Abstract
Intracerebral hemorrhage (ICH) is a severe and sudden medical condition caused by the rupture of blood vessels in the brain, leading to permanent damage to brain tissue and often resulting in functional disabilities or death in patients. Diagnosis and analysis of ICH typically rely on brain CT imaging. Given the urgency of ICH conditions, early treatment is crucial, necessitating rapid analysis of CT images to formulate tailored treatment plans. However, the complexity of ICH CT images and the frequent scarcity of specialist radiologists pose significant challenges. Therefore, we collect a dataset from the real world for ICH and normal classification and three types of ICH image classification based on the hemorrhage ___location, i.e., Deep, Subcortical, and Lobar. In addition, we propose a neural network structure, dual-task vision transformer (DTViT), for the automated classification and diagnosis of ICH images. The DTViT deploys the encoder from the Vision Transformer (ViT), employing attention mechanisms for feature extraction from CT images. The proposed DTViT framework also incorporates two multilayer perception (MLP)-based decoders to simultaneously identify the presence of ICH and classify the three types of hemorrhage locations. Experimental results demonstrate that DTViT performs well on the real-world test dataset.
Similar content being viewed by others
Introduction
Intracerebral hemorrhage (ICH) is a type of severe condition characterized by the formation of a hematoma within the brain parenchyma1,2. Representing 10-15% of all stroke cases, ICH is linked to significant morbidity and mortality rates3. Head computerized tomography (CT) is the standard method to diagnose ICH that can obtain accurate images of the head anatomical structure and detect abnormalities. However, analyses of head CT images for ICH classification and diagnosis usually require skilled radiologists, who are in very short supply in developing countries or regions. This may lead to delays in setting appropriate treatment plans and interventions, which are very crucial for accurate ICH patients.
Furthermore, the diagnosis and analysis of ICH in CT images often require extensive experience and concentrated attention. However, when physicians are overworked or face a high volume of cases, misdiagnoses can inevitably occur. Therefore, the use of assistive medical technologies to aid physicians in diagnosing and analyzing ICH images can enhance the speed of diagnosis and treatment. This not only alleviates the shortage of medical resources but also improves diagnostic efficiency and accuracy, which are of significant importance to both physicians and patients.
In recent years, computer vision methods based on deep learning have played a crucial role in CT imaging diagnostics4. Literature4 raises the challenges, opportunities, and future prospects of using computer vision methods in enhancing diagnostic accuracy, automating image analysis, improving patient outcomes. Neural network models trained on extensive real-world data have demonstrated high accuracy and efficiency on medical images5,6,7,8. There are studies that have utilized computer vision and deep learning techniques for ICH image detection and classification to aid medical professionals and enhance diagnostic efficiency9,10,11. In literature9, a deep learning model based on ResNet and EfficientDet that detects bleeding in CT scans is proposed, offering both classification and region-specific decision insights and achieving an accuracy of 92.7%. Literature10 demonstrates the efficacy of convolutional neural network (CNN)-based deep learning models, particularly CNN-2 and ResNet-50, in classifying strokes from CT images, with future plans to optimize these models for improving diagnostic accuracy and efficiency. In Literature11, an evolutionary-based ensemble learning model from brain tumor classification is proposed, which contributed to the treatment plans for patients with brain tumor. Li et al.12 propose a Unet-based neural network model to detect hemorrhage strokes of CT images, achieving an accuracy of 98.59%.
Datasets are essential for training high-performance neural network models, particularly in the context of medical imaging. However, challenges such as limited access to diverse cultural artifacts, varying image quality standards, and restrictions on image usage rights add complexity to the creation of high-quality ICH image datasets. Several publicly available ICH image datasets exist, such as the brain CT images with intracranial hemorrhage masks published on Kaggle, which includes 2500 CT images from 82 patients, though it is relatively small in size13. Another dataset contains high-resolution brain CT images with 2,192 sets of images for segmentation14. Additionally, the RSNA 2019 public dataset, with 874,035 CT images, represents the largest collection available15.
Compared to existing datasets, our dataset offers unique characteristics. Firstly, our data was collected over the past four years, from 2020 to 2024, allowing it to reflect the symptom characteristics of ICH under recent social conditions. Secondly, our dataset includes 15,936 CT images from 249 patients, with images meticulously filtered by medical physicians to remove meaningless noise. Finally, and most importantly, our ICH CT images are labeled based on hemorrhage ___location, which is critical for diagnosis and treatment planning for ICH patients.
