Abstract
Mammography is the recommended imaging modality for breast cancer screening. Expressions of human epidermal growth factor receptor 2 (HER2), estrogen receptor (ER), and progesterone receptor (PR) are critical to the development of therapeutic strategies for breast cancer. In this study, a deep learning model (CBAM ResNet-18) was developed to predict the expression of these three receptors on mammography without manual segmentation of masses. Mammography of patients with pathologically proven breast cancer was obtained from two centers. A deep learning-based model (CBAM ResNet-18) for predicting HER2, ER, and PR expressions was trained and validated using five-fold cross-validation on a training dataset. The performance of the model was further tested using an external test dataset. Area under receiver operating characteristic curve (AUC), accuracy (ACC), and F1-score were calculated to assess the ability of the model to predict each receptor. For comparison we also developed original ResNet-18 without attention module and VGG-19 with and without attention module. The AUC (95% CI), ACC, and F1-score were 0.708 (0.609, 0.808), 0.651, 0.528, respectively, in the HER2 test dataset; 0.785 (0.673, 0.897), 0.845, 0.905, respectively, in the ER test dataset; and 0.706 (0.603, 0.809), 0.678, 0.773, respectively, in the PR test dataset. The proposed model demonstrates superior performance compared to the original ResNet-18 without attention module and VGG-19 with and without attention module. The model has the potential to predict HER2, PR, and especially ER expressions, and thus serve as an adjunctive diagnostic tool for breast cancer.
Similar content being viewed by others
Introduction
Breast cancer ranks as one of the most prevalent malignant diseases affecting women globally. The International Agency for Research on Cancer (IARC)1 reported that in 2020, there were 19.29 million new cancer cases worldwide. The most significant change is the rapid increase in new breast cancer cases to 2.26 million, making breast cancer the world’s leading cancer for the first time, replacing lung cancer (2.2 million cases) and accounting for 11.7% of all new cancer cases.
The assessment of breast cancer screening involves a three-pronged approach: appropriate imaging, clinical evaluation, and needle aspiration biopsy. Mammography is the preferred imaging technique for screening breast cancer, serving as a crucial tool for detecting lesions and identifying early alterations in breast tissue, particularly before the onset of physical symptoms. Early detection via mammography can significantly enhance the likelihood of recovery2.
Gene expression profiling has emerged as a reliable method for categorizing breast cancers into distinct molecular subtypes based on genetic differences in tumors. Recent advancements indicate the existence of four molecular subtypes, each demonstrating unique prognostic implications in their gene expression patterns: luminal A, luminal B (both HER2 negative and HER2 positive), HER2 positive, and triple-negative3,4,5. These subtypes vary considerably in terms of phenotypic presentation, treatment response, and survival outcomes6. The amplification of the HER2 gene has proven crucial in breast cancer treatment, leading to the development of targeted therapies like trastuzumab, which have shown significant success. Historically, survival rates for patients with HER2-positive breast cancer were low, but the introduction of systemic therapies targeting HER2 has markedly improved survival outcomes7,8. The identification of HER2 gene amplification through fluorescence in situ hybridization (FISH) or HER2 protein overexpression via immunohistochemistry (IHC) is often key to recognizing patients who may benefit from these targeted treatments. Additionally, the identification of estrogen receptors (ER) and progesterone receptors (PR) represents another significant breakthrough in breast cancer therapy. The majority of breast cancers display an overexpression of ER and PR, and the advent of hormone receptor-targeting medications like tamoxifen has considerably enhanced survival rates among patients with hormone receptor-positive breast cancer9,10.
Computer-aided diagnosis (CAD) systems are designed to provide objective insights to assist radiologists in interpreting and diagnosing medical images. These systems are particularly effective in distinguishing between benign and malignant lesions. In the last few years, deep learning11,12,13, especially convolutional neural networks (CNN), has been proven to be very effective in vision tasks. Some recently proposed CAD systems have used well-known deep learning techniques and obtained promising results14,15,16. Within the ___domain of breast cancer, many researchers have explored the application of deep learning and mammography to predict the nature of breast lumps17,18,19,20. However, more intensive research is needed, particularly regarding the use of deep learning to forecast HER2, ER, and PR status based on mammography.
