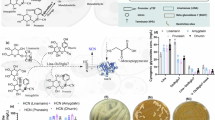
Abstract
The WRINKLED1 (WRI1) transcription factor controls carbon flow in plants through regulating the expression of glycolysis and fatty acid biosynthesis genes. The role of Gossypium hirsutum WRINKLED1 (GhWRI1) in seed-oil accumulation still needs to be explored. Multiple sequence alignment of WRI1 proteins confirmed the presence of two conserved AP2 domains. The amino acid sequence of GhWRI1 exhibited 62% homology with AtWRI1 and phylogenetic analysis showed that GhWRI1 and AtWRI1 originated from common ancestors. Comparison of three dimensional structures of AtWRI1 and GhWRI1 indicated the presence of an altered alpha helix on the C-terminus that harbours spore coat protein ___domain in A. thaliana and other members of Brassicaceae but Kin17 ___domain in G.hirsutum. In the present study, we constructed a novel gene cassette containing AtWRI1 driven by seed-specific promoter and Tobacco Etch Virus enhancer. The transgenic plantlets of G. hirsutum exhibited 35% enhancement in seed oil content and nearly 4-fold increase in oil-bodies in seed endosperm. GC-MS analysis exhibited additional fatty acids i.e. lauric acid methyl ester, 1-dodecanol, palmitoleic acid, margaric acid, stearic acid, linolenic acid, methyl 9,10-methylene-octadecanoate, methyl 18-methylnonadecanoate, 13-docosenoic acid methyl ester, methyl 20-methyl-heneicosanoate, lignoceric acid in the transformants. This is an important study highlighting the enhancement of seed oil content in cotton by seed-specific expression of AtWRI1.
Similar content being viewed by others
Introduction
WRINKLED1 is an ethylene-responsive transcription factor belonging to APETALA2 family harbouring two AP2 DNA-binding domains1 at the N- terminus. This multifunctional transcription factor regulates various plant developmental processes2. The WRINKLED1 gene from Arabidopsis thaliana (AtWRI1) plays a vital role in oil biosynthetic pathway by binding with the promoters of genes involved in glycolysis and fatty acid biosynthesis3. The mutants of AtWRI1 (wri1) exhibit reduced oil content to 80% in comparison with the wild type4. The glycolysis process was compromised due to which the developing seeds were not able to convert sucrose into Triacylglycerides biosynthetic precursors5. Complementation of wri1-3 mutants resulted in the up-regulation of ketoacyl carrier protein synthase, acetyl-CoA carboxylase and pyruvate kinase genes that are involved in fatty acid synthesis and oil accumulation in the plastids6.
Many factors play role in the regulation of WRI1 i.e. cellular sugar - in the presence of optimal levels of cellular sugars, it binds to KINASE 10 and reduces its affinity for GLUTAMATE RECEPTOR IONOTROPIC KAINITE; GRIK7. This results in decreased phosphorylation of the KIN10 activation loop, making WRI1 more stable for the activatation of transcription of genes involved in glycolysis and fatty acid biosynthesis (Fig. 1). Conversely, when the cellular sugar levels are low, GRIK binds tightly to KIN10, phosphorylating its activation loop. Activated KIN10 phosphorylates WRI1 leading to its degradation via ubiquitin-proteasomal pathway and hence reducing Fatty acid synthesis.
Regulation of WRINKLED1 in fatty acid biosynthesis. High levels of sugar activate WRI1 protein by dephosphorylation of Kin10 protein that activates genes involved in fatty acid biosynthesis. During low sugar conditions, Kin10 binds GRIK inhibitor which is responsible for phosphorylation of WRI1 and leads to proteosomal degradation.
