly samples detected with parasitic DNA from different bio-geographical zones of India using Kimura 2-parameter model with bootstrap analysis of 1000 replications and Neighbor-joining statistical method aligned other similar sequences retrieved from BLAST results of NCBI and Sergentomyia iyengari as an outgroup.
Discussion
The NKAEP, aligned with the new NTDs roadmap 2021–2030 by WHO, was aimed to eliminate kala-azar by 20236,10. Integrated vector management and vector surveillance coupled with case detection and management are the key pillars that could play an important role in controlling disease transmission and hence eliminating it from the country. In consideration of this hypothesis, a cross-sectional entomological survey was initiated to conduct molecular xenomonitoring of leishmaniasis among the sand fly population, particularly in Phlebotomus spp., which are proven, potential, or suspected vectors of the disease in India.
A nationwide survey targeted five distinct biogeographical zones in India: eastern, western, northern, southern, and central. Bihar and West Bengal, in the eastern zone, are endemic for VL, with recent PKDL cases6. Rajasthan, in the western zone, has a unique climate and ecosystem with historical CL outbreaks, which have recently declined32. Himachal Pradesh in the north and Kerala in the south are emerging foci for leishmaniasis, with recent indigenous reports of ACL and VL (from Kerala)7,8,9. In both states, Leishmania donovani MON-37 has been identified as the parasite responsible for disease manifestation. However, while P. argentipes has been confirmed as the vector in the southern region, P. longiductus is only suspected to be a potential vector from the Himachal Pradesh. Madhya Pradesh in the central zone has no reported cases in the past decade. The study found Phlebotomus spp. positive for the Leishmania parasite in all zones except Madhya Pradesh. Specifically, P. argentipes from Bihar, West Bengal, and Kerala, P. papatasi and P. sergenti from Rajasthan; and P. longiductus, P. bruneyi, and P. major from Himachal Pradesh were detected with L. donovani parasitic DNA. Through this study, we report for the first time that P. papatasi, P. sergenti, P. bruneyi, and P. major to be detected with L. donovani parasite.
In India, among the 13 proven, potential, or suspected vectors for leishmaniasis, P. argentipes is the only confirmed vector that transmits L. donovani, the parasite responsible for kala-azar/VL19,33,34,35. This species is widely distributed across the country, with records from more than 16 out of 27 states and union territories of India13. Given its status as the primary vector of L. donovani, the density of P. argentipes is expected to be high in VL-endemic regions. In this study, we also found a high vector density of P. argentipes in endemic and emerging endemic areas for kala-azar, specifically Bihar, West Bengal, and Kerala respectively. Phlebotomus papatasi is a primary vector of L. tropica and L. major in the new and Old World13,36,37 which is also of widespread distribution in India. Our study corroborated this distribution, primarily in the plain regions such as Rajasthan, Bihar, and West Bengal. Similarly, P. sergenti, a common vector for L. tropica, L. major, and the Toscana virus in the Old World36,38 is also widely recorded in various Indian states13, but in our study, it was only collected from study sites in Rajasthan. Phlebotomus longiductus, P. bruneyi, and P. major are sand fly species typically found in high-altitude and colder regions like Uttarakhand, Himachal Pradesh, Jammu and Kashmir, and Ladakh13,21. Our study also found a high density of these sand flies in high-altitude regions of Himachal Pradesh. Although Madhya Pradesh is not endemic for leishmaniasis, it had one of the highest densities of P. argentipes. This is consistent with previous reports13, suggesting the prevalence of P. argentipes along with other vector and non-vector sand fly species.
