Abstract
Glioma, the most prevalent primary intracranial malignancy among adults, is distinguished by its high morbidity and recurrence rates, posing a considerable threat to patients’ quality of life and survival prospects. Consequently, the pursuit of efficacious molecular prognostic markers holds paramount importance. The exploration of the role of the KDELR3 kinase family in various neoplastic conditions constitutes an emerging area of research. However, the biological functions of KDELR3 and its prognostic implications in brain tumors remain largely undocumented. This study endeavored to ascertain the potential of KDELR3 as a prognostic indicator for glioma. We integrated a comprehensive dataset encompassing 1127 glioma samples, sourced from our cohort, The Cancer Genome Atlas (TCGA), and the Chinese Glioma Genome Atlas (CGGA), to delve into the expression patterns of KDELR3 in glioma and their associated implications. Notably, KDELR3 was markedly overexpressed in glioma and demonstrated a positive correlation with clinical progression. By utilizing Kaplan-Meier survival analysis and the Cox proportional hazards regression model, we evaluated the prognostic significance of KDELR3, revealing it as an independent predictor of adverse outcomes in glioma patients. Furthermore, gene set enrichment analysis unveiled potential signaling pathways associated with KDELR3 expression in glioma, primarily encompassing Cytokine-cytokine receptor interaction, extracellular matrix (ECM)-receptor interaction, and complement and coagulation cascades. In summation, our findings provide profound insights into the potential role of KDELR3 and its application as a novel and promising prognostic biomarker for glioma.
Similar content being viewed by others
Introduction
Glioma, the most prevalent histological subtype of primary malignant neoplasms affecting the adult central nervous system, arises from glial cells and demonstrates elevated rates of disability and recurrence. Presently, glioma is acknowledged as one of the most formidable malignancies, profoundly influencing patients’ quality of life, posing a substantial threat to their survival, and imposing a considerable burden on individuals, families, and society at large. Glioma constitutes approximately 70.9% of all malignancies within the central nervous system, with glioblastoma multiforme (GBM) being the most prevalent pathological type, accounting for 50.1% of all malignant tumors of the nervous system1,2. According to the 2020 global cancer statistics, neurological cancers ranked as the twenty-first most prevalent cancer worldwide, with an annual incidence of 308,102 novel cases, accounting for 1.6% of all cancer cases, and an annual mortality rate of 251,329 deaths, representing 2.5% of all cancer-related fatalities3. In the 2021 World Health Organization (WHO) classification system, gliomas were categorized into grades 1 to 4, where WHO grades 1 and 2 represented low-grade gliomas with a gradual progression rate, whereas WHO grades 3 and 4 signified high-grade gliomas characterized by rapid progression4.
The advancement in glioma treatment has been incremental and has primarily relied on surgical resection, in conjunction with radiotherapy and chemotherapy5. However, the therapeutic response has been deemed inadequate, primarily due to the infiltrative nature of gliomas, which hinders the achievement of a genuine complete resection. GBM has emerged as the most aggressive form of brain tumor in adults, exhibiting a median survival ranging from 17 to 25 months despite advancements in current therapies and the implementation of tumor-treating fields (TTFields)6,7.
Gliomas of WHO grades 2 to 3 were classified as lower-grade gliomas (LGG)8, demonstrating a substantial disparity in median survival, spanning from 22.8 to 176.4 months9,10. The heterogeneity exhibited by GBM and LGG has contributed to considerable variations in survival outcomes. Beyond traditional histological grading, molecular markers provide profound prognostic insights, emphasizing the crucial importance of tumor genomics studies4,11. Molecular pathology not only elucidates the unique molecular genetic variations present in tumors but also exerts a direct influence on clinical prognosis and the selection of therapeutic interventions. Consequently, the pursuit of more efficacious molecular prognostic markers holds substantial value in the management of glioma patients. Deciphering the pathogenesis and etiology of gliomas could potentially lead to the discovery of advanced therapeutic strategies and effective diagnostic and prognostic biomarkers.
KDELR, a protein-binding receptor primarily localized within the endoplasmic reticulum (ER) and Golgi apparatus, served as a crucial mediator in the transport of ER proteins between these two organelles12,13. The KDELR-orchestrated cycling of proteins from the Golgi to the ER was indispensable for a variety of biological processes, including protein synthesis and folding, as well as lipid metabolism14. Within the Golgi apparatus, KDELR facilitated the transport of proteins from the Golgi to the ER via the COPI pathway, thereby initiating the retrograde transport of vesicles to the ER and maintaining a balanced membrane flux between the two organelles15. Among the substrates involved in KDELR-mediated retrograde transport were protein chaperones, folding chaperones, folding enzymes, enzymes targeting proteins for degradation, and glycosylases14,16, all of which contributed to the maintenance of proper protein processing and the regulation of cellular responses to ER stress17. Furthermore, the expression level of KDELR was modulated by the activation of the unfolded protein response (UPR) signaling pathway, with the aim of mitigating the loss of ER-resident proteins18,19. Additionally, research has demonstrated that the KDELR pathway enhanced autophagy under stress conditions by promoting the formation of autophagosomes and their fusion with lysosomes20.