Transformer, a network architecture based on the self-attention mechanism, has achieved remarkable success in the field of natural language processing (NLP) in recent years16. Building on this success, researchers have extended Transformer to the field of computer vision, introducing Vision Transformer (ViT)17. The ViT model segments images into patches and feeds them into the Transformer, and then, computes attention weights between different pixel blocks, facilitating effective feature extraction from images. Experimental results demonstrate that Vision Transformers achieve superior and more promising performance compared to classical CNN-based neural networks.
Transfer learning is widely used in deep learning to reduce training time and improve training efficiencies. A pre-trained model is firstly built on a large benchmark dataset such as ImageNet18, and common features are captured and saved in the weights of models. When applying the model to a new dataset or different tasks, we can train the model using task-specific datasets on top of the pre-trained model. Transfer learning also reduces the need for large labeled datasets, making deep learning on small or imbalanced datasets possible, which is particularly suitable for medical images as medical images are usually difficult to obtain.
Building on the aforementioned discussions, we first build an image dataset from real-world sources that includes head images of both healthy individuals and ICH patients, categorized into three types-Deep, Lobar, and Subtentorial-based on the ___location of the hemorrhage. Furthermore, a dual-task Vision Transformer (DTViT) is designed to simultaneously classify images of normal individuals and ICH patients, as well as categorize three types of ICH based on the hemorrhage ___location. We conduct experiments using the new dataset and the proposed model, and the results show that our proposed model achieves superior testing accuracy of \(99.88\%\). The contributions of this paper are summarized as follows:
-
We have constructed a real-world ICH image dataset comprising CT images of both normal individuals and patients classified into three types of ICH, addressing issues of insufficient brain hemorrhage image datasets and the lack of datasets for brain hemorrhage ___location, which is of great significance to both medical and computer vision research.
-
Built upon the constructed dataset, we have developed a deep learning model, i.e., dual-task Vision Transformer (DTViT), which is capable of performing dual-classification tasks simultaneously: distinguishing CT images of normal and ICH patients and identifying the type of ICH according to the hemorrhage ___location.
-
Our comparative experiments show that the DTViT model achieves 99.88% accuracy, outperforming existing CNN models and demonstrating the high quality of our dataset.
The remainder of this paper is structured as follows: first, we describe the dataset and data preprocessing techniques. Next, we introduce the architectures of ViT, DTViT, and the application of transfer learning for image classification and diagnosis. Following that, we detail the experimental setup, evaluation metrics, and present the results obtained. We then discuss the limitations of this study and propose directions for future research. Finally, we conclude the paper.
Data collection and image processing methodology
In this section, we introduce the newly collected dataset, the data preprocessing of ICH images, and the construction of DTVIT for image diagnosis.
Dataset
The dataset is sourced from the Department of Neurology at The First Hospital of Yulin. It includes 15,936 CT slices from 249 patients with intracerebral hemorrhage (ICH) collected between 2018 and 2021, and 6,445 CT slices from 199 healthy individuals in 2024. The healthy subjects have one set of CT images, while the ICH patients have two sets, captured within 24 hours and within 72 hours of symptom onset separately. All images were obtained using the GE LightSpeed VCT scanner at Yulin First Hospital. All scans are saved as DCM files, featuring a resolution of 512\(\times\)512 pixels, a slice thickness of 5 mm, and an inter-slice gap of either 5 mm or 1 mm.
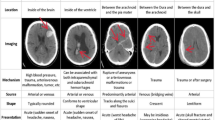
In addition, each group of ICH images is manually classified into three different types according to hemorrhage ___location by an expert physician: Deep ICH, Lobar ICH, and Subtentorial ICH. Figure 1 shows a sample image that includes three types of hemorrhages along with normal images.
To protect patient’s privacy, the dataset only includes patients’ gender and CT images. A detailed data description is listed in Table 1.
Data preprocessing
The data preprocessing process is composed of three stages: morphological treatment, manual filtering (Fig. 2), and data augmentation.
Morphological treatment
When taking CT images, patients are equipped with fixation braces to keep their heads steady, which is also captured by the scanner and may add noise to the image data. Therefore, it is necessary to remove the artifact to purify the data. We apply morphological processing to remove the fixation brace and normalize the images, converting them from DICOM to JPG format. The steps are as follows.