Phlebotomus argentipes, the primary vector for L. donovani in VL, was recently demonstrated to be involved in the transmission of CL in Kerala’s Western Ghats, suggesting its role in both dermatotropic (ACL) and viscerotropic (VL) manifestations8,19. In Bihar, recent VL outbreaks have been attributed to P. argentipes, which was the only suspected vector39. In another study, laboratory-reared P. argentipes exposed to Leishmania-infected individuals tested positive for the parasite using microscopy and qPCR40. Further, the present study also identified P. argentipes with L. donovani parasitic DNA from Bihar, West Bengal, and Kerala, supporting its role as a primary vector. Phlebotomus papatasi and P. sergenti are proven vectors for L. major and L. tropica respectively in other regions of the world41,42. In India, P. papatasi is the primary vector transmitting L. tropica, causing CL13,36. Our study is the first to report the detection of L. donovani parasite DNA in P. papatasi and P. sergenti, highlighting a potential risk for new disease outbreaks and the possible emergence of visceral leishmaniasis (VL) in these areas of Rajasthan. In high-altitude regions, where the breeding of P. argentipes and P. papatasi is limited, other Phlebotomus spp. could act as vectors if L. donovani is circulating. A study in Himachal Pradesh found blood-fed P. longiductus specimens with L. donovani21, suggesting that other species may play a role in transmission. The present study confirms the presence of L. donovani DNA in P. longiductus, P. bruneyi, and P. major, with P. longiductus showing a particularly high level of parasitic DNA. This suggests that parasite circulation could be sustained even in areas with a low density of traditional vectors. The report of Lata et al., 202221 from Himachal Pradesh underscores the potential for multiple species to serve as vectors in areas with varying sand fly populations.
Research from various parts of the world indicates that Phlebotomus spp. has a strong preference for feeding on mammalian hosts35,43,44. Studies focusing on the feeding and resting behavior of P. argentipes confirm that this species tends to feed on a range of mammals and is majorly endophilic (cattle sheds and in and around human dwellings) in nature35,45. Our study found similar results, with a higher number of Phlebotomus spp. resting in cattle sheds compared to human dwellings. Furthermore, all sand flies that tested positive for L. donovani DNA were collected from cattle sheds, either during daytime resting, evening collection, or light trap catches. Despite a high level of Leishmania parasitic DNA among sand flies, there have been no recent outbreaks or reported cases of leishmaniasis in these zones. This suggests that the parasite may be circulating among domesticated animals such as cattle, dogs etc., which is hypothesised to be the reservoir hosts based on serological positivity18. In the absence of known hosts Leishmania transmission could occur to humans46. In many of these areas, domesticated animals often share the same shelters with humans, a common scenario among the underprivileged. Such scenario can increase the risk of disease transmission and outbreaks if infected sandflies bites humans. In such cases, strategies like zooprophylaxis or the use of endectocides to prevent sand fly bites and population control could be effective47,48,49.
Although Madhya Pradesh is currently a non-endemic state for leishmaniasis, it has had sporadic case reports in the past, including imported cases of VL due to migratory populations from Bihar5,50. This raises concerns and suggests a potential risk for disease transmission, given that it has one of the highest densities of P. argentipes and the local population often lives in poor conditions alongside domesticated animals. The combination of a large population of P. argentipes, a sand fly species susceptible to infection, and continued migration from endemic states heightens the risk of a leishmaniasis outbreak. Given these factors, proactive measures are essential to monitor and control the potential spread of the disease.
In conclusion, the current study provides vital information on the molecular xenomonitoring of phlebotomine sand flies through cross-sectional surveillance across various biogeographical zones in India, as the country advances toward eliminating kala-azar in the current year. Through the study, six Phlebotomus species were identified with the presence of L. donovani DNA. Notably, this study is the first to report P. papatasi, P. sergenti, P. bruneyi, and P. major as potential vector detected with L. donovani parasitic DNA. The combination of a high level of Leishmania parasitic DNA in sand flies, poor living conditions, proximity to domesticated animals, migratory populations from endemic to non-endemic states, and high sand fly density underscores the risk of leishmaniasis transmission and the potential for outbreaks under favourable conditions. As India works toward eliminating kala-azar in alignment with the new NTDs roadmap, the program must emphasize IVM and vector surveillance as case management alone will not suffice in this critical phase. All pillars of the NKAEP must work in concert to prevent new outbreaks and achieve final disease elimination. Establishing a robust vector and disease surveillance system will be crucial to avoid a resurgence of kala-azar in the country.