Cotton seed contains 15–20% of oil and about 30–38% of kernel8. Cottonseed oil contains essential fatty acids, antioxidants and some vitamins and minerals9. It contains 55% polyunsaturated fatty acids, 18% monounsaturated fatty acids and 27% saturated fatty acids10 indicating its higher nutritional value11. Previously RNAi technology was utilizesed by researchers to attenuate the expression of genes coding for fatty acid desaturase (FAD2), stearoyl-ACP desaturase 1 (SAD1) and β-ketoacyl-acyl carrier protein synthase (KASII) which altered fatty acid composition and increased the oil content of G.hirsutum12,13,14,15. The knockout mutation of the GhFAD2 (fatty acyl desaturase 2) genes by CRISPR/Cas9 editing system increased the oleic acid level to 77.7% with decrease in linoleic acid (from 58.6 to 6.9%) and palmitic acid (from 23.95 to 13.18%)16. These studies were conducted under the expression of native promoters. But the current study focused on the seed-specific expression of transgene which can only alter the oil content of the seed, posing minumal disturbance to the metabolic processes in other plant parts. Cotton wri1 gene plays role in the development of cotton fiber through regulating the carbon flow17. In G. hirsutum, silencing of WRI1 increased the length of fiber and reduced the seed oil content18. This suggests a positive correlation of WRI1 expression with the fatty acid biosynthetic pathway and a negative correlation with fiber elongation. Ectopic expression of WRI1 from G. barbadense complemented the seed phenotype, germination on sucrose deficient media and expression of oil biosynthesis genes in the wri1-3 mutant of A. thaliana6. As homologues of WRI1 gene in various plants vary a great deal in their sequence and whether they show divergence or convergence in their function is still uncertain19. In this study, we compared various homologues of wri1 genes from diverse plant species and identified differences in their structures and minidomains in C-termini. We constructed a gene cassette containing Atwri1 driven by seed specific promoter and translational enhancer in the binary vector and transformed G. hirsutum to analyze its effect on cotton seed oil content.
Results
Multiple sequence alignment and phylogenetic analysis of WRINKLED-1 proteins from various plant species
The amino acid sequences of WRI-1 homologues from various plant species i.e. Gossypium hirsutum, G. arboreum, G. raimondii, G. barbadense, (A) thaliana, Brassica napus, (B) oleraceae, Camelina sativa, Solanum lycopersicum, Glycine max, Arachis hypogaea, Nicotiana benthamiana and Zea mays were aligned through Clustal W tool in MEGAX software and conserved domains were determined20. Highly conserved regions were observed in the N- termini of these proteins which mainly constitute two AP2 domains while the C- termini were more diverse (Fig. 2). In Gossypium spp. and A. thaliana, the first AP2 ___domain was spanning 57 amino acids (YRGVTRHRWTGRFEAHLWDKSSWNSIQNKKGKQVYLGAYDSEEAAAHTYDLAALKYW) and the second AP2 ___domain 51 amino acids (SKYRGVARHHHNGRWEARIGRVFGNKYLY LGTYNTQEEAAAAYDMAAIEYRG). Both of these domains are involved in direct binding of WRI1 to double stranded DNA thus enabling WRI1 to play the role of transcription factor (Fig. 2).
A phylogenetic tree was constructed with MEGAX software using the maximum likelihood method. It was observed that all the cotton species clustered in the same clade. The clade containing cotton species had shown close relationship with Brassicaceae family (Fig. 3). The amino acid sequence of wri-1 gene between G. hirsutum and A. thaliana had shown 62% of the homology. The WRI1 of (A) thaliana and (B) napus show 75% similarity and so belonged to the same clade. The WRI1 of G. max and A. hypogaea show 62% similarity. The similarity of WRI1 between A. hypogaea and N. benthamina was 53%. The similarity between A. thaliana and N. benthamiana was 57%. Z. mays and S. lycopersicum show 40% homology whereas this clade shows 16% similarity with A. thaliana.
Comparison of conserved domains of WRI1 in various plant species
The conserved motifs/domains predicted by motif search tool found two AP2 DNA binding domains in WRI1 of all species but the ___location of the domains showed some variation (Fig. 4). In all species, the length of first ___domain was from 57 to 60 aa while the second ___domain was 54 to 67 aa long. There was an additional spore coat ___domain (134 aminoacids) in WRI1 of A. thaliana and other members of the Brassicaceae family. The spore coat ___domain started from 243 to 378 aa approximately. In three of the species of cotton, G. hirsutum, G. raimondii and G. arboreum, there was an additional ___domain Kin 17 (Fig. 4). The position of this ___domain was same in all cotton species at 235 to 302 aa and it was 67 aa long. Surprisingly, this ___domain was not present in the wri1 from G. barbadense (Gbwri1) which complements the function of AtWRI121.
3D structure comparison of At WRI1 and Gh WRI1
Through protein sequence alignment it was observed that there were differences in a few residues of conserved DNA binding AP2 domains. To study the effect of difference in these residues, we predicted the secondary and 3D structures of proteins to reveal any differences in the percentage of helices, beta sheets, and coils. In AtWRI1 and GhWRI1, there were 23% and 21% helices, 13% and 12% Beta sheets, and 63% and 66% coils respectively. It was observed that there was no difference in the structures of AP2 domains that were mainly involved in DNA binding and transcriptional regulation (Fig. 5A,B). From the superimposed structures, it was observed that there was a difference in the C-terminus of the protein structures as shown in Fig. 5C. There was an alpha helix on the C-terminus of AtWRI1 that was not present in the GhWRI1. There was another alpha-helix on the C-termini of both AtWRI1 and GhWRI1 that was different in length and position. This alpha helix was 21 residues long (246–266 aa) in AtWRI1 and 32 residues long (280-312aa) in GhWRI1.