Initially, the pixel array is extracted from the DICOM file and duplicated twice. The first duplicate is used to create a mask, while the second is used to generate the final output. We start by binarizing the pixel array to emphasize key features and apply erosion using a disk-shaped element to minimize noise. Next, the edge columns are zeroed out to remove potential artifacts, and flood filling is performed to enhance specific areas, thus producing the mask image.
Subsequently, this mask is applied to the second, normalized pixel array. The resulting image is then converted to 8-bit integers and saved as a JPG file. An instance of the original CT image and the processed result is displayed in Fig. 3. It can be seen that the fixed brace has been successfully removed and the normalized image is much clearer than the original.
Manual filtering
The head CT scan usually starts from the base of the brain (near the neck) and covers the entire brain up to the forehead. This means that only part of CT scans can capture the hemorrhage ___location and present a clear view for diagnoses, while other CT images do not include effective information and are useless for classification. Therefore, medical specialists manually diagnose and select each group of CT images to remove meaningless images, refine the dataset, and reduce noise. This step is critical to constructing the dataset. Figure 2 shows sample images that were removed from the dataset. These images do not exhibit any signs of hemorrhage.
Data augumentation
As presented in Table 1, The number of images for three types of cerebral hemorrhage locations varies significantly. Therefore, we perform data augmentation operations on subtentorial CT images. Firstly, we augmented the dataset of three types of cerebral hemorrhage CT images proportionally to ensure that the distribution was approximately 1:1:1. Secondly, we also augmented the images without cerebral hemorrhage to achieve a roughly 1:1 ratio between the two categories. The augmented dataset consists of 30,222 images in total, including 13,221 normal images and 17,001 ICH images, which are further categorized into 6093 Deep, 5940 Lobar, and 4968 Subtentorial images. Furthermore, to enhance the diversity of the dataset, we applied image transformation on the training dataset, including center cropping to the size of 224x224, random rotation of 15 degrees, random sharpness adjustment with a factor of 2, and normalization with the mean value [0.485, 0.456, 0.406] and standard deviation [0.229, 0.224, 0.225].
DTViT for ICH CT image diagnosis
This section introduces the structure of DTViT. We first introduce the architecture of DTViT and then discuss the transfer learning and details of the pre-trained model. An overall diagram is shown in Fig. 4.
Structure of vision transformer
Vision Transformer (ViT)17 firstly applies the self-attention mechanism to image classification tasks, which has obtained excellent performances. The ViT, as shown in Fig. 4, consists of several key components: patch embedding, position embedding, Transformer encoder, classification head, and other optional components. The processing procedure is as follows. Firstly, a raw 2D image is initially transformed into a sequence of 1D patch embeddings, which mirrors the word embedding technique used in natural language processing. Similarly, the positional embeddings are applied to the constructed patch embeddings to retain information about the ___location of each patch in the original image. Then, the sequences are fed into the encoder of the Transformer, which consists of multi-head self-attention layers and MLP layers, with normalization and residual connections. It enables the model to capture intricate connections among different patch blocks. Finally, the output is processed by the decoder, i.e., the full-connection layer or MLP layer, to output the classification results.
The details of crucial procedures are as follows. When an \(c\times h \times w\) 2D image (c represents the channel number, h represents image heights, and w represents image width) is fed into the model. The first step is patching embedding, where the image is cut into N image pieces, where \(N=hw/p^{2}\), and p is the specified patch size of the image piece. Then, these image pieces are flattened as a vector with size of \(p^{2}c\) and linear projected to a lower-dimensional space as
where \(W\in \mathbb {R}^{D\times p^{2}c}\) is the weight matrix; \(x_{i}\in \mathbb {R}^{p^{2}c}\) represents the flattened image vector, and \(z_{i}\in \mathbb {R}^{D}\) represents the image vector after projection. Then, a token \(z_{class}\in \mathbb {R}^{D}\) for classification and an embedding recording of the positional information are applied to the vector, yielding the final inputs to the Transformer as
where \(pos_{i}\in \mathbb {R}^{D}\) represent positional embeddings to retain spatial information of image pieces.