The current study however reports P. papatasi, P. sergenti, P. bruneyi, P. longiductus, and P. major specimens to be detected with L. donovani parasitic DNA, but has the limitation of confirming those species as vectors involved in the transmission of the parasite. Future studies can focus on confirmation of these species as vectors of L. donovani.
Methods
Study area
Between January 2022 and December 2023, we conducted an entomological survey in five biogeographical zones in India to investigate leishmaniasis (Fig. 1). The zones were selected based on endemicity and case reports. Surveys were done in Bihar (Vaishali) and West Bengal (Malda) in the eastern zone, Rajasthan (Bikaner, Ajmer, Nagaur, Jodhpur) in the western zone, Himachal Pradesh (Shimla, Kinnaur, Kullu, Mandi) in the northern zone, Kerala (Thrissur, Kollam, Malappuram, Thiruvananthapuram) in the southern zone, and Madhya Pradesh (Bhopal, Sagar, Hoshangabad) in the central zone. Bihar and West Bengal were chosen for high VL rates, featuring a subtropical climate and dense population. Rajasthan was selected for its history of CL cases, characterized by arid deserts and semi-arid regions. Himachal Pradesh was included for its varied topography and emergence as an endemic area for ACL. Kerala, a biodiversity hotspot, was surveyed for the coexistence of VL and ACL. Madhya Pradesh, with no recorded leishmaniasis cases, served as a control, offering a contrast to endemic zones.
Sand fly collection
The entomological collections were planned in each zone in consideration of the ecosystem, climate, and rainfall. Sand fly collections from each zone were conducted when sand fly density was expected to be highest, typically just before the start of the rainy season. Five different collection sites were chosen within each study district from different states across the biogeographical zones. The sand flies were sampled at a single time point by cross-sectional sampling approach. Standard methods like mechanical aspirators and modified CDC light traps were used to conduct a complete sand fly collection in both domestic and peri-domestic areas, such as cattle sheds and human dwellings24. The houses (120–140 house per zone) within the study sites were selected randomly to ensure comprehensive representation of the area for sampling. Resting collections with mechanical aspirators were carried out during the morning (0900 to 1200) and evening (1700 to 1900) hours, while trap (35 traps per zone) collections took place from 1800 (the preceding day) to 0600 (the subsequent day). The traps were placed about one meter above ground level in each sampling site24.
Taxonomic identification of sand flies
Sand fly specimens were transported to the Indian Council of Medical Research - Vector Control Research Centre (ICMR-VCRC) Field Station in Kottayam, Kerala, India and preserved in 70% ethanol. For each sand fly, the head and the last three abdominal segments were dissected under a stereo microscope (Weswox Optik-SZM-100, India) and mounted in Hoyer’s medium on microscopic glass slides for morphological species identification. Identification was performed using a Zeiss binocular microscope (Primostar 3, Carl Zeiss Suzhou Co., Ltd., China), following standard taxonomic keys25,26. The remaining parts of the dissected sand fly specimen (the thorax, legs, abdomen and wings) were preserved in 70% ethanol for the detection of parasitic DNA and DNA barcoding for molecular identification of the sand flies.
Genomic DNA extraction from sand flies
Female sand flies were homogenized using a mortar and pestle, and whole genomic DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN, Hannover, Germany) following the manufacturer’s instructions. The DNA extraction process was conducted under aseptic conditions to avoid contamination of the samples. The extracted DNA was eluted in nuclease free water and stored at -40 °C until further analysis.