In mammals, the KDELR family consists of three distinct subtypes: KDELR1, KDELR2, and KDELR3. KDELR3 demonstrated the ability to modulate specific functional properties of proteins within the ER lumen. Notably, its expression in atherosclerotic macrophages was significantly reduced compared to non-atherosclerotic tissues, thereby positioning it as a potential diagnostic biomarker for atherosclerosis21. KDELRs were also abnormally expressed in a variety of tumors, exhibiting a close correlation with prognosis and therapeutic response, including renal cancer, prostate cancer, and hepatocellular carcinoma22,23,24. Furthermore, in melanoma, KDELR3 facilitated metastasis by regulating the glycosylation of the metastasis inhibitor KAI1 and the degradation of gp78, thereby emerging as a prometastatic gene25. However, the expression patterns and clinical implications of KDELR3 in glioma have remained largely unexplored.
In our study, we demonstrated that KDELR3 was overexpressed in a broad range of malignancies, including glioma. Elevated KDELR3 expression was significantly associated with tumor progression, suggesting its potential as a biomarker for glioma diagnosis and prognosis. Moreover, the transcriptional expression of KDELR3 in glioma patients emerged as an independent risk factor influencing patient survival. Additionally, pathway and functional enrichment analyses revealed that the tumorigenesis mechanism mediated by KDELR3 potentially involved cytokine-cytokine receptor interactions, ECM-receptor interactions, and other critical biological processes. Our findings underscored the pivotal role of KDELR3 in glioma prognosis and provide a reliable biomarker for the diagnosis and prognosis of glioma.
Results
Overexpression of KDELR3 in glioma
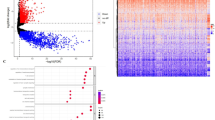
Data acquired from the Cancer Cell Line Encyclopedia (CCLE) repository revealed a substantial expression of KDELR3 in numerous cancer cell lines, particularly GBM and LGG (Fig. 1A). In light of the paucity of studies examining the association between KDELR3 and tumorigenesis, we conducted an exhaustive analysis of 33 tumor types from TCGA. Notably, 14 cancer types exhibited overexpression of KDELR3 (Fig. 1B).
The expression of KDELR3 in different types of cancer, including glioma. (A) The expression of KDELR3 in different types of cancer cells was obtained from the CCLE database. (B) The expression of KDELR3 in different types of cancer was obtained from Tumor Immune Estimation Resource database. *p < 0.05; ***p < 0.001.
Correlation of KDELR3 overexpression with advanced glioma
Subsequently, we investigated the correlation between KDELR3 mRNA levels and clinicopathological parameters in glioma patients. The TCGA database encompassed comprehensive clinical information, including tumor grade, gender, survival status, isocitrate dehydrogenase (IDH) mutation/wild-type status, 1p19q co-deletion status, and therapeutic response assessment. Baseline analysis revealed a statistically significant association between KDELR3 expression levels and various factors, including WHO grade, IDH mutation status, 1p19q deletion, therapeutic response, Overall Survival (OS), Disease-Free Survival (DFS), Progression-Free Interval (PFI), and age (Table S1). As depicted in Fig. 2A, KDELR3 expression was augmented in tumor tissues, demonstrating a positive correlation with tumor grade. Furthermore, elevated KDELR3 expression in the TCGA database was associated with advanced age, IDH wild-type status, and the absence of 1p19q co-deletion. Notably, patients who exhibited progressive disease (PD) after initial treatment displayed higher KDELR3 expression compared to those with complete remission (CR), partial remission (PR), and stable disease (SD) (Fig. 2B-F).
Elevated KDELR3 expression correlates with adverse survival outcomes
Considering the association between high KDELR3 expression in glioma patients and tumor grade as well as therapeutic response, we sought to elucidate its prognostic significance through the Kaplan-Meier survival analysis. As illustrated in Fig. 3, elevated KDELR3 expression levels were significantly associated with poorer OS, DFS, and PFI in TCGA samples (Fig. 3A-C). Subgroup analysis further revealed a significant correlation between higher KDELR3 expression and inferior OS in patients with WHO grade 2 and 3 , both age groups (< 60 years and ≥ 60 years), male and female patients, IDH mutation status, 1p19q co-deletion status, and PD (Fig. 3D-M). In samples from the Chinese Glioma Genome Atlas (CGGA), significantly high levels of KDELR3 expression were notably associated with poorer OS (Fig. S1). In summary, the findings of this study demonstrated a significant correlation between KDELR3 expression and the prognosis of glioma patients, indicating its potential as a biomarker for predicting survival outcomes in this patient cohort.