Previous studies have demonstrated a relationship between molecular features and mammographic features. For instance, luminal subtype tumors typically appear oval on mammography21, while triple-negative tumors often manifest as poorly defined masses or with acanthotic calcifications22. The HER2-positive subtype is characterized by calcification and structural distortion21,23. Previous studies also proved the ability of deep learning model to predict the molecular features of breast masses.
Nonetheless, previous investigations18,24,25,26,27 used manually segmented masses as input, which might not work well with breast cancer with prominent masses and lead to potential bias. Therefore, we hope to use Deep Learning to predict breast cancer HER2, ER, and PR status of breast cancer by whole mammogram images. In this study, aiming to eliminate the pain caused by puncture biopsy and the risk of tumor dissemination along the puncture tract, a CNN model was established to predict molecular features of breast cancer using whole mammogram images. Moreover, the model we constructed does not require manual segmentation of masses, which resulted in significant labor savings and reduced economic costs.
Materials
Study design
The study was a retrospective, multicentre investigation approved by the institutional review boards of all participating hospitals (Huazhong University of Science and Technology Union Shenzhen Hospital, Shenzhen, China, and Second Hospital of Shandong University, Jinan, China). The patients were informed that their de-identified images could be used for research purposes and provided their consent. The patients were sourced from the two local hospitals mentioned above, and the enrolled patients comprised two datasets: (1) a training dataset from Huazhong University of Science and Technology Union Shenzhen Hospital; (2) an independent test dataset from Second Hospital of Shandong University. A deep learning-based model was trained and validated using the training dataset to classify HER2-positive or negative subtypes, ER positive or negative, and PR positive or negative. Subsequently, the developed model was tested using the independent test dataset.
Study population
As shown in Fig. 1, in training dataset, digital imaging and medical communication (DICOM) data of mammogram in 237 patients diagnosed with breast cancer between January 2018 and November 2021 was collected, and all data do not overlap. In test dataset DICOM data of mammography radiographs in 155 patients diagnosed with breast cancer between January 2020 and May 2022 was collected, and equally, all data does not overlap. The inclusion criteria for this study were as follows: (1) breast malignancy confirmed by surgical or puncture pathology; (2) preoperative mammography; and (3) pathological results of ER and PR status using IHC and HER2 status by IHC and FISH. Exclusion criteria were as follows: (1) lack of clinical information; (2) lack of pathological findings; (3) lack of IHC results; (4) lack of FISH results when HER2 was 2 + on IHC results; and (5) mammography images after chemotherapy/targeted/endocrine therapy. In training dataset, 184, 203, and 202 patients were finally included in the training of HER2, ER, and PR status prediction, respectively. In test dataset, 146, 148, and 149 patients were finally included in the test of HER2, ER, and PR status prediction, respectively.
Methodology
Histopathology
HER2, ER, and PR analysis of surgical pathology specimens: Two or more pathologists with more than 10 years of professional experience in breast pathology re-examined all breast cancer samples to confirm HER2, ER, and PR status. According to the current American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines, HER2 positivity was defined as an IHC result of 3+, or a result of 2 + but amplified by FISH (HER2/CEP17 ratio is greater than 2.0), otherwise, it was considered negative28. According to the current guideline in China29,30, the status was considered positive when the hormonal receptors (ER and PR) had a value ≥ 1%, otherwise, it was considered negative.
Image acquisition
Selenia Dimensions(Hologic, Inc.)is the mammographic image acquisition device. It uses an Image Acquisition system that includes a digital image receptor. This receptor, which covers an area of 24 cm* 29 cm, is a direct-capture detector using an amorphous selenium photo conductor. Standard 2D mammograms were acquired in cephalocaudal (CC) and mediolateral oblique (MLO) views with the Combo protocol which can automatically control exposure. Images were stored as 12-bit grayscale in DICOM format with a resolution of 4096*3328 (row*columns) and a pixel spacing of 0.07 cm*0.07 cm.
Image pre-processing
The interference information in the initial DICOM images such as machine edges, orientation labels and empty padding in the original image was removed. The images of right breast are horizontally flipped to align the image. Then the images are further resized to 512*256 (height*width) by nearest neighbor interpolation to obtain the standard image and concatenated horizontally.