This alpha helix harboured a spore coat ___domain in AtWRI1 and Kin17_mid ___domain in GhWRI (Fig. 5D). The spore coat ___domain is involved in the protection of a dormant spore from degrading enzymes lysozyme and from mechanical damage. It also protects from chemicals like hydrogen peroxide22. However, the Kin17_mid ___domain is involved in the activation of WRI1 protein that subsequently regulates the transcription of genes involved in fatty acid biosynthesis23.
The metabolic role of spore coat and kin17 domains in seed oil accumulation and biosynthesis is still unexplored. However, experimental data suggests that overexpression of AtWRI1 enhanced seed oil content in G. max24 while GhWRI1 has been reported to be involved in fiber elongation17 and seedoil accumulation25. Moreover, native GhWRI1 was already present in the untransformed plant so we decided to proceed with the isolation and transformation of AtWRI1 in order to enhance the seed oil content in G.hirsutum.
Transient expression of AtWRI1 in N. benthamiana
In order to confirm that the WRI1 transcription factor isolated from A. thaliana was functional and expressing inside the nucleus, we conducted transient expression experiments before their stable transformation. pMDC83 plasmid was used as cloning vector (Fig. 6A) to construct 35 S:: AtWRI1:GFP through gateway cloning system. AtWRI1:GFP was found to be expressed transiently in the epidermal cells of N. benthamiana leaf, with strong expression in the nucleus as well as in the cytoplasm (Fig. 6B).
This experiment validated the successful construction of recombinant plasmids for the spatial and temporal expression of the 35 S:: AtWRI1:GFP cassette.
Overexpression of Atwri1 in G. hirsutum transformants
Construction of SSP: TEV:AtWRI1gene cassette in pCAMBIA1301 vector (Fig. 7A) was confirmed through PCR with the gene specific primers (Table 1) showing band size of 2.6 kb (Fig. 7B).
To transform G. hirsutum, a total of 12,000 seeds were delinted. Approximately 9000 seeds (75%) were germinated and 7000 embryos (58%) were isolated from the sterile cotton seeds and excision was made on the apical part of the embryos and then co-cultured with the A. tumefaciens transformed with pCAMBIA1301- gene cassette. Nearly 2000 transformed embryos (16%) were plated on MS medium. 1000 germinating embryos (8.3%) were transferred to hygromycin selection medium after propagation of roots. Nearly 200 embryos (1.6%) were able to survive on the selection media. About 100 (0.83%) of plantlets that survived were then transferred to autoclaved soil (Fig. 8). The transformation efficiency was 1.4%. Many of the transformed plantlets couldn’t survive on the soil and faced fungal contamination issues. However, ten transgenic lines were successfully maintained in pots and 6 plants showed stable integration of transgene by PCR (Figure S1).
Morphological and anatomical features of WT and transformant seeds
Seeds of WT and transformant cotton plants containing AtWRI1 gene (Figure S1) were compared with respect to morphological and anatomical characters. It was observed that there was no significant colour difference between the seeds of wild type and transformants, but the surfaces of the outer coat had shown a difference in smoothness. The surface of WT seeds was smooth but the surface of transformants was more rough (Fig. 9A). The seeds of wild type (un transformed control) and transformant plants were taken in triplicates (n = 3) and dissected into two equal halves, yellow coloured oil bodies spreading throughout the seeds were counted (Fig. 9B). It was found that there was a significantly higher number of oil bodies in the transformants 39 ± 1.52 as compared to the wild type 10 ± 2.0. There was a statistically significant enhancement in the number of oil bodies between WT and Transformant 1, 2 and 3 (Fig. 9C) as the P- value was less than 0.05 in each case. Moreover, for measurement of total seed oil content, untransformed control and transformed seeds were taken in triplicate (n = 3) and each replicate were having 7 cotton seeds. The higher volume of total seed oil content was observed in the transformant 1, 2 and 3 in comparison with the WT/ untransformed control (Fig. 9D). For seed weight, total of 10 seeds of untransformed control and transformed were taken in triplicates (n = 3). A significant increase in the seed weight was observed in Transformant 3 in comparison with the WT (Fig. 9E). There was no significant difference in the seed weight between WT and Transformant 1 and 2 .