Survival analysis of KDELR3 in gliomas. (A) OS Kaplan-Meier analysis of KDELR3 in gliomas. (B) DSS Kaplan-Meier analysis of KDELR3 in gliomas. (C) PFI Kaplan-Meier analysis of KDELR3 in gliomas. (D−M) Kaplan-Meier analysis of overall survival of KDELR3 in WHO grade 2 and 3, Age>60, Age ≤ 60 , Female, Male, IDH Mut, 1p19q codel, 1p19q non-codel, primary therapy outcome PD gliomas.
KDELR3 correlates with immune infiltration in glioma
As depicted in Fig. 4A, KDELR3 exhibits a positive correlation with the infiltration levels of macrophages, T helper type 2 (Th2) cells, neutrophils, eosinophils, immature dendritic cells (iDCs), T cells, activated dendritic cells (aDCs), cytotoxic cells, and Natural killer (NK) CD56bright cells. Conversely, it displayed a negative correlation with plasmacytoid dendritic cells (pDCs) and gamma delta T cells (γδT cells). Statistically significant differences in enrichment scores were observed between the high and low KDELR3 expression groups for the aforementioned immune cell populations (p < 0.001) (Fig. 4B, Fig. S2). Specifically, significant positive correlations were identified for macrophages (r = 0.670, p < 0.001), Th2 cells (r = 0.573, p < 0.001), neutrophils (r = 0.563, p < 0.001), eosinophils (r = 0.559, p < 0.001), iDCs (r = 0.516, p < 0.001), T cells (r = 0.499, p < 0.001), aDCs (r = 0.465, p < 0.001), and cytotoxic cells (r = 0.408, p < 0.001). Conversely, negative correlations were observed for pDCs (r = -0.461, p < 0.001) and γδT cells (r = -0.441, p < 0.001). These findings suggest the potential role of KDELR3 as an immunomodulator in glioma.
Functional enrichment analysis
To elucidate the functional roles and signaling pathways of genes co-expressed with KDELR3, enrichment analyses were conducted using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. The results of the GO analysis indicated that the co-expressed genes were predominantly associated with biological processes such as cytokine-cytokine receptor interaction, extracellular matrix (ECM)-receptor interaction, and complement and coagulation cascades (Fig. 5A). The KEGG analysis revealed that the co-expressed genes were primarily enriched in pathways such as Reactome cd22-mediated B-cell receptor (BCR) regulation, Reactome signaling via the B-cell receptor, Reactome FcεRI-mediated NF-κB activation, and Reactome FcγRIIIA-mediated IL-10 synthesis (Fig. 5B).
Co-expression network of KDELR3 in glioma
To gain insights into the potential biological functions mediated by KDELR3 in glioma, we utilized the GeneMANIA database, which encompassed physical interactions, co-expression, predicted interactions, co-localization, genetic interactions, pathway membership, and shared protein domains. Through this comprehensive analysis, we identified 19 proteins that interact with KDELR3, including KDELR2, CGREF1, EXOC2, ARHGAP6, RALBP1, ARHGAP4, CHN1, NCF2, ASAP1, ARFGAP1 (twice mentioned, presumably a typographical error and should be considered once), COPA, SLC31A1, ARAP1, ARAP2, ARFGAP3, ASAP2, and XBP1. Based on these interactions, a detailed network diagram was constructed (Fig. 6).
Elevated KDELR3 expression identified as an independent prognostic factor in glioma patients
A univariate Cox regression analysis was conducted to assess the prognostic significance of KDELR3 expression in glioma patients. The analysis revealed that high KDELR3 expression was significantly associated with poorer overall survival (OS) (hazard ratio [HR] = 1.125, 95% confidence interval [CI]: 1.061 to 1.193, p < 0.001). Additionally, other variables that were found to be associated with adverse survival outcomes included age, tumor grade, IDH mutation status, 1p19q codeletion, and treatment-naive response (Table S2). Based on these findings, a nomogram prognostic evaluation model was developed to predict the 1-, 3-, and 5-year OS rates. The total score, obtained by summing the individual scores, corresponded to the predicted OS rates for each patient (Fig. 7A). Furthermore, to evaluate the diagnostic significance of KDELR3 in glioma, a Receiver Operating Characteristic (ROC) curve was constructed using RNA-sequencing data from The Cancer Genome Atlas (TCGA). Notably, the areas under the ROC curve at 1-year, 3-year, and 5-year intervals were 0.836, 0.829, and 0.732, respectively, demonstrating substantial diagnostic accuracy (Fig. 7B).