Deep learning analysis
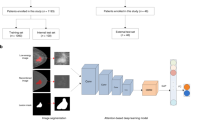
As shown in Fig. 2, the input image is a pre-processed normalized image without tumor segmentation.
The proposed model utilizes ResNet-18 as its backbone. ResNet-18 starts with a 7*7 convolutional layer followed by a 3*3 max-pooling layer. It includes four residual blocks, each containing four convolutional layers, with skip connections applied every two layers. The features extracted by these convolutional layers are reduced in dimensionality using average pooling layers. Finally, a linear layer with a sigmoid activation function maps the output to a single floating-point number in the range [0,1], representing the prediction of the target variable. To improve the performance on unsegmented images, Convolutional Block Attention Module (CBAM)31 is inserted between two adjacent residual blocks. CBAM is a lightweight attention mechanism that can be integrated into existing convolutional neural networks. It consists of two sequential sub-modules: the channel attention module (CAM) and the spatial attention module (SAM). First, the CAM computes an attention map by applying average pooling and max pooling along the spatial dimension (height and width), followed by two fully connected layers and a sigmoid function. This attention map is used to recalibrate the channel-wise features. Next, the SAM generates an attention map by applying average pooling and max pooling along the channel dimension (CC and MLO), followed by a 7*7 convolutional layer with a sigmoid function. This map highlights important spatial regions in the feature maps. Together, these two modules enhance the representational power of the model by focusing on both important channels and spatial regions in the input features. In the training dataset, data enhancement approaches such as random clipping and random salt and pepper noise are applied. Similarly, early stopping strategy32 was also applied to prevent overfitting. We employed the binary cross-entropy loss function and the Adam optimizer for model training. To fine-tune the hyperparameters, we used 5-fold cross-validation. The hyperparameter search space is detailed in Table 1. Once the hyperparameters are determined, the model would be trained on the whole training dataset and test on our independent test dataset. The learning rate was controlled using a cosine annealing schedule.
Models comparison
For comparison we also developed original ResNet-18 without attention module and VGG-19 with and without CBAM. The Visual Geometry Group (VGG) convolutional neural network structure is a well-known approach in the field of image recognition. It was initially proposed by the Visual Geometry Group at the University of Oxford. VGG19 comprises a total of 19 layers, including 16 convolutional layers and the final 3 fully connected layers. The prediction is conducted through the use of the sigmoid function. Study by Praharsha et al.33 demonstrated the unique classification performance of their proposed CBAM VGG architecture. For a fair comparison, all the models were trained and tested with the same datasets, hyperparameters and random seeds.
Statistical analysis
The area under the Receiver operating characteristic curve (AUC) was used as the primary metric to assess the ability of our model in predicting HER2 status (HER2-positive vs. HER2-negative), ER status (ER-positive vs. ER-negative), and PR status (PR-positive vs. PR-negative) with 95% CIs were calculated for each molecular subtype. In addition, we calculated specificity, accuracy, precision, recall and F1-score. To visualized the significance of the attention module we proposed, the class activation maps (CAM) before and after each attention block are represented in the Fig. 2, where red indicates regions more important for prediction (HER2, ER, PR-positive regions), these feature maps suggest that attention blocks can learn to focus on HER2, ER, and PR status related regions, including tumor and peritumor regions. All the deep learning development was conduct with the Python Package, PyTorch. Statistical analysis was performed with Statistical Package for the Social Sciences (SPSS, version 24, Chicago, IL, USA) and R package (v. 4.2.0; R Foundation for Statistical Computing, Vienna, Austria). DeLong test was used to compare the performance between different approaches. Statistical tests were all two-sided, and a P-value less than 0.05 was considered to indicate a statistically significant difference.
Results
Patient population and lesion characteristics
The flowcharts for the datasets are presented in Fig. 1. The fundamental demographic and clinicopathological characteristics of the patients included in the training dataset and test dataset are presented in Table 2.
Deep learning analysis
Figure 3; Table 3 illustrate the outcomes of the model training for HER2, ER, and PR. With regard to HER2, the mean AUC following 5-fold cross-validation was 0.752. For ER, the mean AUC following 5-fold cross-validation was 0.733. With respect to PR, the mean AUC following 5-fold cross-validation was 0.756.