Fatty acid profiling of WT and transformants by GC-MS
It was observed that there was significant difference in the GC-MS profiles of WT and transformed plants (Figure S2). A higher number of compounds was detected in transformants as compared to WT (Table 2) under same identification parameters. There were four kinds of fatty acid compounds which were identical in WT and transformants at specific retention times, RT (3.1, 4.5. 6.9 and 7.5). These compounds included saturated fatty acids i.e. myristic acid (c14), palmitic acid (c16), oleic acid (c18) and linoleic acid (C18).
However, the over-expression of AtWRI1 in transformants had shown more detected compounds at different RTs in comparison with the wild type. These compounds included lauric acid methyl ester (c12), 1- dodecanol (c12), palmitoleic acid (c16), margaric acid (c17), stearic acid (c18), linolenic acid (c18), methyl 9,10-methylene-octadecanoate (c20), methyl 18-methylnonadecanoate (c21), 13-docosenoic acid methyl ester (c22), methyl 20-methyl-heneicosanoate (c23), lignoceric acid (c24). There was a variation of fatty acid compounds in between the transformants. In transformant 1, all compounds were detected except two lauric acid methyl ester (c12) and 13-docosenoic acid methyl ester (c22). In Transformant 3 however, six compounds were not detected: lauric acid methyl ester (c12), 1- dodecanol (c12), linoleic acid (c18), methyl 20-methyl-heneicosanoate (c23), 13-docosenoic acid methyl ester (c22) and lignoceric acid (c24). Transformant 2 therefore accumulated more fatty acid compounds than Transformants 1 and 3.
Discussion
Genetic studies have shown that WRI1 is necessary for the development of normal ovules and seeds in plants26. The required levels of fatty acids are maintained by the activation of WRI1 in the presence of excess sugar leading to the fatty acid biosynthesis, while low levels of sugar lead to the proteosomal degradation of WRI1 (Fig. 1). This regulation of WRI1 involves Kin10, GIRK, SnRK1 and other proteins27. Motif and ___domain search analysis revealed the presence of two conserved AP2 domains in the N-termini of all thirteen plant species studied (Fig. 2). However, the C-termini were largely diverse containing mini motifs and domains which may be involved in protein binding, post-translational modification and transport28. We located a spore coat ___domain in the C-terminus of WRI1 that protects the spores (seeds) from mechanical disruption and was only found in the members of Brassicaceae family29. This ___domain protects the spore from mechanical damage29, but is missing in cotton species and may at least partly explain why the surface of cotton transformants was rough and wrinkled. In the cotton species, an additional ___domain was Kin 17 found in a family of eukaryotic nuclear proteins. This ___domain is responsible for activation and in-activation of WRI1 protein (Fig. 3)21,23. Phylogenetic analysis revealed the ancestral relationship and close homology between the Brassicacceae family and cotton species (Fig. 4).
The 3D structures of AtWRI1 and GhWRI1 were compared and revealed the presence of similar DNA binding motifs but different alpha helices at the C terminal. This different helix may create the sporecoat ___domain in A. thaliana and Kin 17 ___domain in G. hirsutum (Fig. 5). AtWRI1 gene is responsible for oil biosynthesis in plants but GhWRI-1 also has a role in cotton fiber length6. To modulate the cotton seed oil content, AtWRI1 (with established function in regulating seed oil content in Arabidopsis) was selected and tobacco transient expression confirmed nuclear expression (Fig. 6). A gene cassette was constructed consisting of AtWRI1 expressed under a seed specific promoter (Fig. 7). AtWRI1-expressing cotton transformants exhibited a significant increase in the number of oil bodies and oil content compared to wild type (Fig. 9B, D), with novel fatty acid profiles (Figure S2 and Table 2).
In oilseeds, the plastids contain short chain saturated and monounsaturated fatty acids which travel towards the cytosol for other modifications and also for producing polyunsaturated fatty acids, mainly oleic acid (18:1), while little stearic acid and palmitic acid accumulate in the cytosol30. Other studies have also reported the enchancement of oleic acid in sunflower with oleate (80–90%) by molecular techniques31. In the present study lauric acid methyl ester (c12), 1- dodecanol (c12), palmitoleic acid (c16), margaric acid (c17), stearic acid (c18), linolenic acid (c18), methyl 9,10-methylene-octadecanoate (c20), methyl 18-methylnonadecanoate (c21), 13-docosenoic acid methyl ester (c22), methyl 20-methyl-heneicosanoate (c23), lignoceric acid (C24) were produced only in the transformants (Table 2).