KDELR3 expression is also elevated in our cohort
To further validate the expression pattern of KDELR3 in our cohort, the expression levels of KDELR3 were assessed in clinical specimens. The results revealed a significant upregulation of KDELR3 in tumors of grades 3 and 4 (Fig. 8A). Moreover, within our cohort, increased KDELR3 expression levels were concurrently associated with decreased OS (Fig. 8B). A comprehensive clinical correlation analysis demonstrated a positive correlation between KDELR3 expression and tumor grade (Fig. 8C).
Discussion
Glioma, a highly aggressive intracranial malignancy, demonstrates a progressively worsening prognosis as its grade increases. Despite the elusive etiology of glioma, two notable risk factors have been identified: exposure to high doses of ionizing radiation and inherited mutations in high-penetrance genes associated with rare syndromes26,27. However, as our comprehension of tumor genomics, immunity, and the microenvironment has deepened, the mechanisms underlying glioma initiation and progression have become increasingly apparent. This has facilitated the refinement of pathological classification, the enhancement of clinical diagnostic accuracy, and the improvement of clinical prognosis assessments. Nonetheless, due to the inherent heterogeneity and aggressiveness of glioma, the current clinical treatment outcomes remain suboptimal. In the instance of GBM, the prognosis is particularly dire, with only 3–5% of patients achieving a five-year survival rate. Although the prognosis for LGG is comparatively better, there are still instances of LGG with poor prognoses and short survival durations. Notably, approximately 70% of patients with LGG experience high-grade transformation within ten years. In recent years, numerous studies have identified potential biomarkers associated with glioma survival through gene expression profiling28,29. Consequently, the diligent pursuit of early diagnostic markers and precise prognostic biomarkers is imperative for improving the prognosis of glioma.
KDELR, a seven-transmembrane protein, primarily functioned in the retention of chaperones harboring the carboxy-terminal Lys-Asp-Glu-Leu (KDEL) sequence within the luminal protein of the endoplasmic reticulum (ER) membrane. This process prevented the misfolding and misassembly of peptide chains, thereby contributing to the regulation of cellular function30. KDELR3, a member of the ER protein retention receptor family, was postulated to be an unfolded protein response (UPR) gene activated via the inositol-requiring enzyme 1 (IRE1) pathway14. Furthermore, the loss of KDELR3 expression was observed to concurrently elevate the phosphorylation levels of protein kinase RNA-like ER kinase (PERK) and eukaryotic translation initiation factor 2α (EIF2α) in cells, thus triggering the UPR process25. KDELR was involved in the degradation of the extracellular matrix (ECM) of cancer cells through Src activation and ASAP1 phosphorylation, thereby fostering tumor metastasis and aggressive growth31. Additionally, KDELR3 promoted high metastasis in melanoma via the metastatic inhibitor KAI125. However, the diagnostic and prognostic significance of KDELR3 in cancer remains unclear. Our findings offer valuable insights into its pathological role in enhancing tumor growth and invasion, as well as its potential as a diagnostic and prognostic biomarker for glioma.
KDELR3 played a pivotal role in promoting the metastasis and invasion of melanoma25. Moreover, KDELR3, a UPR-associated gene, was shown to predict survival, immune microenvironment, and prognosis in bladder cancer patients32. Consequently, it can be speculated that the expression of KDELR3 may influence patient survival through the progression of tumor cells. Nevertheless, there is still a lack of consensus regarding the definition of KDELRs and KDELR3 expression in cancer, as well as crucial aspects such as tumor cell progression. KDELR1 was an independent prognostic factor for overall survival (OS) and correlates with immunity in glioma patients33. KDELR2 was reported to facilitate breast cancer proliferation through histone deacetylase 3 (HDAC3)-mediated cell cycle progression, indicating its potential as a target for combination therapy34. KDELR2, a downstream target gene of hypoxia inducible factor 1α (HIF1α), fostered the onset and progression of GBM via the mammalian target of rapamycin (mTOR) signaling pathway, potentially emerging as a therapeutic target for the treatment of GBM patients35. Knockdown of KDELR2 not only triggered apoptosis in glioma cells through the C/EBP homologous protein (CHOP) and c-Jun N-terminal kinase/p38 (JNK/p38) pathways but also enhanced their sensitivity to temozolomide36. Studies by Hui Mao et al. have also shown that KDELR2 was highly expressed in human glioma tissues and cell lines, regulated cellular functions by targeting cyclin D1 (CCND1), and that high expression of KDELR2 was associated with poor prognosis37. Given the similarities in their gene family, it remains to be explored whether KDELR3, another member, possesses analogous functional implications in the development of glioma.