Then, the input is processed by the Transformer’s encoder. As shown in Fig. 4, the encoder consists of \(L\) identical blocks, each containing normalization layers, a multi-head attention (MHA) layer, and an MLP layer. The normalization layer is applied before and after the MHA layer, represented as
where \(\mu\) represents the mean of the vector, \(\sigma\) represents the variance of the vector, and \(\epsilon\) is a small constant for numerical stability. Subsequently, the MHA operation is applied to the vector as
where \(X\) is the input after the layer normalization; \(W_{Q}\), \(W_{K}\), \(W_{O}\), and \(W_{V}\) are learnable weight matrices; \(Q\), \(K\), and \(V\) represent the calculated queries, keys, and values. Additionally, the attention calculation formula is as follows:
where \(d\) is the scaling factor, which is the dimension of the query, key, and value vectors. Finally, the MHA output is added to the initial input, followed by a layer normalization, and then fed into the MLP layer to produce the output. This process is repeated iteratively for L layers to obtain the final output.
Structure of DTViT
Given the high-density shadows, variable scales, and diverse locations characteristic of ICH CT images, we have adapted the ViT encoder for use in the DTViT. Additionally, we have integrated two MLP-based decoders into the DTViT to facilitate dual-task classification. As illustrated in Fig. 4, both decoders utilize the feature extraction results from the encoder. The first MLP classifier determines whether a hemorrhage is present in the image, while the second MLP classifier identifies the type of hemorrhage based on the ___location information extracted by the encoder. Correspondingly, the loss function is defined as
where \(loss_1\) represents the loss of classifier 1 and \(loss_2\) represents the loss of classifier 2. This dual-classification approach leverages shared features to enhance the accuracy and efficiency of hemorrhage detection and classification.
Transfer learning
Transfer learning refers to training a model that has already been pre-trained on another large dataset, such as ImageNet18. This approach can significantly save time and data size requirements, and enhance training efficiency. It allows the model to leverage previously learned features and knowledge, which is especially useful when dealing with similar but new tasks. Therefore, we train the DTViT based on a pre-trained Vision Transformer encoder19 as the backbone and fine-tune the model’s parameters.
Specifically, there are three types of pre-trained ViT models: ViT-Base, ViT-Large, and ViT-Huge. The ViT-Base model, with its 12 blocks, is suitable for datasets of moderate complexity. The ViT-Large model, which has 24 blocks and larger embedding dimensions, offers better performance but requires more computational resources. The ViT-Huge is the largest one of the three models, taking 32 blocks and being suitable for high-complexity tasks. For our work with DTViT, we use the ViT-Large as the pre-trained model20, which is trained on ImageNet-1K18. The configuration of DTViT is shown in Table 2.
The accuracy and loss curves in training and validating processes on datasets with and without augmentation. (a) Training and validating losses with augmentation. (b) Training and validating accuracies with augmentation. (c) Training and validating losses without augmentation. (d) Training and validating accuracies without augmentation.
Experiments
In this section, we conduct experiments using the new dataset with and without data augmentation to validate the dataset and evaluate the performance of the DTViT. The experiment environment and parameters used in the training process are first given. Then, we introduce several evaluation metrics used in experiments. Further, we present the accuracy and losses for methods evaluated on the dataset. In addition, we apply classical vision models to the dataset to better validate the dataset and the proposed model.
Environments and parameters
The experimental hardware setup is as follows: The CPU used is an AMD Ryzen 5975WX with 32 cores, and the GPU is an NVIDIA RTX 4090 equipped with 24 GB of memory. The operating system is Ubuntu 20.04. CUDA version 12.3 was utilized for computation. All experiments were conducted using Python version 3.8.18 and PyTorch version 2.3.0.
Additionally, we utilized the AdamW optimizer21 with an initial learning rate of \(2 \times 10^{-5}\) and a weight decay rate of 0.01. The batch size is set as 8 for the training data without data augmentation, 32 for the training data with data augmentation, 32 for the validating data, and 4 for the testing data. The model was trained over ten epochs. We employed the cross-entropy function as the loss function to optimize the model’s performance.
Evaluation indices
We have conducted various experiments and adopted multiple models to evaluate the performance of DTViT. Firstly, the correct classification of images, i.e., the accuracy, is the criterion of the model performance, which is presented as
where TP represents true positive instances; TN represents true negative instances; FP represents false positive instances; FN represents false negative instances. The precision metric represents the proportion of correctly predicted positive instances out of all the instances predicted as positive in a class. Precision is determined using the following equation:
The recall metric quantifies the proportion of positive instances that are correctly identified, and the corresponding formula to calculate recall is presented as
In addition, the F1-score metric is the harmonic mean of precision and recall. It is computed as
In medical fields, specificity is a good evaluation index to guarantee that individuals who do not have the disease are not wrongly diagnosed, thereby avoiding unnecessary treatment and anxiety. The calculation of specificity is presented as
We take these metrics to evaluate the performances of DTViT on the newly collected dataset with and without data augmentation.