In rapeseed, mutants have been identified with altered fatty acid content for increasing the crop’s nutritional value32. In addition to increase in the oil percentage, there was reduction in fuzzy lint on the surface of seed, which increased oil extraction efficiency33. Vernolic acid production in transgenic cotton seed oil has been achieved by overexpression of a fatty acid desaturase (FAD2) that resulted in an increased content of linoleic acid12,34. Since WRINKLED-1 transcription factor directs carbon flow from sugar biosynthesis towards fattyacid biosynthesis, so the assessment of fiber yield and quality of transgenic cotton plants with reference to non-transgenic controls is logical. In our case, due to small sample size of the transgenic plants from a single event, the fiber analysis were not performed.
Conclusion
There is a need to develop varieties of cotton that can produce increased oil content, given pressures on land use and increasing demand. By introducing the AtWRI1 gene under the seed specific promoter in cotton, it was found that the oil content in transgenic cotton plants was significantly increased (35%) as compared to the wild type plant. This approach should be helpful in increasing the oil yield not only in cotton but also in other oil producing crops.
Materials and methods
Multiple sequence alignment and phylogenetic analysis
The amino acid and coding DNA sequences (CDS) of WRINKLED1 genes (WRI1) were retrieved from thirteen different species of plants through NCBI Genbank (https://www.ncbil.nlm.nih.gov). Online motif search software (https://www.genome.jp/tools/motif/) was used to identify conserved motifs in different species. These sequences were aligned and % homology was analyzed on MEGAX by using the CLUSTALW tool under default settings. Multiple sequence alignment was then used to prepare a phylogenetic tree by using MEGAX software (https://www.megasoftware.net/). Un-rooted maximum likelihood tree on a Jones-Taylor-Thornton matrix based model was generated with bootstrap value of 500.
3D structure analysis of WRI1
3D structures of GhWRI1 and AtWRI1 were predicted by using the alphafold protein structure database (https://alphafold.ebi.ac.uk/). To visualize the similarities and differences between these structures, we used PyMOL software (https://pymol.org/). RaptorX (http://raptorx6.uchicago.edu/) was used to identify the percentage of secondary structures of proteins35.
Transient expression of AtWRI1 in tobacco leaf
We constructed a gene cassette in which AtWRI1was fused with green florescent protein (GFP) cloned in pMDC83 vector under constitutive promoter Cauliflower Mosaic Virus 35 S promoter. Primers were designed for the Gateway cloning method36 (Table 1).
Plasmid containing the gene of interest was used to transform Agrobacterium tumefaciens by using a freeze thaw method37. Nicotiana benthamiana leaf epidermal cell transfection using A. tumefaciens was performed. Overnight cultures were set up using the transformed A. tumefaciens cells with pro35S:: AtWRI1:GFP and p19 protein with antibiotic selection marker. The cultures were incubated at 30 ℃ for 18 h with constant shaking at 220 rpm. Cells were harvested from the overnight culture and washed 3 times with the infiltration buffer (40 mM MES, 2.5 mM Na2PO3, 2.8 mM glucose, 100 mM acetosyringone) and were suspended in 1 mL of infiltration buffer. P19 protein and A. tumefaciens cells with the construct were mixed in 1:1 and then used for infiltration of N. benthamiana. The lower surface of the 4-week old N. benthamiana plant was gently infiltrated with the cell suspension. Plants were incubated for 3 days to allow protein to be expressed. GFP fluorescence in the epidermal cells was observed using confocal microscopy.
Expression of AtWRI1 in transgenic G. hirsutum
RNA was extracted from young leaves of A. thaliana (Col-0) through RNeasy mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. Random hexamer primers and SuperScript™ II RT First Strand cDNA Synthesis kit (Invitrogen) was used for reverse transcription of RNA. Sequence specific primers were designed from coding region having adaptors for the restriction sites through Primer3 online software (http://bioinfo.ut.ee/primer3-0.4.0/) and were used for amplification of wri1 gene (Table 1). For the isolation of seed specific Promoter, SSP (GenBank accession no. M19389), sequence specific primers and G. hirsutum DNA template were used (Table 1). 5’ UTR from Tobacco Etch Potyvirus (TEV) was synthesized that is known to enhance translation of genes38. The promoter, enhancer and gene was ligated together to form a gene cassette (SSP: TEV:AtWRI1) that was cloned in plant expression binary vector pCambia 1301. The construct was transformed in Agrobacterium by electroporation to transform G. hirsutum plants.
