Abstract
During a radiotherapy (RT) course, geometrical variations of target volumes, organs at risk, weight changes (loss/gain), tumor regression and/or progression can significantly affect the treatment outcome. Adaptive RT has become the effective methods along with technical advancements in imaging modalities including cone-beam computed tomography (CBCT). Planning CT (pCT) can be modified via deformable image registration (DIR), which is applied to the pair of pCT and CBCT. However, the artifact existed in both pCT and CBCT is a vulnerable factor in DIR. The dose calculation on CBCT is also suggested. Missing information due to the artifacts hinders the accurate dose calculation on CBCT. In this study, we aim to develop a deep learning-based metal artifact reduction (MAR) model to reduce the metal artifacts in CBCT for head and neck cancer RT. To train the proposed MAR model, we synthesized the kV-CBCT images including metallic implants, with and without metal artifacts (simulated image data pairs) through sinogram image handling process. We propose the deep learning architecture which focuses on both artifact removal and reconstruction of anatomic structure using a dual-encoder architecture. We designed four single-encoder models and three dual-encoder models based on UNet (for an artifact removal) and FusionNet (for a tissue restoration). Each single-encoder model contains either UNet or FusionNet, while the dual-encoder models have both UNet and FusionNet architectures. In the dual-encoder models, we implemented different feature fusion methods, including simple addition, spatial attention, and spatial/channel wise attention. Among the models, a dual-encoder model with spatial/channel wise attention showed the highest scores in terms of peak signal-to-noise ratio, mean squared error, structural similarity index, and Pearson correlation coefficient. CBCT images from 34 head and neck cancer patients were used to test the developed models. The dual-encoder model with spatial/channel wise attention showed the best results in terms of artifact index. By using the proposed model to CBCT, one can achieve more accurate synthetic pCT for head and neck patients as well as better tissue recognition and structure delineation for CBCT image itself.
Similar content being viewed by others
Introduction
Recently, an image-guided radiotherapy (IGRT) or adaptive radiotherapy (RT) has become the effective methods along with technical advancements in imaging modalities such as magnetic resonance imaging or cone-beam computed tomography (CBCT)1,2,3,4. During RT, geometrical variations of target volumes, organs at risk, weight changes (loss/gain), tumor regression and/or progression can significantly affect the treatment outcome5. The patients’ anatomy can vary along the time that leads to an inaccurate dose delivery (or accumulated dose calculation) which is driven from the static (stationary) treatment planning6. The adaption by daily accumulation can resolve the problem of previous independent computed tomography (CT) scan methods7.
Within this, the synthetic CT generation using CBCT image for adaptive radiotherapy (RT) is also prevalent. Particularly, using the deformable image registration (DIR), it is possible to generate the synthetic CT from planning CT (pCT) and daily CBCT image. By adopting the original plan on pCT to this synthetic CT image, the change of dose delivery compared to the original plan is calculated and some left partial plans can be modified considering the daily change of the anatomy. Furthermore, the structure set contoured on pCT can be modified with respect to the same DIR applied to the pair of pCT and CBCT. However, the artifact existed in both pCT and CBCT is a vulnerable factor in DIR8,9,10,11,12. On the other hand, an attempt has made for the CBCT to directly calculate the patient dose. The dose calculation on CBCT is also suggested by using a deep learning method and has shown a great feasibility. During the conversion process of Hounsfield unit (HU)-electron density curve to CBCT, missing information due to the CBCT artifacts hinders the accurate dose calculation on CBCT13.
There are a lot of methods for removing artifacts in CBCT, but they are mostly for scattering, motion, and cupping artifacts. However, for the head and neck RT, strong metal artifacts significantly degrade the accuracy of dose calculation and the performance of DIR, as well as the image quality of CBCT. In contrast to the CT scanner for pCT, to our knowledge, CBCTs attached to linear accelerator (LINAC) have no dedicated software to reduce metal artifacts.
Existing metal artifact reduction (MAR) methods for a typical dental CBCT utilize a passive inpainting method to prevent destroying tissue parts rather than reducing the artifacts14,15. Recently, a deep learning method was used for MAR in dental CBCT16. As the generative adversarial network has shown the effective result, and the lack of data acquisition, the attempt to solve the MAR problem as unsupervised method using the cycleGAN17 has been also proposed. The method using dual ___domain18, sinogram and image19,20 has been also proposed. The CBCT image data used in aforementioned studies were acquired from dental CBCT. More recently, several studies have presented deep learning-based MAR for CBCT attached to LINAC21,22. In those studies, images from MV-CBCT were used to synthesize the target kV-CBCT images having no metal artifacts17,23,24.