Molecular xenomonitoring and characterization ofLeishmania parasite
Detection of Leishmania parasitic DNA
Phlebotomus spp. are known vectors for leishmaniasis in India, making it essential to assess their infection status. Samples of P. argentipes and P. papatasi were analyzed individually, while those of other Phlebotomus spp. (P. longiductus, P. bruneyi, P. major, and P. sergenti) were pooled in groups of up to six for DNA extraction. The samples were then tested for the presence of Leishmania parasites using real-time polymerase chain reaction (qPCR)27. The qPCR was conducted on a Light Cycler® 96 system (Roche Life Science), following the protocol outlined by Vitale et al., 200428,29 (Table 2). The results were analyzed using LightCycler® 96 software to determine the quantification cycle (Cq).
Characterization of Leishmania parasite
Characterization of the Leishmania parasite among real-time PCR-positive samples were achieved through conventional PCR amplification of the internal transcribed spacer 1 (ITS1) region (Table 2), followed by restriction digestion of the amplicon with the HaeIII enzyme30. PCR products were separated on an ethidium bromide (EtBr) stained 1.5% agarose gel. The resulting 320 bp amplicons from positive samples were purified using the QIAquick gel extraction kit (QIAGEN, CA, USA), following the manufacturer’s protocol. These purified amplicons were then custom sequenced.
Restriction fragment length polymorphism (RFLP) of ITS1
The ITS1-positive samples were subjected to a restriction fragment length polymorphism (RFLP) assay using the HaeIII restriction endonuclease enzyme (NEW ENGLAND Biolabs®) to identify Leishmania species, following the protocols described by Schönián et al., 200330. The reaction setup followed the manufacturer’s protocol with the required components. Subsequently, the digested products were visualised on a 2.5% agarose gel.
Molecular confirmation of sand fly species detected with Leishmania parasitic DNA
DNA extracted from the legs of individual sand fly species detected with the Leishmania parasitic DNA was subjected to barcode PCR targeting the mitochondrial cytochrome oxidase I (COI) gene (~ 720 bp), following the protocol outlined by Kumar et al., 201231 (Table 2). The resulting amplicons were purified and custom sequenced.
GenBank submission
All the sequence generated (ITS1 and COI gene) were edited and analysed to generate contigs using Chromas 2.6.6 and MEGA 7.0.26 software. Later sequences were submitted in National Center for Biotechnology Information, GenBank.
Phylogenetic analysis
The nucleotide sequences obtained in this study underwent BLAST analysis using NCBI’s non-redundant (nr) database. Relevant homologous sequences for the ITS1 and COI gene for related Leishmania species and sand flies were retrieved from GenBank, then aligned using MEGA 7.0.26. The optimal DNA substitution model for phylogenetic tree construction was identified with MEGA. The maximum likelihood method with 1,000 bootstrap replications was carried out to obtain the genetic lineage of Leishmania, while the Neighbor-joining method confirmed the identity of sand fly species detected with Leishmania DNA using the Kimura 2-parameter model. Out-groups of related species and genera validated the phylogenetic results, offering robust insights into disease transmission.
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on request. Sequence generated in this study is available from NCBI GenBank with the accession number: PP481745 - PP481750, PP062796, PP062797, PP495525 - PP495531 and PP501171). https://doi.org/10.6084/m9.figshare.27170847.v1All authors approved and gave their consent to publish the manuscript.
References
Cecílio, P., Cordeiro-da-Silva, A. & Oliveira, F. Sand flies: Basic information on the vectors of leishmaniasis and their interactions with Leishmania parasites. Commun. Biol. 5, 305 (2022).
Desjeux, P. The increase in risk factors for leishmaniasis worldwide. Trans. R. Soc. Trop. Med. Hyg. 95, 239–243 (2001).
WHO & Leishmaniasis (2024). https://www.who.int/news-room/fact-sheets/detail/leishmaniasis
Alvar, J. et al. Leishmaniasis Worldwide and global estimates of its incidence. PLoS ONE. 7, e35671 (2012).
WHO. Leishmaniasis - India. https://www.who.int/india/health-topics/leishmaniasis (2024).
National Center for Vector Borne Diseases Control (NCVBDC). Kala-azar. https://ncvbdc.mohfw.gov.in/index1.php?lang=1&level=1&sublinkid=5774&lid=3692 (2024).