Seeds of cultivated variety of G. hirsutum (FH-490) were delinted by using concentrated sulphuric acid with the ratio 100 mL kg-1. Delinted seeds was sterilized by 30 mL of autoclaved water along with 1 mL of 10% SDS and 2 mL of HgCl2. Seeds were washed with the autoclaved water until all foam removed. Seeds were incubated at 30 ℃ for 72 h39.
Testa and cotyledons were removed from germinated seeds and embryos were isolated under sterile conditions. A cut was made on the hypocotyl with a sharp sterile blade and co-cultured them in the A. tumefaciens inoculum at 30 ℃ for 2 h with 250 rpm shaking40. After incubation, embryos were dried on filter paper and then transferred on MS media plates at 30 ℃ for 4 days with 16 h light and 8 h of dark. Plantlets were transferred to sterile test tubes containing MS selection media supplemented with hygromycin (25 mgL-1), cefotaxime (250 µgmL-1) and B5 vitamins41. Plantlets were incubated in the growth chamber for 6 weeks until roots and shoots develop. Rooted plants were washed with autoclaved water and dipped in IBA (1 mgmL-1) before planting in soil mixture-filled pots42.
Microscopic analysis
A disc of leaf was cut, near the point of infiltration of tobacco plant. The leaf disc was placed upside down on a glass slide in water with cover slip. SP5 confocal microscope (Leica, Wetzlar and Germany) was used to image by using x63 water objective. GFP fluorescence was visualized by sample excitation with Argon laser at 488 nm and the spectral detection was set at 500–548 nm. Final images were prepared by using ImageJ software (https://imagej.nih.gov/ij/). Morphology of delinted seeds and anatomy of wild type (WT) and transformants was observed under the stereomicroscope. WT cotton seeds and transformants were arranged in parallel on the glass slide and images were captured by digital photo camera.
Statistical analysis
Statistical analysis were performed by using GraphPad prism (https://www.graphpad.com/features). One way analysis of variance (ANOVA)43 was used to compare the means of WT/ Control and transformants. P- value less than 0.05 indicated significant differences between mean values with the confidence interval of 95% while the P-values of 0.01 indicated highy significant differences with the confidence interval of 99%, between untransformed control and transformants. Bonferroni’s multiple comparison method was applied as a post-hoc test to find the difference between individual groups44.
Determination of seed-oil content and fatty acid profiling by GC-MS
The outer coat of the seeds was removed and seeds were ground to fine powder using a pestle and mortar. 0.5 g powder of each seed was weighed, and 4 ml of 1 M HCl in MeOH and 300 µl of hexane were added. Samples were seal topped and incubated at 80 °C for 3 h, then cooled to room temperature. 4 ml of 0.9% NaCl was added and the layers allowed to separate. 250 µl of the oil layer at the top was used for gas chromatography- mass spectrometry analysis (GC-MS)45.
The instrument used for GC-MS analysis was GCMS QP 2010 Plus (Shimadzu, Kyoto and Japan) where DB-23, 30 m x 0.25 mm ID x 0.15 μm FT column (Agilent, Calinfornia, United states) was fitted for the separation of oil constituents. The programme used for GC separation and identification of fatty acid methyl esters was that the temperature raised at a rate of 2 °C/min to 160 °C and held for 2 min, then it was raised at a rate of 4 °C/min to 200 °C and held for 2 min and finally it was raised to 224 °C at a rate of 6 °C /min and held for 2 min. Injector temperature was employed at 250 °C whereas the ion source temperature was 200 °C. Helium gas was used as mobile phase. Oil dissolved in n-hexane introduced in GCMS46 The pressure was about 105.5 kPa. The total flow rate was 114.2 ml/min and column flow was 1.10 ml/min. The linear velocity was set upto 40 cm/sec and purge flow was 3mL/min where m/z range was 45–500.These conditions were used to calculate their linear retention indices (RI). Identification of metabolites was performed by matching their mass spectra with the NIST-08 library and comparing RI with standards available in the literature47.
Data availability
Original data is available in the article and supplementary material and further information can be requested from the corresponding author.
References
Vogel, P. A. et al. Expression of the Arabidopsis WRINKLED 1 transcription factor leads to higher accumulation of palmitate in soybean seed. Plant Biotechnol. J. 17, 1369–1379 (2019).
To, A. et al. WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant. Cell. 24, 5007–5023 (2012).
Baud, S. et al. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50, 825–838 (2007).
Focks, N. & Benning, C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118, 91–101 (1998).