The results of the model tests for HER2, ER, and PR are presented in Fig. 4; Table 4. AUC with 95% confidence intervals for the HER2 test dataset were 0.708 (0.609, 0.808), 0.785 (0.673, 0.897) for the ER test dataset, and 0.706 (0.603, 0.809) for the PR test dataset. Table 4 also presents the specificity, accuracy, precision, recall rate, and F1-score results and Table 5 shows the AUC of original ResNet-18 without attention module and VGG-19 with and without attention module.
Discussion
HER2 positivity in breast cancer patients, i.e. overexpression and/or amplification of HER2, is associated with a more aggressive tumor phenotype, malignant transformation of cells, poorer clinical outcomes34, and increased resistance to endocrine therapy35. Historically, survival rates for patients with HER2-positive breast cancer were low. However, the advent of systemic therapy with HER2-targeted agents has led to a significant improvement in survival outcomes8. Similarly, the discovery of ER and PR, and the development of medicines targeting these hormone receptors, such as tamoxifen acetonide, has resulted in a notable enhancement in the survival of patients with hormone receptor-positive breast cancer9,10. It is therefore imperative that the status of HER2, ER, and PR be determined in order to ensure an optimal treatment plan for breast cancer patients.
The clinicopathological characteristics of a tumor are critical in determining the best treatment. Analysis through deep learning allows for rapid, non-invasive study of the entire tumor and each detected lesion. The purpose of this retrospective study was to determine if deep learning models can be used to characterize malignant breast lesions’ pathological characteristics on mammography.
Previous research on mammography and deep learning has focused on developing CAD systems for classifying breast masses as benign or malignant, and achieved good diagnostic efficacy. The study by Moraru et al.36 shows that the DeepLabV3 + has significantly better segmentation performance and outperforms the U-Net network. Shen et al.27 used four models, Resnet-Resnet, Resnet-VGG, VGG-VGG, and VGG-Resnet, and on an independent test set of digitized film mammograms from the Digital Breast Screening Database (CBIS-DDSM), the best single model had an AUC per image of 0.88, and the average of the four models improved the AUC to 0.91 (sensitivity: 86.1%, specificity: 80.1%); on an independent test set of full-field digital mammography (FFDM) images from the INbreast database, the best single model had a per-image AUC of 0.95, and the average of the four models improved the AUC to 0.98 (sensitivity: 86.7%, specificity. 96.1%). Al-Antari et al.18 classified the detected and segmented masses as benign or malignant by You-Only-Look-Once (YOLO) in the INbreast database with an AUC of 94.78%. Ribli et al.26 classified the detected and segmented masses as benign or malignant by Faster R-CNN in the INbreast database and obtained an AUC of 0.95. A CAD system developed by Ribli, D. et al. using three deep learning models, VGG16, ResNet50, and Iception v320, was tested on an independent database (MIAS) and obtained an accuracy of 98.23% and an AUC of 0.99.
Further, studies on mammography and deep learning for HER2, ER, and PR status prediction are scarce, or the results are unsatisfactory. Ueda et al.25 used a deep learning model to classify breast cancer receptor expression from mammography with an AUC of 0.67 for ER positive or negative, 0.61 for PR positive or negative, and 0.75 for HER2 positive or non-HER2 positive in the assay dataset. According to Zhou et al.37, the logistic regression model AUC for assessing HER2 status based on quantitative radiomic features were 0.846 in the training set and 0.787 in the test set, but the study used only IHC to determine HER2 status and based on a small sample (n = 91), the results may have been biased. Dominique, C. et al. used deep learning analysis based on contrast-enhanced spectral mammography (CESM) to identify histotype majority factors in malignant breast tumors, with good results for identifying ER receptor positive or negative (AUC = 0.83), triple-negative breast cancers (0.90), however, it was not effective in detecting HER2 status (AUC = 0.59), and its model relies on CESM, which is not very generalizable.
It should be noted that in the above studies, suspicious masses of the mammography images needed to be manually segmented before input to the models, whereas the model we constructed did not require manual segmentation of masses and obtained a more desirable AUC, especially in describing ER status (For HER2, ER and PR, the AUCs were 0.708, 0.785, and 0.706, respectively), which means that our model is more objective and has higher time and economic benefits. Further, our results show that breast cancer samples can be classified into molecular subtypes from mammograms.