In this study, a thorough examination of KDELR3 expression levels across a variety of tumor types within the TCGA database was performed. Our results indicated that KDELR3 was overexpressed in bladder cancer, mirroring the elevated expression levels observed in cholangiocarcinoma, colorectal cancer, lung cancer, thyroid cancer, and endometrial cancer. Notably, KDELR3 overexpression was also implicated in GBM. Currently, there were limited reports concerning the biological functions and tumorigenic mechanisms underlying KDELR3. KDELR3 has been demonstrated to facilitate metastasis in melanoma by modulating the protein abundance and post-translational modification of the metastatic suppressor KAI1 via gp78-mediated E3 ubiquitin ligase. In this study, the KDELR3-associated signaling pathways activated in glioma appeared to be predominantly enriched in cytokine signaling, cytokine-cytokine receptor interaction, and extracellular matrix (ECM)-receptor interaction pathways, hinting at its potential role in further augmenting tumor proliferation and invasion (Fig. 5A). We further assessed the potential of KDELR3 as a diagnostic and prognostic biomarker for glioma. The ROC curve analysis revealed a high diagnostic accuracy of KDELR3 expression in glioma (Fig. 7B). Additionally, the Kaplan-Meier survival analysis demonstrated a statistically significant correlation between elevated KDELR3 mRNA expression and poor OS in glioma patients (Figs. 3A and 8B, S1). Furthermore, univariate Cox regression analysis indicated that high KDELR3 expression was a predictor of poor OS in glioma patients (Table S2). In summary, our study not only highlights the overexpression of KDELR3 in glioma but also its association with more aggressive tumor phenotypes and adverse prognostic outcomes. Our findings suggest that KDELR3 may represent a promising biomarker for glioma diagnosis and prognosis.
In conclusion, we established potential prognostic and diagnostic profiles for glioma patients using two databases (TCGA and CGGA) and clinical specimens. This methodology effectively stratified glioma patients into two distinct groups with differing survival outcomes. Additionally, we identified the potential signaling pathway of KDELR3 in glioma patients. KDELR3 overexpression was indicative of malignant progression and poor survival in glioma patients, while also showing a strong correlation with biological processes such as cytokine-cytokine receptor interaction, ECM-receptor interaction, and complement and coagulation cascades in glioma. Essentially, the aforementioned results propose KDELR3 as a potential therapeutic target in glioma. Inhibition of KDELR3 may represent a feasible therapeutic strategy to impede the progression of glioma.
Materials and methods
Data acquisition and processing
The acquisition of gene expression data and clinical information pertaining to glioma was facilitated through the utilization of TCGA database (http://cancergenome.nih.gov/) and the CGGA database (http://www.cgga.org.cn/). From the TCGA database, we retrieved raw mRNA sequencing (mRNAseq) data and 699 RNAseq datasets encompassing clinical details from 706 glioma samples. Specifically, we obtained 174 RNAseq expression datasets and 168 RNAseq datasets with clinical information for GBM, along with 532 RNAseq expression datasets and 531 RNAseq datasets with clinical information for LGG. The analysis of these datasets was conducted using the edgeR software package (version 4.0.2). Furthermore, we procured sample information from the CGGA_mRNAseq_325 dataset in the CGGA database and organized the gene expression profiles and corresponding clinical data for each sample. Notably, the RNA-seq data from the CGGA database was directly expressed as Fragments Per Kilobase of transcript per Million mapped reads (FPKM).
Patient information and ethical considerations
This study adhered to the ethical guidelines approved by the Ethics Committee of the 900th Hospital of the Joint Logistics Support Force. All patients signed informed consent forms. A cohort of 135 neurosurgical patients was collected between January 2016 and December 2021. All patients were newly diagnosed with glioma based on the WHO 2007 and 2016 criteria, and possessed comprehensive clinical data. Of these, 103 patients had detailed follow-up data suitable for analytical purposes. The mutational status of IDH was investigated using Sanger sequencing, while fluorescence in situ hybridization was employed to analyze the heterozygous deletion of 1p/19q in glioma samples. The ethical approval for this study was granted by the Ethics Committee of the 900th Hospital of the Joint Logistics Support Force, under Grant No. 2023-062.
Immunohistochemical analysis
A comprehensive collection of 135 cases of glioma was procured from glioma patients at the 900th Hospital of the Joint Logistics Support Force, encompassing 50 cases of GBM, 85 cases of LGG, and 3 instances of normal brain tissue adjacent to grade 1 glioma. The surgical specimens underwent fixation with 40 g/L formaldehyde solution, followed by routine paraffin embedding to yield 4 μm thick sections. These sections were then subjected to hematoxylin and eosin (HE) staining. Immunohistochemical staining for KDELR3 was executed using the EliVision method, with subsequent microscopic examination. The Anti-KDELR3 polyclonal antibody (1:50) was sourced from Boster Inc., China (Catalog No. A14682-1). Additionally, the non-biotin universal two-step immunohistochemistry kit (mouse/rabbit enhanced polymer detection system, PV-9000) was acquired from Wuxi Aorui Dongyuan Biotechnology Co., Ltd.