DTViT performances
Figure 5 illustrates the evolution of losses and accuracies during training and validating processes, both with and without data augmentation. As depicted in Fig. 5a, training and validating losses decrease steadily throughout the process, eventually stabilizing at approximately 0.01. Correspondingly, both training and validating accuracies show steady increases, converging approximately to 0.997 by the end of the training period. By contrast, results from the non-augmented dataset display inconsistency: while training loss decreases continuously to a desirable value, the validating loss fails to converge to the same level, ending around 0.08 and indicating overfitting. Similarly, the validating accuracy does not reach the level of the training accuracy, demonstrating that the model is already sufficiently trained.
Further, we evaluate the trained model on the test dataset to obtain the DTViT’s performance on the real-world untrained data, which is composed of 1266 CT images, including 459 for normal images and 807 ICH images with Deep 595, Lobar 163, and Subtentorial 49 images. Confusion matrixes are shown in Fig. 6, where Fig. 6a shows the result of Task 1 for normal and ICH patients classification, and Fig. 6b shows the results of Task 2 for three types of ICH classification. It can be seen that almost all positive and negative cases on the test dataset are classified correctly, except one normal image is misclassified as an ICH image. Similarly, the classification of ICH types also achieved excellent results with a testing accuracy of 0.996. In detail, evaluation indices of precision, recall, F1 score, and specificity are displayed in Table 3.
Comparative experiments
To better validate the effectiveness of the collected dataset and demonstrate the performance of DTViT, we conducted comparative experiments using classical vision backbone models, including ResNet1822, AlexNet23, SqueezeNet24, DenseNet25, and CvT26 on the original dataset.
In the experiments, several models are tested with their encoders adopted and an MLP classifier as the decoder, similar to the setup of DTViT. All models are trained for ten epochs using transfer learning, where pre-trained weights are fine-tuned under identical hyperparameters, including the optimizer settings. As shown in Table 4, all models achieve high overall accuracies on Task 1, with all models exceeding 0.97 accuracy. On Task 2, performance declines across all models, with accuracy dropping slightly in the best-performing DTViT model 0.9937 and more noticeably in others, such as SqueezeNet, which achieves only 0.9207 accuracy. This difference in performance can be attributed to the increased complexity of Task 2, which requires capturing and differentiating finer image features. In comparison to Task 1, Task 2 involves more intricate image structures that challenge the models’ ability to generalize, as subtle variations in these images demand a more nuanced feature extraction process. The decline is particularly evident in models like ResNet18 and CvT, which, although effective in Task 1, struggle to maintain similar levels of precision and recall in Task 2. Also, it is worth mentioning that our main contribution lies in collecting a high-quality dataset, which enables strong performance across models, and that, although current accuracy improvements are marginal, DTViT’s ability to capture long-range dependencies positions it well for scaling to larger, more complex datasets and enhancing robustness in clinical applications.
Additionally, we have taken sensitivity analysis experiments by choosing different learning parameters, batch size, and patch size of the model training, and analysis and discuss how the chosen of these parameters affects model’s performance. Specifically, we first choose different learning parameters of \(1\times 10^{-3}\), \(1\times 10^{-4}\), and \(5\times 10^{-6}\) to train the model. The training results are shown in Fig. 7. It can be seen that the DTViT is sensitive with the learning rate, especially with the large value. When choosing a relative large learning rate of \(1\times 10^{-3}\), the accuracies are maintaining at \(80\%\) averagely and the learning performance fluctuated throughout the entire process, which are not satisfied. The corresponding losses are high as well. When choosing the learning parameter of \(1\times 10^{-4}\), which is closer to the parameter we used in the paper (\(2\times 10^{-5}\)), the accuracies and losses performance are greatly improved, but still not very satisfied. By contrast, when choosing a relative small value of learning rate, \(5\times 10^{-6}\), the validation accuracy and loss are are as good as training accuracy and loss though they are improved slowly, indicating that the model does not learning the characteristic of the dataset. To conclude, the learning rate is crucial to the learning process, where large value lead to the task failure and small value may slow down the learning process.