The study by Praharsha et al.33 illustrates the distinctive classification capabilities of their proposed CBAM VGG architecture. Therefore, we also developed original ResNet-18 without attention module and VGG-19 with and without attention module for comparison. The results of the comparison demonstrate that the proposed model demonstrates superior performance compared to the original ResNet-18 without attention module and VGG-19 with and without attention module.
There are several limitations of our study. Firstly, it is theoretically possible for a patient to present with multiple lesions in the same breast, each expressing a different molecular isoform. However, our study lacks an analysis of this circumstance, as we do not have a record of any patients who have presented with such a condition. Nevertheless, even in such cases, the Grad CAM has the potential to effectively visualize the prediction of our model, as illustrated in Fig. 2. This could enable the visualization of the region associated with each isoform. Moreover, in accordance with the established guidelines, if a patient is diagnosed with breast cancer and presents with different molecular isoforms, it is recommended that they adhere to the prescribed treatment plan for each lesion. This approach does not influence the selection of appropriate therapeutic options. Furthermore, the sample size of the study is relatively modest, although data augmentation was employed to address this limitation.
Conclusion
Our study demonstrates that convolutional neural network (CNN) models derived from mammography can be employed to estimate the expression of human epidermal growth factor receptor 2 (HER2), estrogen receptor (ER), and progesterone receptor (PR) in breast cancer. This approach holds promise for providing a noninvasive and cost-effective method for assessing tumor biology prior to the initiation of initial treatment. It is important to note that this technique can only be applied for the purpose of clinical guidance with regard to treatment once further high-level evidence has been produced demonstrating its safety. Core needle biopsy (or vacuum biopsy) remains the gold standard for pre-treatment molecular subtype classification. Further prospective studies are required to more fully elucidate the correlation between imaging characterization and cancer biosignatures. In the future, we intend to incorporate additional clinical data on breast cancer, including breast MRI and ultrasound, into our CNN models to enhance their accuracy in estimating the expression of HER2, ER, and PR in breast cancer. This approach has the potential to become a non-invasive and cost-effective method for assessing tumor biology prior to the initial treatment of breast cancer.
In conclusion, our results suggest that deep learning analysis of mammography has the potential to determine HER2, PR, and especially ER status and serve as an adjunctive diagnostic tool for breast cancer.
Data availability
The original data used in this study were approved by the institutional review boards of all participating hospitals, further inquiries can be directed to the corresponding author.
Abbreviations
- AUC:
-
Area under the curve
- ASCO:
-
American Society of Clinical Oncology
- CAD:
-
Computer aided diagnosis
- CAM:
-
Class Activation Maps
- CAP:
-
College of American Pathologists
- CBAM:
-
Convolutional Block Attention Module
- CC:
-
Craniocaudal
- CESM:
-
Contrast-enhanced spectral mammography
- CNN:
-
Convolutional Neural Networks
- DICOM:
-
Digital Imaging and Medical Communication
- DES:
-
Dual-energy subtracted
- ER:
-
Estrogen receptor
- FFDM:
-
Full-field digital mammography
- FISH:
-
Fluorescence in situ hybridization
- HER2:
-
Human epidermal growth factor receptor 2
- HIS:
-
Hospital Information System
- IARC:
-
International Agency for Research on Cancer
- IDC:
-
Infiltrative ductal carcinoma
- IHC:
-
Immunohistochemical
- ILC:
-
Invasive lobular carcinoma
- ISH:
-
In situ hybridization
- MLO:
-
Media-lateral oblique
- PR:
-
Progesterone receptor
- ROC:
-
Receiver operating characteristic
- TNBC:
-
Triple-negative breast cancers
- VGG:
-
Visual Geometry Group
References
Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249. https://doi.org/10.3322/caac.21660 (2021).
Schreer, I. Dense breast tissue as an important risk factor for breast Cancer and implications for early detection. Breast Care (Basel). 4(2), 89–92. https://doi.org/10.1159/000211954 (2009).
Prat, A. et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J. Clin. Oncol. 31(2), 203–209. https://doi.org/10.1200/jco.2012.43.4134 (2013).