The interpretation of the results adhered to the following criteria: KDELR3 positivity was indicated by the presence of brownish-yellow granules in the nucleus or cytoplasm. Cells with dark brown nuclei or cytoplasm were deemed to possess strong expression; cells exhibiting yellow or brown nuclei or cytoplasm displayed moderate intensity; and those with pale yellow nuclei or cytoplasm, or faint staining, were classified as weak-intensity cells. Absence of staining in the nucleus or cytoplasm was considered negative. Quantification of KDELR3 expression was achieved through the application of the histochemical scoring (H-score) method, calculated as H-score = (percentage of weak-intensity cells × 1) + (percentage of moderate-intensity cells × 2) + (percentage of strong-intensity cells × 3).
KDELR3 expression and statistical analysis
The Wilcoxon signed-rank test was employed to assess the expression of KDELR3. Subsequently, the correlation between clinicopathological features and KDELR3 expression was examined utilizing the Wilcoxon signed-rank test. The survival ROC software package within the R statistical environment was utilized to generate the ROC curve, which was employed to evaluate the diagnostic efficacy of KDELR3 expression. The area under the curve served as an indicator of the diagnostic value. Kaplan-Meier curves for each clinical feature were constructed using the Survival and Survminer software packages in R, and the OS between the high-expression and low-expression groups was contrasted through Kaplan-Meier analysis. Univariate Cox regression analysis was performed to ascertain whether KDELR3 expression could serve as a prognostic indicator for OS in glioma patients. A p-value < 0.05 was considered statistically significant, with a Hazard Ratio (HR) value > 1 indicating a risk factor, and an HR value < 1 suggesting a protective factor. All data analyses were conducted using R software (version 4.0.2) and Adobe Photoshop CC.
Gene set enrichment analysis (GSEA)
Gene Set Enrichment Analysis (GSEA) was employed to elucidate the biological pathways underlying differentially expressed genes between KDELR3 high and low expression groups38. RNA-seq data (level 3) and corresponding clinical information pertaining to glioma were retrieved from the TCGA database and subjected to analysis using the GSVA package in R software, with the parameter method=’ssgsea’ selected. Subsequently, the correlation between genes and pathway scores was examined using Spearman correlation analysis. A p-value of < 0.05 was deemed statistically significant. Additionally, functional enrichment analysis employing GO terminology was performed to ascertain the functional roles of differentially expressed genes, providing novel insights into the biological effects of KDELR3. Genes exhibiting a significant correlation with KDELR3 expression (Pearson absolute correlation coefficient > 0.5 and p < 0.05) were identified as risk score-related genes, and their potential biological functions and pathways were investigated. The GO pathway analysis was conducted using the ggplot2 software package in R, with significance thresholds set at p-value and q-value < 0.05.
Analysis of immune infiltration
The level 3 HTSeq-FPKM RNA-seq data from GBMLGG samples in the TCGA database was utilized to convert the RNA-seq data from FPKM format to Transcripts Per kilobase of exon model per Million mapped reads (TPM) format and underwent log2 transformation. Duplicate samples were excluded, and tumor-immune interaction characteristics were assessed using the GSVA package in R software. Furthermore, the correlation between KDELR3 expression levels and the abundance of 24 tumor-invasive immune subsets in GBMLGG was quantified.
PPI network construction
The potential genes interacting with KDELR3 were retrieved from the GeneMANIA database (https://genemania.org/), which facilitated the subsequent identification of the co-expression network.
Nomogram diagram construction
Utilizing the outcomes of both univariate and multivariate Cox regression models, a nomogram prediction model was formulated to foretell the 1-, 3-, and 5-year overall survival rates for GBMLGG patients. Incorporating the independent factors influencing OS, as determined by the multivariate Cox regression, the Nomogram model was established for predicting OS in GBMLGG patients. The R software’s survival package was employed for proportional hazard hypothesis testing and Cox regression analysis, while the rms package was utilized for nomogram analysis and visualization. Statistical significance was ascribed to differences with a p-value < 0.05.
Statistical methods
The overall survival rates of the immunohistochemically high-expression and low-expression groups were comparatively analyzed using the Kaplan-Meier method. Significant differences between the groups were subsequently detected via the Wilcoxon signed-rank test. Potential risk factors were identified through univariate Cox regression analysis. Statistical significance was ascribed to findings with a P value less than 0.05. All data were processed utilizing the statistical software R (version 4.0.2).
Data availability
All relevant data are within the manuscript and its additional files, and the sequencing data used in this article were public TCGA (http://cancergenome.nih.gov/) and CGGA (http://www.cgga.org.cn/) databases.
References
Ostrom, Q. T. et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro-Oncology 24, v1-v95. https://doi.org/10.1093/neuonc/noac202 (2022).
Salari, N. et al. The global prevalence of primary central nervous system tumors: a systematic review and meta-analysis. Eur. J. Med. Res. 28, 16. https://doi.org/10.1186/s40001-023-01011-y (2023).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 68, 394–424. https://doi.org/10.3322/caac.21492 (2018).