In this study, we aim to develop a deep learning-based MAR model to reduce the metal artifacts in CBCT for head and neck cancer RT. To train the proposed MAR model, we synthesized the kV-CBCT images including metallic implants, with and without metal artifacts (simulated image data pairs) through sinogram image handling process. We propose the deep learning architecture which only uses image ___domain, and focuses on both artifact removal and reconstruction of anatomic structure.
Methods
Ethics declarations
All procedures performed in study involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The experimental protocols were approved by the Seoul National University Hospital Institutional Review Board (IRB approval No. H-2111-198-1280) and Written informed consent by the patients was waived due to a retrospective nature of our study.
Generation of synthetic dataset
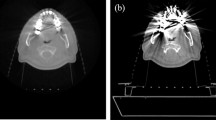
Since the acquisition of pairs of metal artifact CBCT image and artifact free CBCT slices in one patient is not available, we synthesized the CBCT dataset following a previous work25. The metal artifact image and metal artifact free image were selected randomly in our database, as shown in Table 1. The CBCT images were obtained between 2019 and 2020 from the C-arm CBCT attached to the VitalBeam LINAC (Varian Medical Systems/ Siemens Healthineers, CA, USA) in Seoul National University Hospital.
The overall synthetic data generation process is illustrated in Fig. 1. The segmentation of metal region was done through HU thresholding for extraction of metal region. We assumed the HU value of the metal is 8000. The erosion filter and Gaussian blur were applied for boundary smoothing. The position of metals segmented from the metal artifact image were slightly changed according to the anatomical structure of the clean (artifact free) image. Then the position-corrected metal images were converted into synthetic metal artifact CBCT images, as encompassing Radon transformation and inverse Radon transformation. During sinogram handling processing, various artifact strengths can be generated by multiplying values in an arbitrary range during the radon transform and inverse radon transform process. Position-corrected metal images were synthesized into clean images, and the synthesized images would be target images. Synthetic metal artifact images would be input images for the proposed MAR model. In this regard, we synthesized a paired data of metal artifact free CBCT images (i.e., ground truth) and metal artifact CBCT images. We generated the synthesized dataset focusing on producing the case of containing the oral cavity, since it is the most complex part and difficult for restoration due to its complex anatomy structure.
Architecture of MAR models
Single-encoder models
The first model is based on the UNet26 encoder (Fig. 2(a)), which is built for artifact removal, rather than the realistic restoration of the artifact-corrupted region. It consists of convolution layers and maxpooling layers. Maxpooling shows the effectiveness in detecting the artifact region and removing the artifact since the pixel values in artifact region is unusually high compared to the tissue and bone, which enabled maxpooling to detect and remove those regions well.
The second model is based on the FusionNet27 encoder (Fig. 2(b)), but several structures are modified for restoration of anatomic structures. For better restoration, the residual connection is added to each convolution layers, and the maxpooling is changed to the strided convolution layer. In this architecture, the characteristic of restoration is highly increased which makes a small-negative effect in artifact reduction, the model tries to preserve information in input as many as possible. The purpose of the second model is on anatomy restoration, not a radical artifact reduction. The decoder consists of upsampling layer and convolution layer with residual connection.
The third model is based on the modified UNet encoder by adding residual convolutional block to the first model (denoted as UNet/L, which indicates large encoder of UNet) (Fig. 2(c)).
The fourth model is based on the modified FusionNet by adding maxpooling layer and spatial attention layer to the second model (denoted as FusionNet/L, which indicates large encoder of FusionNet) (Fig. 2(d)). The third and fourth models are designed to merge the strengths of the UNet (i.e., artifact removal) and FusionNet (i.e., restoration of anatomic structure) into the single-encoder models. Since these models are combinations of UNet and FusionNet, convolutional and attentional features are extracted and linked to decoder through skip connections.
Dual-encoder models
Our proposed model architecture is based on the dual encoder–decoder architecture. The first encoder (encoder1 of Fig. 3) is based on the UNet manipulated for the artifact removal, while the second encoder (encoder2 of Fig. 3) is based on the FusionNet manipulated for the restoration of metal artifact region. The basic structures for each encoder are the same as the UNet and FusionNet described as the first model and second model, respectively.