Lata, S., Kumari, S., Das, R., Pasi, S. & Dhiman, R. C. Typical and atypical cutaneous leishmaniasis in Himachal Pradesh (India). Heliyon. 7, e07282 (2021).
Saini, P. et al. Cutaneous and visceral Leishmaniasis caused by the same zymodeme of Leishmania Donovani in Kerala, India. Am. J. Trop. Med. Hyg. 110, 59–63 (2024).
Thakur, L. et al. Leishmania donovani infection atypicaly cutaneous a manifestationsations, Himachal Pradesh, India, 2014–2018. Emerg. Infect. Dis. 26, 1864–1869 (2020).
WHO. Ending the neglect to attain the Sustainable Development Goals: A road map for neglected tropical diseases 2021–2030. https://www.who.int/publications/i/item/9789240010352 (2021).
Killick-Kendrick, R. The biology and control of Phlebotomine sand flies. Clin. Dermatol. 17, 279–289 (1999).
Alkan, C. et al. Sandfly-borne phleboviruses of Eurasia and Africa: Epidemiology, genetic diversity, geographic range, control measures. Antiviral Res. 100, 54–74 (2013).
Shah, H., Fathima, P., Kumar, N., Kumar, A. & Saini, P. Faunal richness and checklist of sandflies (Diptera: Psychodidae) in India. Asian Pac. J. Trop. Med. 16, 193 (2023).
Aara, N. et al. Clinco-epidemiologic study of cutaneous leishmaniasis in Bikaner, Rajasthan, India. Am. Soc. Trop. Med. Hyg. 89, 111–115 (2013).
Davami, M. H. et al. First microscopical and molecular-based characterization of Leishmania major within naturally infected Phlebotomus Salehi (Diptera; Psychodidae) in Fars Province, southern Iran. Ann. Trop. Med. Parasitol. 105, 485–491 (2011).
Izri, M. A. & Belazzoug, S. Phlebotomus (Larroussius) perfiliewi naturally infected with dermotropic Leishmania infantum at Tenes, Algeria. Trans. R. Soc. Trop. Med. Hyg. 87, 399 (1993).
Aït-Oudhia, K., Harrat, Z., Benikhlef, R., Dedet, J. P. & Pratlong, F. Canine Leishmania infantum enzymatic polymorphism: A review including 1023 strains of the Mediterranean area, with special reference to Algeria. Acta Trop. 118, 80–86 (2011).
Jambulingam, P., Pradeep Kumar, N., Nandakumar, S., Paily, K. P. & Srinivasan, R. Domestic dogs as reservoir hosts for Leishmania Donovani in the southernmost Western ghats in India. Acta Trop. 171, 64–67 (2017).
Srinivasan, R., Kumar, N. P. & Jambulingam, P. Detection of natural infection of Leishmania donovani (Kinetoplastida: Trypanosomatidae) in Phlebotomus argentipes (Diptera: Psychodidae) from a forest ecosystem in the western ghats, India, endemic for cutaneous leishmaniasis. Acta Trop. 156, 95–99 (2016).
Ranasinghe, S., Rogers, M. E., Hamilton, J. G. C., Bates, P. A. & Maingon, R. D. C. A real-time PCR assay to estimate Leishmania chagasi load in its natural sand fly vector Lutzomyia Longipalpis. Trans. R. Soc. Trop. Med. Hyg. 102, 875–882 (2008).
Lata, S., Kumar, G., Ojha, V. P. & Dhiman, R. C. Detection of Leishmania Donovani in Wild-Caught Phlebotomine Sand flies in endemic focus of Leishmaniasis in Himachal Pradesh, India. J. Med. Entomol. 59, 719–724 (2022).
Muñoz, C. et al. Molecular xenomonitoring and host identification of Leishmania sand fly vectors in a Mediterranean periurban wildlife park. Transbound. Emerg. Dis. 66, 2546–2561 (2019).