David, N. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-Oncology 23, 1231–1251. https://doi.org/10.1093/neuonc/noab106 (2021).
Schaff, L. R. & Mellinghoff, I. K. Glioblastoma and other primary brain malignancies in adults: a review. JAMA 329, 574–587. https://doi.org/10.1001/jama.2023.0023 (2023).
Jiang, H. et al. Super-early initiation of temozolomide prolongs the survival of glioblastoma patients without gross-total resection: a retrospective cohort study. J. Neurooncol. 144, 127–135. https://doi.org/10.1007/s11060-019-03211-1 (2019).
Ballo, M. T. et al. Correlation of tumor treating fields dosimetry to survival outcomes in newly diagnosed glioblastoma: a large-scale numerical simulation-based analysis of data from the phase 3 EF-14 randomized trial. Int. J. Radiat. Oncol. Biol. Phys. 104, 1106–1113. https://doi.org/10.1016/j.ijrobp.2019.04.008 (2019).
Reifenberger, G., Wirsching, H. G., Knobbe-Thomsen, C. B. & Weller, M. Advances in the molecular genetics of gliomas—Implications for classification and therapy. Nat. Rev. Clin. Oncol. 14, 434–452. https://doi.org/10.1038/nrclinonc.2016.204 (2016).
Bell, E. H. et al. Comprehensive genomic analysis in NRG oncology/RTOG 9802: A phase III trial of radiation versus radiation plus procarbazine, lomustine (CCNU), and vincristine in high-risk low-grade glioma. J. Clin. Oncol. 38, 3407–3417. https://doi.org/10.1200/jco.19.02983 (2020).
Mair, M. J., Geurts, M., van den Bent, M. J. & Berghoff, A. S. A basic review on systemic treatment options in WHO grade II-III gliomas. Cancer Treat. Rev. 92, 8. https://doi.org/10.1016/j.ctrv.2020.102124 (2021).
Belén, D. M. & Miguel Ángel, M. Advances in the knowledge of the molecular biology of glioblastoma and its impact in patient diagnosis, stratification, and treatment. Adv. Sci. 7, 20. https://doi.org/10.1002/advs.201902971 (2020).
Raykhel, I. et al. A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 179, 1193–1204. https://doi.org/10.1083/jcb.200705180 (2007).
Lewis, M. J. & Pelham, H. R. B. Sequence of a second human KDEL receptor. J. Mol. Biol. 226, 913–916. https://doi.org/10.1016/0022-2836(92)91039-r (1992).
Trychta, K. A., Bäck, S., Henderson, M. J. & Harvey, B. K. KDEL receptors are differentially regulated to maintain the ER proteome under calcium deficiency. Cell Rep. 25, 1829–1840e1826. https://doi.org/10.1016/j.celrep.2018.10.055 (2018).
Bard, F., Mazelin, L., Péchoux-Longin, C., Malhotra, V. & Jurdic, P. Src regulates golgi structure and KDEL receptor-dependent retrograde transport to the endoplasmic reticulum. J. Biol. Chem. 278, 46601–46606. https://doi.org/10.1074/jbc.M302221200 (2003).
Bikard, Y. et al. The KDEL receptor has a role in the biogenesis and trafficking of the epithelial sodium channel (ENaC). J. Biol. Chem. 294, 18324–18336. https://doi.org/10.1074/jbc.RA119.008331 (2019).
Lewis, M. J. & Pelham, H. R. B. Ligand-induced redistribution of a human KDEL receptor from the golgi complex to the endoplasmic reticulum. Cell 68, 353–364. https://doi.org/10.1016/0092-8674(92)90476-s (1992).
Wires, E. S., Trychta, K. A., Kennedy, L. M. & Harvey, B. K. The function of KDEL receptors as UPR genes in disease. Int. J. Mol. Sci. 22, 14. https://doi.org/10.3390/ijms22115436 (2021).
Andrew, S., Kohei, K. & Takeshi, O. Improvement of intracellular traffic system by overexpression of KDEL receptor 1 in antibody-producing CHO cells. Biotechnol. J. 15, 9. https://doi.org/10.1002/biot.201900352 (2020).
Tapia, D. et al. KDEL receptor regulates secretion by lysosome relocation- and autophagy-dependent modulation of lipid-droplet turnover. Nat. Commun. 10, 17. https://doi.org/10.1038/s41467-019-08501-w (2019).
Huang, H. et al. Identification of biomarkers in macrophages of atherosclerosis by microarray analysis. Lipids Health Dis. 18 https://doi.org/10.1186/s12944-019-1056-x (2019).
Li, K. et al. Development of a phagocytosis-dependent gene signature to predict prognosis and response to checkpoint inhibition in clear-cell renal cell carcinoma. Front. Immunol. 13, 20. https://doi.org/10.3389/fimmu.2022.853088 (2022).