After each encoder produces convolutional feature and attentional feature, two features are mixed in regional proper way using the feature fusion. The convolutional feature is produced through convolution operation in a convolution block, and the attentional feature is produced through spatial and channel-wise attention28,29 operations on the feature maps that are operated from the convolution block.
Our purpose is to add the anatomy restoration detail in radically-removed feature map. Therefore, three dual-encoder models are designed with respect to feature fusion methods in the decoder. The first dual-encoder model uses two convolutional features with a simple addition method (Fig. 3(a)). The second dual-encoder model uses one convolutional feature and one attentional feature with the spatial attention method (Fig. 3(b)). The third dual-encoder model reflects the convolutional feature and spatial attention feature to the decoder through channel wise attention operation (denoted as channel wise attention block in Fig. 3(c)). This is designed to provide high restoration performance by providing sufficient local (focusing where) and global (focusing what) information for the decoder restoration process through the attention operation of the information extracted from each encoder. The third model dual-encoder model is our final proposed model (ours) with the best performance in this task.
Since we proposed and experimented four single-encoder models and three dual-encoder models, the model structures were complicated. Therefore, we summarized the characteristics of each model in Table 2.
Loss function
Since we have the synthesized metal artifact dataset including metal artifact free CBCT images for labels, the supervised way of training is applied. Loss function is the combination of mean squared error (MSE) loss, structural-similarity index (SSIM) loss, and auxiliary loss. MSE and SSIM losses are described in following Eqs. (1) and (2):
The FusionNet based encoder2 for anatomy restoration has over-preserved the metal artifact, so the direct loss between the middle encoder feature and the target (artifact removed) image using MSE as the auxiliary loss is added. This auxiliary loss has given the effect of leaving the metal artifact region while preserving the proper features. For this, we took a convolution layer to each layer’s encoder feature, and directly calculated the loss between the target and the convolution output.
Experiments
The total numbers of synthesized dataset for training, validation, and test are 29104, 3795, and 7201, respectively. The training was done for 100 epoch with batch size 32, and the random crop was used for data augmentation.
Quantitative evaluation
The following four evaluation indicators were used to quantitatively evaluate the performance of MAR. For each evaluation metrics, MSE is utilized to measure the difference in pixel values between input and output images, as Eq. (3). It is particularly useful for highlighting significant errors.
The SSIM assesses the quality of MAR by comparing the luminance, contrast, and structural information between the input and output images, as Eq. (4). This metric is effective in evaluating how well essential details are preserved during artifact reduction.
Peak signal-to-noise ratio (PSNR) evaluates the performance of MAR by measuring the ratio between the maximum possible signal and the noise introduced by the reduction process, as Eq. (5). It used for quantifying the extent to which the output image retains the quality of the target image after artifact removal.
and Pearson correlation coefficient (PCC) was used to check the linear relationship between images, as Eq. (6):
Artifact index method
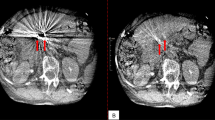
To evaluate the performance of MAR, the artifact index30 calculation was conducted. The artifact index calculation, as indicated by the green and red circles in Fig. 5 of the qualitative results, was performed using below Eq. (7):
The standard deviation (SD) of the green circle represents regions with high standard deviation, indicating significant metal artifacts and image distortion. In contrast, the SD of the red circle represents regions with the lowest standard deviation, suggesting minimal artifacts in each CT image.
Results
Quantitative evaluation
The quantitative evaluation was performed with our synthesized dataset. Quantitative result has been shown on Table 3. Our model, having dual-encoder and spatial and channel wise attention, showed the best result compared to the other single-encoder and dual-encoder models. The residual connection has the effect of over-preserving, so the metrics of FusionNet is slightly lower than the UNet in single-encoder models.
In addition, the single-encoder models with large encoder architecture (i.e., UNet/L and FusionNet/L) did not show effective results. The single-encoder models having similar structures with dual-encoder models in a single merged encoder were observed to have low quantitative values compared to the dual-encoder models.
We also compared the feature fusion methods. The dual-encoder with simple addition showed the in-between performance of single-encoder UNet and FusionNet. And the method using spatial attention showed slightly higher numerical performance improvement compared to the method using only simple addition. By adding spatial and channel-wise attention, it can be observed that the performance increases. This model achieved the highest numerical evaluation results among all the models.