Then, we take experiments of different batch size of 8 and 32 to see the influence of batch size to the model’s performance. Overall, performances are not influenced too much, while the training time is around 4419 seconds for batch 32 and 5223 seconds for batch 8 on Nvidia A10 GPU. As for the performance, there is no significance performance differences on these two values. It is worth noting that the memory required by the model is positively correlated with the batch size. The DTViT cannot be trained on a Nvidia A10 GPU (24GB) with a batch size of 64 due to insufficient memory.
Finally, we trained the DTViT model with patch sizes of 16 and 32. Overall, there was no significant difference in performance, but the training time varied between the two parameters. The larger patch size of 32 required fewer computational resources, significantly reducing the training time. The best validation accuracy for the 32-pixel patch size was \(98.58\%\) over 10 epochs, closely matching the results obtained with the 16-pixel patch size in the manuscript. In terms of training time, the 32-pixel patch required only 1,544 seconds on an Nvidia A10, compared to 4,451 seconds for the 16-pixel patch.
Accuracies and losses of choosing different learning parameters over the training process. (a) Training and validation accuracies of learning rate \(1\times 10^{-3}\). (b) Training and validation accuracies of learning rate \(1\times 10^{-4}\). (c) Training and validation accuracies of learning rate \(5\times 10^{-6}\). (d) Training and validation losses of learning rate \(1\times 10^{-3}\). (e) Training and validation losses of learning rate \(1\times 10^{-4}\). (f) Training and validation losses of learning rate \(5\times 10^{-6}\).
Discussion
In this paper, we first collect CT images from intracerebral hemorrhage (ICH) patients and normal people, which are sourced from real-world patient data from Yulin First Hospital. Furthermore, medical specialists put effort into data filtering and labeling of three types of ICH haemorrhage.
Based on the new dataset, we propose the dual-task vision transformer (DTViT) model based on the vision transformer for dual-task classification. The proposed model is composed of an encoder to extract information from CT images and two decoders for different classification tasks, i.e., classification of normal and ICH images and classification of three types of ICH based on the ___location of the hematoma. The proposed DTViT achieves 99.7% of the training accuracy and 99.88% of the testing accuracy on the augmented dataset. We also compared the proposed model with multiple state-of-the-art models, i.e., Resnet18, SqueezeNet, and Alexnet, and the results show that our model achieves the best performance over these models.
There is some previous research focusing on ICH classification or detection9,10,27,28. For example27, proposes an ICH classification and localization method using a neural network model, achieving an accuracy of 97.4%, while the input signals are microwave signals and the hardware requirements are relatively high. Also, a CT-image-based deep learning method is proposed in9, which is based on the EfficientDet and achieves an accuracy of 92.7%. Similar work using computer vision methods for ICH detection and classification includes29,30, where29 achieves ICH classification using a CNN-based architecture called EfficientNet30; uses ResNet-18 for ICH classification with the accuracy 95.93%.
However, to the best of our knowledge, there is neither such a dataset describing the ___location of hematoma of ICH CT images nor such classification model simultaneously determining whether a CT image is of a cerebral hemorrhage or normal and classifies the three types of cerebral hemorrhage.
The primary aim of creating the Intracerebral Hemorrhage (ICH) CT image dataset and developing the classification model is to harness state-of-the-art computer vision technology to assist physicians in diagnosing and treating patients with cerebral hemorrhage. Cerebral hemorrhage is a severe, acute medical condition affecting approximately two million individuals annually, often associated with higher mortality and morbidity and limited treatment options. Early diagnosis and tailored treatment strategies based on the hemorrhage ___location are crucial for patient outcomes, as different hemorrhage sites require varied treatment approaches. Therefore, the classifier developed in this study, which categorizes hemorrhage based on its ___location, is clinically significant as it aids healthcare professionals in quickly and accurately determining treatment plans.
Limitations
This study faces several limitations. First, the dataset, derived from real clinical data, is challenging to obtain and inherently imbalanced, which may limit the model’s accuracy and generalizability. Future efforts will focus on expanding and balancing the dataset through continued data collection. Second, the current data augmentation technique involves mere replication of the dataset. Future improvements will explore the use of generative models, such as diffusion models, for data enhancement.