Daniel, C. K. et al. Comprehensive molecular portraits of human breast tumours. Nature 490(7418), 61–70. https://doi.org/10.1038/nature11412 (2012).
Prat, A. et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 24(Suppl 2), S26–35. https://doi.org/10.1016/j.breast.2015.07.008 (2015).
Tsoutsou, P. G., Vozenin, M. C., Durham, A. D. & Bourhis, J. How could breast cancer molecular features contribute to locoregional treatment decision making? Crit. Rev. Oncol. Hematol. 110, 43–48. https://doi.org/10.1016/j.critrevonc.2016.12.006 (2017).
Hayes, D. F. HER2 and breast cancer - a phenomenal success story. N Engl. J. Med. 381(13), 1284–1286. https://doi.org/10.1056/NEJMcibr1909386 (2019).
Wilson, F. R. et al. Herceptin® (trastuzumab) in HER2-positive early breast cancer: a systematic review and cumulative network meta-analysis. Syst. Rev.7(1), 191. https://doi.org/10.1186/s13643-018-0854-y (2018).
Fisher, B. et al. Influence of tumor estrogen and progesterone receptor levels on the response to tamoxifen and chemotherapy in primary breast cancer. J. Clin. Oncol. 1(4), 227–241. https://doi.org/10.1200/jco.1983.1.4.227 (1983).
Fisher, B. et al. Treatment of primary breast cancer with chemotherapy and tamoxifen. N Engl. J. Med. 305(1), 1–6. https://doi.org/10.1056/nejm198107023050101 (1981).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521(7553), 436–444. https://doi.org/10.1038/nature14539 (2015).
Schmidhuber, J. Deep learning in neural networks: an overview. Neural Netw. 61, 85–117. https://doi.org/10.1016/j.neunet.2014.09.003 (2015).
Bengio, Y., Courville, A. & Vincent, P. Representation learning: a review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35(8), 1798–1828. https://doi.org/10.1109/tpami.2013.50 (2013).
Shin, H. C. et al. Deep convolutional neural networks for computer-aided detection: CNN Architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 35(5), 1285–1298. https://doi.org/10.1109/tmi.2016.2528162 (2016).
Chen, H. et al. Standard plane localization in fetal ultrasound via ___domain transferred deep neural networks. IEEE J. Biomed. Health Inf. 19(5), 1627–1636. https://doi.org/10.1109/jbhi.2015.2425041 (2015).
Ciompi, F. et al. Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out-of-the-box. Med. Image Anal. 26(1), 195–202. https://doi.org/10.1016/j.media.2015.08.001 (2015).
Carneiro, G., Nascimento, J. & Bradley, A. P. Automated analysis of unregistered multi-view mammograms with deep learning. IEEE Trans. Med. Imaging. 36(11), 2355–2365. https://doi.org/10.1109/tmi.2017.2751523 (2017).
Al-Antari, M. A., Al-Masni, M. A., Choi, M. T., Han, S. M. & Kim, T. S. A fully integrated computer-aided diagnosis system for digital X-ray mammograms via deep learning detection, segmentation, and classification. Int. J. Med. Inf. 117, 44–54. https://doi.org/10.1016/j.ijmedinf.2018.06.003 (2018).
Dhungel, N., Carneiro, G. & Bradley, A. P. A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med. Image Anal. 37, 114–128. https://doi.org/10.1016/j.media.2017.01.009 (2017).
Chougrad, H., Zouaki, H. & Alheyane, O. Deep convolutional neural networks for breast cancer screening. Comput. Methods Progr. Biomed. 157, 19–30. https://doi.org/10.1016/j.cmpb.2018.01.011 (2018).
Wu, M. & Ma, J. Association between Imaging Characteristics and different molecular subtypes of breast Cancer. Acad. Radiol. 24(4), 426–434. https://doi.org/10.1016/j.acra.2016.11.012 (2017).
Luck, A. A. et al. Breast carcinoma with basal phenotype: mammographic findings. AJR Am. J. Roentgenol. 191(2), 346–351. https://doi.org/10.2214/ajr.07.2659 (2008).
Seo, B. K. et al. Correlation of HER-2/neu overexpression with mammography and age distribution in primary breast carcinomas. Acad. Radiol. 13(10), 1211–1218. https://doi.org/10.1016/j.acra.2006.06.015 (2006).