Phuphisut, O. et al. Sand fly identification and screening for Leishmania spp. in six provinces of Thailand. Parasites Vectors. 14, 352 (2021).
Alexander, B. Sampling methods for phlebotomine sandflies. Med. Vet. Entomol. 14, 109–122 (2000).
Kalra, N. & Bang, Y. Manual on Entomology in Visceral Leishmaniasis (World Health Organization, 1988).
Lewis, D. The Phlebotomine Sandflies (Diptera: Psychodidae) of the Oriental Region.. Vol. 37 (Bulletin of the British Museum (Natural History), 1978).
Castelli, G. et al. Molecular diagnosis of Leishmaniasis: quantification of parasite load by a real-time PCR assay with high sensitivity. Pathogens. 10, 865 (2021).
Vitale, F. et al. TaqMan-Based detection of Leishmania infantum DNA using canine samples. Ann. N. Y. Acad. Sci. 1026, 139–143 (2004).
El Tai, N. O., Osman, O. F., Fari, E., Presber, M., Schönian, G. & W. & Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania Donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans. R. Soc. Trop. Med. Hyg. 94, 575–579 (2000).
Schönian, G. et al. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn. Microbiol. Infect. Dis. 47, 349–358 (2003).
Kumar, N. P., Srinivasan, R. & Jambulingam, P. DNA barcoding for identification of sand flies (Diptera: Psychodidae) in India. Mol. Ecol. Resour. 12, 414–420 (2012).
Rajni, E. et al. Cutaneous leishmaniasis in Bikaner, India: Clinicoepidemiological profile; parasite identification using conventional, molecular methods and CL Detect™ rapid test, a new Food and Drug Administration-approved test. Trop. Parasitol. 9, 115 (2019).
Sumova, P. et al. PpSP32-like protein as a marker of human exposure to Phlebotomus argentipes in Leishmania Donovani foci in Bangladesh. Int. J. Parasitol. 51, 1059–1068 (2021).
Roy, L. et al. The ongoing risk of Leishmania Donovani transmission in eastern Nepal: An entomological investigation during the elimination era. Parasites Vectors. 16, 404 (2023).
Poché, D. M. et al. Bionomics of Phlebotomus argentipes in villages in Bihar, India with insights into efficacy of IRS-based control measures. PLoS Negl. Trop. Dis. 12, e0006168 (2018).
Karmaoui, A., Sereno, D., Jaafari, E., Hajji, L. & S. & A systematic review and global analysis of the seasonal activity of Phlebotomus (Paraphlebotomus) sergenti, the primary vectors of L. Tropica. PLoS Negl. Trop. Dis. 16, e0010886 (2022).
Jahanifard, E., Navidpour, S. & Vazirianzadeh, B. Study on phlebotominae of two big marshlands of Khoozestan province, Iran. J. Exp. Zool. India. 12, 407–408 (2009).
Es-Sette, N. et al. Phlebotomus sergenti a common vector of Leish tropicaropica and Toscana virus in Morocco. J. Vector Borne Dis. 51, 86–90 (2014).
Priyamvada, K. et al. Visceral leishmaniasis outbreaks in Bihar: Community-level investigations in the context of elimination of kala-azar as a public health problem. Parasites Vectors. 14, 52 (2021).
Singh, O. P. et al. Xenodiagnosis to evaluate the infectiousness of humans to sandflies in an area endemic for visceral leishmaniasis in Bihar, India: A transmission-dynamics study. Lancet Microbe. 2, e23–e31 (2021).
Kykalová, B., Tichá, L., Volf, P. & Loza Telleria, E. Phlebotomus papatasi antimicrobial peptides in Larvae and females and a gut-specific defensin upregulated by Leishmania major infection. Microorganisms. 9, 2307 (2021).
Al-Bajalan, M. M. M., Niranji, S. S., Al-Jaf, S. M. & Kato, H. First molecular identification of Leishmania major in Phlebotomus papatasi in an outbreak cutaneous leishmaniasis area in Iraq. Acta Trop. 215, 105807 (2021).