Xu, Z. et al. A glycolysis-related five-gene signature predicts biochemical recurrence-free survival in patients with prostate adenocarcinoma. Front. Oncol. 11, 11. https://doi.org/10.3389/fonc.2021.625452 (2021).
Tang, Y. et al. Identification of hypoxia-related prognostic signature and competing endogenous RNA regulatory axes in hepatocellular carcinoma. Int. J. Mol. Sci. 23, 20. https://doi.org/10.3390/ijms232113590 (2022).
Marie, K. L. et al. Melanoblast transcriptome analysis reveals pathways promoting melanoma metastasis. Nat. Commun. 11, 18. https://doi.org/10.1038/s41467-019-14085-2 (2020).
Edick, M. J. et al. Lymphoid gene expression as a predictor of risk of secondary brain tumors. Genes Chromosomes Cancer 42, 107–116. https://doi.org/10.1002/gcc.20121 (2004).
Chen, H. et al. Diet and risk of adult glioma in eastern Nebraska, United States. Cancer Causes Control. 13, 647–655. https://doi.org/10.1023/a:1019527225197 (2002).
Pan, Y. et al. Whole tumor RNA-sequencing and deconvolution reveal a clinically-prognostic PTEN/PI3K-regulated glioma transcriptional signature. Oncotarget 8, 52474–52487. https://doi.org/10.18632/oncotarget.17193 (2017).
Hsu, J. B. K., Chang, T. H., Lee, G. A., Lee, T. Y. & Chen, C. Y. Identification of potential biomarkers related to glioma survival by gene expression profile analysis. BMC Med. Genom. 11, 18. https://doi.org/10.1186/s12920-019-0479-6 (2019).
Munro, S. & Pelham, H. R. B. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907. https://doi.org/10.1016/0092-8674(87)90086-9 (1987).
Ruggiero, C. et al. A golgi-based KDELR-dependent signalling pathway controls extracellular matrix degradation. Oncotarget 6, 3375–3393. https://doi.org/10.18632/oncotarget.3270 (2014).
Zhu, K., Xiaoqiang, L., Deng, W., Wang, G. & Fu, B. Identification of a novel signature based on unfolded protein response-related gene for predicting prognosis in bladder cancer. Hum. Genom. 15, 73. https://doi.org/10.1186/s40246-021-00372-x (2021).
Yuan, Y. F. et al. KDELR1 is an independent prognostic predictor and correlates with immunity in glioma. Front. Oncol. 12, 15. https://doi.org/10.3389/fonc.2022.783721 (2022).
Wei, H. et al. KDELR2 promotes breast cancer proliferation via HDAC3-mediated cell cycle progression. Cancer Commun. 41, 904–920. https://doi.org/10.1002/cac2.12180 (2021).
Liao, Z. et al. KDELR2 promotes glioblastoma tumorigenesis targeted by HIF1a via mTOR signaling pathway. Cell Mol. Neurobiol. 39, 1207–1215. https://doi.org/10.1007/s10571-019-00715-2 (2019).
Zhang, G. et al. KDELR2 knockdown synergizes with temozolomide to induce glioma cell apoptosis through the CHOP and JNK/p38 pathways. Transl. Cancer Res. 10, 3491–3506. https://doi.org/10.21037/tcr-21-869 (2021).
Hui, M., Nian, J., Wang, Z., Li, X. J. & Huang, C. H. KDELR2 is an unfavorable prognostic biomarker and regulates CCND1 to promote tumor progression in glioma. Pathol. Res. Pract. 216, 10. https://doi.org/10.1016/j.prp.2020.152996 (2020).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Funding
This research was funded by the Natural Science Foundation of Fujian Province (Grant Nos. 2022J011094, 2023J011368 and 2024J011155), the Research Project of the 900th Hospital of the Joint Logistic Support Force (Grant No. 2022MS12), Medical Key Specialty (Neurosurgery) Project of PLA Joint Logistics Team (Grant No. LQZD-SW), and Science and Technology Innovation Platform Project of Fujian Science and Technology Plan (Grant No.2022Y2017).
Author information
Authors and Affiliations
Contributions
HZ and ZHC made substantial contributions to the conception and design of the work, and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved; JF, LYF and LZ drafted the work and revised it critically for important intellectual content, and wrote the main manuscript text; JPZ, XPW and DYM made the acquisition, analysis, and interpretation of data; MCS and BQX prepared figures and table. All authors reviewed the manuscript and approved the version to be published.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, J., Zhao, L., Fu, L. et al. KDELR3 overexpression as a novel prognostic and diagnostic biomarker in glioma: comprehensive bioinformatic analysis insights. Sci Rep 14, 30783 (2024). https://doi.org/10.1038/s41598-024-80991-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-80991-1