Future directions
In subsequent work, we aim to develop a multimodal diagnostic dataset for cerebral hemorrhage, integrating clinical data such as blood pressure, lipid profiles, and bodily element levels to enhance diagnostic accuracy. Furthermore, we plan to create a cerebral hemorrhage classification and diagnosis model based on Artificial Intelligence-Generated Content (AIGC), which will improve diagnostic efficiency for physicians.
To successfully integrate the Dual-Task Vision Transformer (DTViT) into clinical workflows, three crucial areas will be addressed:
-
1.
User Interface Development: We will design a user-friendly graphical user interface (GUI) or web-based platform that allows medical specialists to upload CT slice images, receive classification results, and view processed images seamlessly.
-
2.
Optimization for Resource-Limited Environments: Recognizing the need for wider accessibility, we plan to develop lightweight versions of the model optimized for resource-constrained settings, with reduced computational requirements and faster inference speeds. This will be inspired by efficient models like EdgeNeXt31 and MobileViT32.
-
3.
Continuous Improvement and Data Collection: We will collect new data under appropriate licensing and refine the model based on clinician feedback. This iterative process will enhance the model’s performance and further tailor it to clinical needs.
Finally, it is important to note that the classification results produced by the model, particularly during the initial development and testing phases, are intended solely as a reference and should not yet be used as the sole basis for clinical diagnosis or treatment. We appreciate the reminder of this crucial distinction.
Conclusion
In this paper, we have constructed a dataset for Intracerebral Hemorrhage (ICH) based on real-world clinical data. The dataset comprises CT images that have been initially processed and manually labeled by medical experts into categories of normal and ICH images. Moreover, the ICH images have been categorized into three types, Subcortical and Lobar, according to the hemorrhage’s ___location. Additionally, we have introduced a dual-task vision Transformer (DTViT) aiming at the classification of Intracerebral hemorrhage. This innovative neural network model incorporates an encoder, which utilizes the cutting-edge vision Transformer architecture, and two decoders that are designed to classify images as ICH or normal and to determine the hemorrhage type. Our experiments have demonstrated that the DTViT has achieved remarkable accuracies on both tasks on the test dataset. We have also compared the DTViT with other models such as Resnet18, SqueezeNet, Alexnet, and CvT. To our knowledge, this research is the first to have developed both a specialized dataset and a neural network model tailored for hemorrhage ___location classification in a clinical context. This contribution holds substantial potential for enhancing clinical diagnosis and treatment planning.
Data availibility
The constructed dataset used in this study can be accessed at https://www.kaggle.com/datasets/louisfan/dataset-for-dtvit. The source code for this study can be accessed at https://github.com/JFan1997/DTViT.
References
Gebel, J. M. & Broderick, J. P. Intracerebral hemorrhage. Neurol. Clin. 18, 419–438 (2000).
de Oliveira Manoel, A. L. Surgery for spontaneous intracerebral hemorrhage. Crit. Care 24, 45 (2020).
Wan, Y., Holste, K. G., Hua, Y., Keep, R. F. & Xi, G. Brain edema formation and therapy after intracerebral hemorrhage. Neurobiol. Dis. 176, 105948 (2023).
Anwar, R. W., Abrar, M. & Ullah, F. Transfer learning in brain tumor classification: Challenges, opportunities, and future prospects. In 2023 14th International Conference on Information and Communication Technology Convergence (ICTC), 24–29, https://doi.org/10.1109/ICTC58733.2023.10392830 (2023).
Kim, H. E. et al. Transfer learning for medical image classification: A literature review. BMC Med. Imaging 22, 69 (2022).
Jiang, H. et al. A review of deep learning-based multiple-lesion recognition from medical images: Classification, detection and segmentation. Comput. Biol. Med. 157, 106726 (2023).
Butoi, V. I. et al. Universeg: Universal medical image segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 21438–21451 (2023).
Yuan, F., Zhang, Z. & Fang, Z. An effective CNN and transformer complementary network for medical image segmentation. Pattern Recogn. 136, 109228 (2023).
Cortés-Ferre, L., Gutiérrez-Naranjo, M. A., Egea-Guerrero, J. J., Pérez-Sánchez, S. & Balcerzyk, M. Deep learning applied to intracranial hemorrhage detection. J. Imaging 9, 37 (2023).
Chen, Y.-T. et al. Deep learning-based brain computed tomography image classification with hyperparameter optimization through transfer learning for stroke. Diagnostics 12, 807 (2022).