Cernac, A. & Benning, C. WRINKLED1 encodes an AP2/EREB ___domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40, 575–585 (2004).
Imran, M. et al. Ectopic expression of fiber related Gbwri1 complements seed phenotype in Arabidopsis thaliana, (2020).
Soto-Burgos, J. & Bassham, D. C. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PloS One. 12, e0182591 (2017).
Rashid, U., Anwar, F. & Knothe, G. Evaluation of biodiesel obtained from cottonseed oil. Fuel Process. Technol. 90, 1157–1163 (2009).
Onukwuli, D. O., Emembolu, L. N., Ude, C. N., Aliozo, S. O. & Menkiti, M. C. Optimization of biodiesel production from refined cotton seed oil and its characterization. Egypt. J. Petroleum. 26, 103–110 (2017).
Eevera, T. & Pazhanichamy, K. Cotton seed oil: A feasible oil source for biodiesel production. Energy Sour. Part a Recover. Utilization Environ. Eff. 35, 1118–1128 (2013).
Pouvreau, B. et al. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant. Physiol. 156, 674–686 (2011).
Liu, F., Zhao, Y. P., Zhu, H., Zhu, Q. H. & Sun, J. Simultaneous silencing of GhFAD2-1 and GhFATB enhances the quality of cottonseed oil with high oleic acid. J. Plant Physiol. 215, 132–139 (2017).
Liu, Q. et al. Genetic Enhancement of Palmitic acid Accumulation in Cotton seed oil through RNA i down-regulation of Gh KAS 2 Encoding β-ketoacyl-ACP Synthase II (KASII), Plant Biotechnol, pp. 15132–143 (2017).
Liu, Q., Singh, S. & Green, A. High-oleic and high-stearic cottonseed oils: Nutritionally improved cooking oils developed using gene silencing. J. Am. Coll. Nutr. 21, 205S–211S (2002).
Cui, Y. et al. Genome-wide characterization and analysis of CIPK gene family in two cultivated allopolyploid cotton species: Sequence variation, association with seed oil content, and the role of GhCIPK6. Int. J. Mol. Sci. 21, 863 (2020).
Chen, Y. et al. High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR/Cas9 system. Plant Biotechnol. J. 19, 424 (2021).
Qaisar, U., Akhtar, F., Azeem, M. & Yousaf, S. Studies on involvement of Wrinkled1 transcription factor in the development of extra-long staple in cotton. Indian J. Genet. Plant. Breed. 77, 298–303 (2017).
Qu, J. et al. Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant. Physiol. 160, 738–748 (2012).
Tan, Q. et al. The molecular mechanism of WRINKLED1 transcription factor regulating oil accumulation in developing seeds of castor bean. Plant. Divers. 45, 469–478 (2023).
Qaisar, U. et al. Identification, sequencing and characterization of a stress induced homologue of fructose bisphosphate aldolase from cotton. Can. J. Plant. Sci. 90, 41–48 (2010).
Imran, M. et al. Ectopic expression of Fiber related Gbwri1 complements seed phenotype in Arabidopsis thaliana. Int. J. Agric. Biol. 25, 123–130 (2021).
Driks, A. & Eichenberger, P. The Bacterial Spore: From Molecules to Systems (Wiley, 2020).
Garcia-Molina, A., Xing, S. & Huijser, P. A conserved KIN17 curved DNA-binding ___domain protein assembles with SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE7 to adapt Arabidopsis growth and development to limiting copper availability. Plant Physiol. 164, 828–840 (2014).
Vogel, P. A. et al. E.B. Cahoon, t. Clemente, expression of the Arabidopsis WRINKLED 1 transcription factor leads to higher accumulation of palmitate in soybean seed. Plant. Biotechnol. J., (2019).
Zhao, Y., Liu, Z., Wang, X., Wang, Y. & Hua, J. Molecular characterization and expression analysis of GhWRI1 in Upland cotton. J. Plant. Biol. 61, 186–197 (2018).
Okamuro, J. K., Caster, B., Villarroel, R., Van Montagu, M. & Jofuku, K. D. The AP2 ___domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. 94, 7076–7081 (1997).
Shigyo, M., Hasebe, M. & Ito, M. Molecular evolution of the AP2 subfamily. Gene 366, 256–265 (2006).
Ramzan, A., Rehman, T., But, M. S., Imran, M. & Qaisar, U. Biofilm formation and its regulation by extracellular appendages in Pseudomonas aeruginosa. World J. Biology Biotechnol. 7, 1–5 (2022).