Dominique, C. et al. Deep learning analysis of contrast-enhanced spectral mammography to determine histoprognostic factors of malignant breast tumours. Eur. Radiol. 1–11. https://doi.org/10.1007/s00330-022-08538-4 (2022).
Ueda, D. et al. Training, validation, and test of Deep Learning models for classification of receptor expressions in breast cancers from mammograms. JCO Precis Oncol. 5, 543–551. https://doi.org/10.1200/po.20.00176 (2021).
Ribli, D., Horváth, A., Unger, Z., Pollner, P. & Csabai, I. Detecting and classifying lesions in mammograms with deep learning. Sci. Rep. 8(1), 4165. https://doi.org/10.1038/s41598-018-22437-z (2018).
Shen, L. et al. Deep learning to improve breast cancer detection on screening mammography. Sci. Rep. 9(1), 12495. https://doi.org/10.1038/s41598-019-48995-4 (2019).
Wolff, A. C. et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 31(31), 3997–4013. https://doi.org/10.1200/jco.2013.50.9984 (2013).
Yang, W. & Bu, H. Guidelines for estrogen and progesterone receptor immunohistochemistry for breast cancer. CHIN. J. Pathol. 44(4), 237–239. https://doi.org/10.3760/cma.j.issn.0529-5807.2015.04.005 (2015).
Jiang, Z. et al. Chinese society of clinical oncology (CSCO) breast Cancer guidelines 2022. Transl. Breast Cancer Res. 3 (2022).
Woo, S., Park, J., Lee, J. Y. & Kweon, I. S. CBAM: Convolutional Block Attention Module (Springer, 2018).
Prechelt, L. Early stopping—but when?. In Neural Networks: Tricks of the Trade (Montavon, G. et al.) 2nd edition (Springer, 2012) 53–67. https://doi.org/10.1007/978-3-642-35289-8_5.
Praharsha, C. H. & Poulose, A. CBAM VGG16: an efficient driver distraction classification using CBAM embedded VGG16 architecture. Comput. Biol. Med. 180, 108945. https://doi.org/10.1016/j.compbiomed.2024.108945 (2024).
Tai, W., Mahato, R. & Cheng, K. The role of HER2 in cancer therapy and targeted drug delivery. J. Control Release. 146(3), 264–275. https://doi.org/10.1016/j.jconrel.2010.04.009 (2010).
Engel, R. H. & Kaklamani, V. G. HER2-positive breast cancer: current and future treatment strategies. Drugs 67(9), 1329–1341. https://doi.org/10.2165/00003495-200767090-00006 (2007).
Nastase, I-N., Moldovanu, S. & Moraru, L. Deep learning-based segmentation of breast masses using convolutional neural networks. J. Phys. 2701, 012005. https://doi.org/10.1088/1742-6596/2701/1/012005 (2024).
Zhou, J. et al. Evaluating the HER-2 status of breast cancer using mammography radiomics features. Eur. J. Radiol. 121, 108718. https://doi.org/10.1016/j.ejrad.2019.108718 (2019).
Author information
Authors and Affiliations
Contributions
Peng Gong and Shun Zeng designed experiments; Hongyu Chen, Rui Jing, Wenzhuo Yang and Ligong He wrote manuscript; Peng Liu, Bo Liang, Tianle Zou, Dan Shi and Wenhao Wu carried out experiments; Qiusheng Lin, Zhenyu Ma and Jinhui Zha analyzed experiments results; Yonghao Zhong, Xianbin Zhang and Guangrui Shao revised the manuscript, figures and tables.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Institutional Review Board of The Second Hospital of Shandong University [Approval No.: KYLL-2020(KJ)P-0195] and the Ethics Committee of Huazhong University of Science and Technology Union Shenzhen Hospital (Approval No.: KY-2021-044-01). All methods mentioned in this study were carried out in accordance with relevant guidelines and regulations, and, all experimental protocols were approved by a named institutional.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zeng, S., Chen, H., Jing, R. et al. An assessment of breast cancer HER2, ER, and PR expressions based on mammography using deep learning with convolutional neural networks. Sci Rep 15, 4826 (2025). https://doi.org/10.1038/s41598-024-83597-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-83597-9