Ullah, F. et al. Evolutionary model for brain cancer-grading and classification. IEEE Access 11, 126182–126194. https://doi.org/10.1109/ACCESS.2023.3330919 (2023).
Li, L. et al. Deep learning for hemorrhagic lesion detection and segmentation on brain CT images. IEEE J. Biomed. Health Inform. 25, 1646–1659 (2020).
Vbookshelf. Computed tomography (CT) images. https://www.kaggle.com/datasets/vbookshelf/computed-tomography-ct-images (2024). Accessed: 2024-07-25.
Wu, B. et al. BHSD: A 3D multi-class brain hemorrhage segmentation dataset. In International Workshop on Machine Learning in Medical Imaging, 147–156 (Springer, 2023).
Flanders, A., Prevedello, L., Shih, G. et al. Construction of a machine learning dataset through collaboration: The RSNA 2019 brain CT hemorrhage challenge. Radiol. Artif. Intell.2 (2020).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst.30 (2017).
Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. (2020).
Deng, J. et al. Imagenet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition, 248–255 (IEEE, 2009).
Wightman, R. Pytorch image models. https://github.com/rwightman/pytorch-image-models, https://doi.org/10.5281/zenodo.4414861 (2019).
Touvron, H. et al. Training data-efficient image transformers & distillation through attention. In International Conference on Machine Learning, 10347–10357 (PMLR, 2021).
Loshchilov, I. & Hutter, F. Decoupled weight decay regularization. (2017).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 770–778 (2016).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst.25 (2012).
Iandola, F. N. et al. Squeezenet: Alexnet-level accuracy with 50x fewer parameters and \(<\) 0.5 MB model size. (2016).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 4700–4708 (2017).
Wu, H. et al. CvT: Introducing convolutions to vision Transformers (2021).
Li, Q. et al. Classification and ___location of cerebral hemorrhage points based on sem and SSA-GA-BP neural network. IEEE Trans. Instrum. Meas. (2024).
Raposo, N. et al. A causal classification system for intracerebral hemorrhage subtypes. Ann. Neurol. 93, 16–28 (2023).
Phaphuangwittayakul, A. et al. An optimal deep learning framework for multi-type hemorrhagic lesions detection and quantification in head CT images for traumatic brain injury. Appl. Intell. 1–19 (2022).
Altuve, M. & Pérez, A. Intracerebral hemorrhage detection on computed tomography images using a residual neural network. Phys. Med. 99, 113–119 (2022).
Maaz, M. et al. EdgeNeXt: Efficiently amalgamated CNN-Transformer architecture for mobile vision applications. In International Workshop on Computational Aspects of Deep Learning at 17th European Conference on Computer Vision (CADL2022) (Springer, 2022).
Mehta, S. & Rastegari, M. MobileViT: Light-weight, general-purpose, and mobile-friendly vision Transformer (2022).
Acknowledgements
We would like to express our deepest gratitude to the Department of Neurology, Yulin Hospital, The First Affiliated Hospital of Xi’an Jiaotong University, Yulin, Shaanxi, China, and the Department of Neurology, The First Hospital of Yulin, Yulin, Shaanxi, China, for their invaluable support and collaboration throughout this research project. Their expertise and resources have been instrumental in the successful completion of this work.
Funding
This work was supported by Yulin Science and Technology Planning Project under YF-2021-34.
Author information
Authors and Affiliations
Contributions
J.F. was responsible for manuscript writing and methodology design. X.F. contributed to the conceptualization, data collection, preprocessing, and manuscript revision. C.S. was responsible for data collection, preprocessing, and annotation. X.W. also contributed to data collection, preprocessing, and annotation. B.F. provided funding resources and participated in data collection. L.L. was responsible for figure visualization. G.L. contributed to methodology design, provided computational resource support, and participated in manuscript revision. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Medical Ethics Committee of the First Hospital of Yulin (2023-003). In addition, We have carefully considered the ethical and privacy implications of using sensitive medical data in this study. Patients provide informed consent agreeing that their data may be used for research purposes. All patient data was de-identified, ensuring that no personally identifiable information can be traced back to individuals.
Competing interests
The author(s) declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fan, J., Fan, X., Song, C. et al. Dual-task vision transformer for rapid and accurate intracerebral hemorrhage CT image classification. Sci Rep 14, 28920 (2024). https://doi.org/10.1038/s41598-024-79090-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-79090-y