Driks, A. & Eichenberger, P. The spore coat, the bacterial spore: From molecules to systems, 179–200. (2016).
Bates, P. D. & Browse, J. The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci. 3, 147 (2012).
Murphy, D. J. Using Modern Plant Breeding to Improve the Nutritional and Technological Qualities of oil Crops21D607 (OCL, 2014).
Gupta, S. K. Breeding Oilseed Crops for Sustainable Production: Opportunities and Constraints (Academic, 2015).
Shockey, J. et al. Naturally occurring high oleic acid cottonseed oil: Identification and functional analysis of a mutant allele of Gossypium barbadense fatty acid desaturase-2. Planta 245, 611–622 (2017).
Kouser, S., Mahmood, K. & Anwar, F. Variations in physicochemical attributes of seed oil among different varieties of cotton (Gossypium hirsutum L). Pak J. Bot. 47, 723–729 (2015).
Shad, M., Hussain, N., Usman, M., Akhtar, M. W. & Sajjad, M. Exploration of computational approaches to predict the structural features and recent trends in α-amylase production for industrial applications. Biotechnol. Bioeng. 120, 2092–2116 (2023).
Esposito, D., Garvey, L. A. & Chakiath, C. S. Gateway cloning for protein expression. High. Throughput Protein Expression Purification: Methods Protocols, 31–54. (2009).
Xu, R. & Qingshun, L. Q. Protocol: Streamline cloning of genes into binary vectors in Agrobacterium via the Gateway® TOPO vector system. Plant. Methods. 4, 1–7 (2008).
Nicolaisen, M., Johansen, E., Poulsen, G. B. & Borkhardt, B. The 5′ untranslated region from pea seedborne mosaic potyvirus RNA as a translational enhancer in pea and tobacco protoplasts. FEBS Lett. 303, 169–172 (1992).
Gould, J. H. & Magallanes-Cedeno, M. Adaptation of cotton shoot apex culture to Agrobacterium-mediated transformation. Plant. Mol. Biology Report. 16, 283–283 (1998).
Din, S. et al. Development of broad-spectrum and sustainable resistance in cotton against major insects through the combination of Bt and plant lectin genes. Plant Cell Rep. 40, 707–721 (2021).
Shad, M. et al. Enhancing the resilience of transgenic cotton for insect resistance. Mol. Biol. Rep., 1–9. (2022).
Shad, M. et al. A brief overview for the development of herbicide-resistant sugarcane transformation approaches. NUST J. Nat. Sci., 6 (2021).
Ross, A., Willson, V. L., Ross, A. & Willson, V. L. One-way anova, Basic and advanced statistical tests: Writing results sections and creating tables and figures, 21–24. (2017).
Lee, S. & Lee, D. K. What is the proper way to apply the multiple comparison test? Korean J. Anesthesiol.. 71, 353–360 (2018).
Liu, J. et al. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant. Physiol. Biochem. 48, 9–15 (2010).
Mihai, A. L., Negoiță, M. & Belc, N. Evaluation of fatty acid profile of oils/fats by GC-MS through two quantification approaches. Romanian Biotechnol. Lett. 24, 973–985 (2019).
Eisenstecken, D., Stanstrup, J., Robatscher, P., Huck, C. W. & Oberhuber, M. Fatty acid profiling of bovine milk and cheese from six European areas by GC-FID and GC‐MS. Int. J. Dairy Technol. 74, 215–224 (2021).
Acknowledgements
We acknowledge the School of Biological Sciences, University of the Punjab, Lahore, Pakistan for providing the infrastructure and resources for research. We appreciated Higher Education Commission of Pakistan (HEC) for providing International Research Support Initiative Program (IRSIP) opportunity and Durham University, UK for allowing infrastructure for some of the experiments in Department of Biosciences. We express our gratitude to the laboratory fellows Julien Agneessens and Samina Yousaf for their help and support.
Funding
This research was financially support by School of Biological Sciences, University of the Punjab, Lahore Pakistan and Pakistan Science Foundation (PSF) .
Author information
Authors and Affiliations
Contributions
M.B. and U.Q. planned the experiments, analyzed data and drafted the manuscript. M.B prepared all the figures and tables. M.B. and M.I. performed the mainstream experiments. K.L., T.R. and A.I. analyzed the data and revised the manuscript. Manuscript read and approved by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Butt, M., Imran, M., Rehman, T. et al. Seed-specific expression of AtWRI1 enhanced the yield of cotton seed oil. Sci Rep 14, 30750 (2024). https://doi.org/10.1038/s41598-024-80684-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-80684-9