Artifact index
The artifact index demonstrated significant improvement across all models compared to the input image as shown in Fig. 4. Due to the varying characteristics of each deep learning model, the numerical differences were minimal. Models based on FusionNet with residual connection blocks exhibited a wide range of values due to their over-preserving nature, while the single UNet-based models showed a relatively narrow value distribution. Our proposed model, utilizing spatial and channel-wise attention methods, effectively removed artifacts, resulting in the lowest mean index and the narrowest distribution.
Qualitative evaluation
The comparison was conducted between areas frequently subjected to adaptive RT within the oral cavity and regions with high occurrences of metal insertion in dental areas. Tissue restoration performance is indicated by the orange arrow, over-preservation of tissue is highlighted by the red arrow, and reduction of heavy streak artifacts is denoted by the purple arrow. Considering the high intensity of metal artifacts in the input images due to the insertion of high-density implants, The UNet removed the artifact region well but mostly makes the grayish and blurry tissue region (see orange arrows in Fig. 5). Unlike UNet, the FusionNet over-preserved the input image’s infromation (see red arrows in Fig. 5) and did not remove the metal artifact effectively, while preservation and restoration of artifact-corrupted region is better than UNet. In addition, the UNet/L and FusionNet/L models showed some improvements compared to the UNet and FusionNet model mentioned above, but they did not demonstrate effective results. The proposed architecture, which used dual encoder did two positive effect from each encoder, which effectively removes metal artifacts but also restores the tissue without blurring or making the new greish region (see in Fig. 5(a)). In particular, the tooth area indicated by the yellow arrow in Fig. 5(b) for patient 2 demonstrates noticeably good performance.
Figure 6 presents the comparison results for different feature fusion methods of dual-encoder models. Observing the red arrows in Fig. 6, the model that simply added convolutional features in the decoder failed to effectively removed streak artifacts from the input image. In contrast, the model using spatial attention demonstrates more effective streak artifacts removal. And as indicated by the orange arrow in Fig. 6(a) using spatial attention improved tissue restoration performance better than simple addition.
Our proposed model, which utilizes both spatial and channel-wise attention, demonstrated superior performance in streak artifact reduction and tissue restoration. The tissue area marked by the orange arrow was realistically restored, and streak artifacts (see red arrow in Fig. 6(a)) were also effectively removed. Particularly for severe metal artifacts case in Fig. 6(b), the models with simple addition and spatial attention left substantial streak artifacts (purple arrows in Fig. 6(b)) and failed to restore the tissue areas adequately (orange arrows in Fig. 6(b)). However, our proposed model effectively restored the tissue area (orange arrow in Fig. 6(b)) and exhibited high-performance severe streak artifacts reduction capabilities (purple arrow in Fig. 6(b)).
Discussion
We developed and trained a synthetic dataset exclusively with kV-CBCT data. Our proposed model, which uses dual-encoder instead of a single-encoder, exhibited superior performance in reducing metal artifacts. The dual-encoder facilitates more nuanced data processing and integration. Each encoder is tailored to different aspects of the imaging data, enhancing artifact reduction synergistically. The first encoder extracts fine details from images, while the second encoder focuses on identifying and mitigating noise and artifacts. This specialized approach ensures a thorough analysis of both the image structure and potential disruptions. Additionally, by employing an attention mechanism, we selectively utilized essential information, thereby improving the performance in MAR. This method has demonstrated potential to enhance the quality of the output images and provide benefits in clinical settings.
Despite the synthetic pCT’s accuracy via DIR, the deformation of structures in the oral cavity is discouraged. We noted geometric discrepancies between the deformed structures, attributed to the reduction of metal artifacts in CBCT. Moreover, we observed that the volumetric similarity between the deformed structures (from both the original and the MAR-enhanced CBCT) and the ground truth contour was actually less than that between the structure on the original pCT and the ground truth contour. Although the commercial MAR technique (O-MAR, Philips Healthcare, Netherlands) was applied to the pCT, pronounced streak artifacts remained. These residual metal artifacts on the pCT could hinder the generation of accurate deformed structures.
In future research, we plan to employ deep learning-based MAR techniques on both pCT and CBCT to create synthetic CT and deformed structures. Furthermore, future studies should explore direct dose calculation on CBCT, which was not addressed in our current study, to observe potential dose differences between the original and MAR-enhanced CBCT.