Yousefi, S. et al. Determination of the feeding behavior of Phlebotomus sergenti using multiplex PCR and tent-baited traps in a new focus of anthroponotic cutaneous leishmaniasis in the southeast of Iran. Exp. Parasitol. 244, 108426 (2023).
Leonel, J. A. F. et al. Species, natural Leishmania spp. detection and blood meal sources of phlebotomine sandflies (Diptera: Psychodidae: Phlebotominae) in Peridomiciles from a Leishmaniases endemic area of Brazil. Transbound. Emerg. Dis. 1–10 (2024).
Dinesh, S., Ranjan, A. & Palit, A. Seasonal and nocturnal landing/biting behaviour of Phlebotomus argentipes (Diptera: Psychodidae). Ann. Trop. Med. Parasitol. 95, 197–202 (2001).
Glidden, C. K. et al. Phylogenetic and biogeographical traits predict unrecognized hosts of zoonotic leishmaniasis. PLoS Negl. Trop. Dis. 17, e0010879 (2023).
Senanayake, S. S. C., Abeyewicreme, W., Dotson, E. M. & Karunaweera, N. D. Characteristics of phlebotomine sandflies in selected areas of Sri Lanka. Southeast. Asian J. Trop. Med. Public. Health. 46, 994–1004 (2015).
Poché, D. M., Wang, H. H. & Grant, W. E. Visceral leishmaniasis on the Indian subcontinent: Efficacy of fipronil-based cattle treatment in controlling sand fly populations is dependent on specific aspects of sand fly ecology. PLoS Negl. Trop. Dis. 14, e0008011 (2020).
Miglianico, M. et al. Repurposing isoxazoline veterinary drugs for control of vector-borne human diseases. Proc. Natl. Acad. Sci. USA 115 (2018).
Nandedkar, S., Malukani, K. & Varma, A. Maiden visit of visceral leishmaniasis to Malwa region. J. Commun. Dis. 43, 233–235 (2011).
Acknowledgements
We are thankful to the Director, Directorate of Scheduled Tribe Development Department, and the Chief Conservator of Forest and Chief Wild Life Warden, Government of Kerala for allowing us to carry out the survey in the tribal hamlets of Western Ghats. We would also like to acknowledge Dr. Bidyut Nandi (Malda College, West Bengal), Mr. Krishna Prasad, (ICMR-VCRC), Ms. Lanza Achu Thomas (ICMR-VCRC), Mr. Chaman Kaushal (HP University), Mr. Ranglal Meena (ICMR- NIIRNCD, Jodhpur), Mr. Biju (ICMR- RMRIMS, Patna), Mr. Niranjan (ICMR- NIREH, Bhopal) for their support and coordination in collection of sand fly samples during the study.
Funding
This study is supported by the Indian Council of Medical Research, New Delhi (Grant no. 6/9 − 7(331)/2020/ECD-II).
Author information
Authors and Affiliations
Contributions
PS contributed to the conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, review and editing draft. HSK contributed to the data curation, formal analysis, investigation, methodology, software, writing original, review and editing draft. FPA contributed to the data curation, investigation, methodology, review and editing draft. APM contributed to the data curation, formal analysis, investigation, methodology, review and editing draft. AK (Ashish Kumar), AR, MST, SSM, DKS and KP contributed to the methodology and, review and editing draft. AK (Ashwani Kumar) and MR contributed to the methodology, project administration and, review and editing draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shah, H.K., Fathima, P.A., Ajithlal, P.M. et al. Nationwide cross-sectional surveillance of Leishmania donovani in phlebotomine sand flies and its impact on national kala-azar elimination in India. Sci Rep 14, 28455 (2024). https://doi.org/10.1038/s41598-024-78915-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-78915-0
Keywords
This article is cited by
-
AI-driven analysis by identifying risk factors of VL relapse in HIV co-infected patients
Scientific Reports (2025)