This study has some limitations to be discussed. Firstly, the dataset used in this study was acquired from a single institution. The performance of the model developed in this study was evaluated fairly using patient data not used in training, and it is expected to perform well even when data from other institutions is utilized. Although a sufficient number of data was used to evaluate the model’s performance, utilizing a larger dataset or data from other institutions in the future would greatly assist in enhancing the model’s performance. Secondly, various experiments were conducted to find the optimal model structure for this study using different CNN-based architectures, but the latest structures introduced recently, such as Diffusion-based models, were not applied. As shown in Table 3, our model demonstrated exceptionally good performance metrics sufficient for clinical use. However, if there is a desire to further improve the MAR performance, it would be beneficial to apply the latest AI model structures in future work.
Conclusion
As the daily CBCT image acquisition became popular, the importance of CBCT images in adaptive RT became bigger. Acquisition of high-quality CBCT images, and artifact correction makes the more accurate synthetic planning CT and deformed structures. In addition, the accurate dose calculation on CBCT can be also available. In this study, we developed a dual encoder deep learning model for MAR for CBCT in head and neck RT It was shown that our proposed deep learning-based method has dramatically improved the image quality of CBCT. By using the proposed MAR model to CBCT, it can achieve more accurate synthetic pCT for head and neck patients as well as better tissue recognition and structure delineation. For further improvements in adaptive RT for head and neck cancer patients, the implementation of deep learning-based MAR model to both the pCT and CBCT is required.
Data availability
The datasets generated during and/or analysed during the current study are not publicly available due to policy of the Institutional Review Board of the Seoul National Hospital (SNUH) but are available from the corresponding author on reasonable request.
Abbreviations
- CBCT:
-
Cone-beam computed tomography
- CT:
-
Computed tomography
- DIR:
-
Deformable image registration
- HU:
-
Hounsfield unit
- IGRT:
-
Image-guided radiotherapy
- LINAC:
-
Linear accelerator
- MAR:
-
Metal artifact reduction
- MSE:
-
Mean squared error
- O-MAR:
-
Metal artifact reduction for orthopedic implants
- PCC:
-
Pearson correlation coefficient
- pCT:
-
Planning computed tomography
- PSNR:
-
Peak signal-to-noise ratio
- RT:
-
Radiotherapy
- SD:
-
Standard deviation
- SSIM:
-
Structural similarity index
References
Hussain, A. & Muhammad, W. Treatment planning in radiation therapy. In An Introduction to Medical Physics 63–129 (2017).
Son, J., Lee, S. Y., Choi, C. H., Park, J. M. & Kim, J. I. Impact of 0.35 T magnetic field on dose calculation for non-small cell lung cancer stereotactic radiotherapy plans. J. Radiat. Prot. Res. 48, 117–123 (2023).
Abuhaimed, A. & Martin, C. J. Assessment of organ and size-specific effective doses from cone beam CT (CBCT) in image-guided radiotherapy (IGRT) based on body mass index (BMI). Radiat. Phys. Chem. 208, 110889 (2023).
Hirose, T. et al. Observer uncertainties of soft tissue-based patient positioning in IGRT. J. Appl. Clin. Med. Phys. 21, 73–81 (2020).
Giacometti, V., Hounsell, A. R. & McGarry, C. K. A review of dose calculation approaches with cone beam CT in photon and proton therapy. Phys. Med. 76, 243–276 (2020).
Veiga, C. et al. Toward adaptive radiotherapy for head and neck patients: feasibility study on using CT-to‐CBCT deformable registration for dose of the day calculations. Med. Phys. 41, 031703 (2014).
Brock, K. K. Seminars in Radiation Oncology 181 (NIH Public Access).
Ritter, L. et al. The influence of body mass index, age, implants, and dental restorations on image quality of cone beam computed tomography. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 108, e108–e116 (2009).
Fontenele, R. C., Nascimento, E. H., Vasconcelos, T. V., Noujeim, M. & Freitas, D. Q. Magnitude of cone beam CT image artifacts related to zirconium and titanium implants: impact on image quality. Dentomaxillofac. Radiol. 47, 20180021 (2018).
Serban, M., Heath, E., Stroian, G., Collins, D. L. & Seuntjens, J. A deformable phantom for 4D radiotherapy verification: design and image registration evaluation. Med. Phys. 35, 1094–1102 (2008).
Sonke, J. J. & Belderbos, J. Seminars in Radiation Oncology 94–106 (Elsevier).
Fusella, M., Loi, G., Fiandra, C. & Lanzi, E. Impact of image quality on deformable image registration performances in pelvis patients. Phys. Med. Eur. J. Med. Phys. 32, 24 (2016).
Barateau, A. et al. Comparison of CBCT-based dose calculation methods in head and neck cancer radiotherapy: from Hounsfield unit to density calibration curve to deep learning. Med. Phys. 47, 4683–4693 (2020).
Hegazy, M. A., Cho, M. H., Cho, M. H. & Lee, S. Y. Metal artifact reduction in dental CBCT images using direct sinogram correction combined with metal path-length weighting. Sensors 23, 1288 (2023).
Pua, R., Wi, S., Park, M., Lee, J. R. & Cho, S. An image-based reduction of metal artifacts in computed tomography. J. Comput. Assist. Tomogr. 40, 131–141 (2016).
Hyun, C. M. et al. Deep learning method for reducing metal artifacts in dental cone-beam CT using supplementary information from intra-oral scan. Phys. Med. Biol. 67, 175007 (2022).
Cao, Z., Gao, X., Chang, Y., Liu, G. & Pei, Y. A novel approach for eliminating metal artifacts based on MVCBCT and CycleGAN. Front. Oncol. 12, 1024160 (2022).
Tang, H. et al. A new dental CBCT metal artifact reduction method based on a dual-___domain processing framework. Phys. Med. Biol. 68, 175016 (2023).
Zhang, S., Jiang, B. & Shi, H. J. Phys. Conf. Ser. 012033.
Bayaraa, T., Hyun, C. M., Jang, T. J., Lee, S. M. & Seo, J. K. A two-stage approach for beam hardening artifact reduction in low-dose dental CBCT. IEEE Access. 8, 225981–225994 (2020).
Li, Z. et al. Using RegGAN to generate synthetic CT images from CBCT images acquired with different linear accelerators. BMC Cancer 23, 828 (2023).
Amirian, M., Barco, D., Herzig, I. & Schilling, F. P. Artifact reduction in 3D and 4D cone-beam computed tomography images with deep learning—A review. IEEE Access (2024).
Ni, X. et al. Metal artifacts reduction in kV-CT images with polymetallic dentures and complex metals based on MV-CBCT images in radiotherapy. Sci. Rep. 13, 8970 (2023).
Jihong, C. et al. CBCT-based synthetic CT generated using CycleGAN with HU correction for adaptive radiotherapy of nasopharyngeal carcinoma. Sci. Rep. 13, 6624 (2023).
Lee, J. M. AI-Based Metal Artifact Reduction and Multi-organ Segmentation in Computed Tomography for Radiotherapy. Ph.D. Thesis, Seoul National University (2021).
Ronneberger, O., Fischer, P. & Brox, T. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, Proceedings, Part III 18 234–241 (Springer, 2015).
Quan, T. M., Hildebrand, D. G. C., Jeong, W. K. & Fusionnet A deep fully residual convolutional neural network for image segmentation in connectomics. Front. Comput. Sci. 3, 613981 (2021).
Woo, S., Park, J., Lee, J. Y. & Kweon, I. S. Proceedings of the European Conference on Computer Vision (ECCV) 3–19.
Li, X. et al. Multi-level feature fusion network for nuclei segmentation in digital histopathological images. Vis. Comput. 39, 1307–1322 (2023).
Gomi, T., Sakai, R., Hara, H., Watanabe, Y. & Mizukami, S. Usefulness of a metal artifact reduction algorithm in digital tomosynthesis using a combination of hybrid generative adversarial networks. Diagnostics 11, 1629 (2021).
Funding
This work was supported in part by the Korea Health Technology Research and Development Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea, under Grant HI21C1161; in part by an Institute of Information & Communications Technology Planning & Evaluation (IITP) grant funded by the Korean government (MSIT) (No.RS-2020-II201336, Artificial Intelligence Graduate School Program(UNIST)); and in part by the National Research Foundation of Korea (NRF) funded by the MSIT under Grant NRF-2020R1F1A1073430, Grant NRF-2021R1F1A1057818, and Grant RS-2024-00344958.
Author information
Authors and Affiliations
Contributions
J.K. and J.M.L wrote the main manuscript text. W.L. prepared figures. S.J. and J.L. reviewed and edited the draft. J.K. analysed the data. J.H.K. and H.J. provided study materials. S.J. and J.L. designed and supervised the study. S.J. and J.L. contributed equally as corresponding authors. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ki, J., Lee, J.M., Lee, W. et al. Dual-encoder architecture for metal artifact reduction for kV-cone-beam CT images in head and neck cancer radiotherapy. Sci Rep 14, 27907 (2024). https://doi.org/10.1038/s41598-024-79305-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-79305-2