Abstract
Thyroid nodules are a common thyroid disorder, and ultrasound imaging, as the primary diagnostic tool, is susceptible to variations based on the physician’s experience, leading to misdiagnosis. This paper constructs an end-to-end thyroid nodule detection framework based on YOLOv8, enabling automatic detection and classification of nodules by extracting grayscale and elastic features from ultrasound images. First, an attention-weighted DCN is introduced to enhance superficial feature extraction and capture local information. Next, the CPCA mechanism is employed to reduce the interference of redundant information. Finally, a feature fusion network based on an aggregation-distribution mechanism is utilized to improve the learning capability of fine-grained features, enhancing the performance of early nodule detection. Experimental results demonstrate that our method is accurate and effective for thyroid nodule detection, achieving diagnostic rates of 89.3% for benign and 90.4% for malignant nodules based on tests conducted on 611 clinical ultrasound images, with a mean Average Precision at IoU = 0.5 (mAP@50) of 95.5%, representing a 6.6% improvement over baseline models.
Similar content being viewed by others
Introduction
The butterfly-shaped thyroid is an essential endocrine gland in the neck, and thyroid hormones play a crucial role in regulating metabolism, heart rate, and calorie burning1. Cancerous cells, known as thyroid nodules, can grow on the surface of the thyroid. Various conditions, including simple excessive growth, diffuse cysts, infections, or tumors, can form nodules (benign or malignant). Approximately 6% of women and 1%–2% of men are affected by thyroid nodules, making this a common nodular disease2,3,4. Therefore, strengthening the detection of thyroid nodules, particularly improving the screening capabilities for early-stage nodules, and facilitating timely intervention are of significant clinical importance.
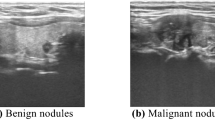
Ultrasound (US) imaging is a non-invasive method for soft tissue imaging and has become the primary clinical tool for assessing thyroid nodules5. Figure 1 shows a thyroid ultrasound image, with nodules indicated by red arrows and other tissues marked by white arrows. In Fig. 1a, structures such as blood vessels, trachea, and muscles are present, making it difficult to distinguish the nodule from surrounding tissues, which affects detection accuracy. Figure 1b highlights the characteristics of early nodules, which are typically small in size and exhibit indistinct features.
During the diagnostic process, physicians analyze thyroid nodules by identifying features such as increased blood flow, punctate echogenic foci, irregular borders, and height-to-width ratios6. However, this process presents several challenges: first, the variability in shape, size, and margins of nodules complicates the distinction of different types of abnormalities on thyroid ultrasound; second, the inherent speckle noise of ultrasound can reduce contrast resolution, hindering visual quality and obscuring important information in the diagnostic process, thereby affecting the radiologist’s interpretation7,8,9. Although intelligent diagnostic systems can alleviate medical pressures and reduce human errors, they still face the following challenges:
-
Superficial feature extraction: Superficial features (such as size, shape, texture, and aspect ratio) contain rich information and are crucial for enhancing model detection performance. Therefore, extracting high-quality superficial features from ultrasound images is a primary concern.
-
Early detection: Early identification is critical for diagnosing and treating thyroid nodules. However, the small size and indistinct features of nodules make enhancing early detection performance the second major challenge.
-
Background interference: The background of thyroid ultrasound images is complex, containing various tissues and substantial noise interference, significantly impacting detection accuracy. Therefore, suppressing background interference to enhance the overall detection performance of the model is the third research challenge.
To address the critical issues in our research, we conducted a literature review and experimental analysis, proposing a detection model specifically for nodules in thyroid ultrasound images. First, we collected substantial ultrasound image data from collaborating hospitals and performed comparative analyses using various advanced algorithms. After balancing detection efficiency and performance, we selected the YOLOv8-L model as our core model, which demonstrated the best overall performance. The main contributions of this study are as follows:
-
Proposed a thyroid nodule detection framework based on YOLOv8-L, enabling rapid localization of nodules in ultrasound images and distinguishing between benign and malignant cases, thereby improving diagnostic efficiency and alleviating medical burdens.
-
We improved the baseline model for the thyroid nodule detection task by integrating various modules that enhance representation capabilities into the backbone network, considering the unique characteristics of ultrasound backgrounds and nodules. To boost the diagnostic ability for early nodules, we introduced the Gather and Distribute Mechanism (GD)23 during the feature fusion stage, facilitating the interaction and fusion of global and multi-level features through convolution and self-attention mechanisms. This approach enhanced multi-scale feature fusion capability and improved the model’s detection performance for early nodules through fine-grained feature extraction.
Related work
Computer-aided diagnostic tools can effectively improve radiologists’ efficiency when detecting thyroid nodules. They mainly include manually annotated feature methods based on machine learning and automatic feature extraction methods based on deep learning. The former primarily uses shallow features such as image texture10, geometric morphology11, and statistical distribution12 and then classifies the target through a classifier, but its accuracy and efficiency are not as good as deep learning. Deep learning, represented by convolutional neural networks, can automatically extract deep semantic features in images, avoiding the need for manual feature design and extraction. It has become a research hotspot in diagnosing thyroid nodules on ultrasound images.
The target detection algorithm based on deep learning is constantly evolving in the ultrasound field and is widely used to detect thyroid nodule ultrasound images, achieving excellent performance. Zheng et al. proposed the Cascade Mask RCNN model, which optimizes Region of Interest (ROI) classification and bounding box correction by designing a more effective detector and improving the loss function to balance easy and complex samples, thereby improving the detection effect13. Fang et al. introduced a feature pyramid, spatial re-mapping, and anchor box redesign into the Faster R-CNN framework, significantly improving detection accuracy and speed14. Song et al. optimized the SSD model and used the MC-CNN framework of the spatial pyramid network to capture the multi-level features of the target for accurate detection15. Wang et al. fused YOLOv2 and ResNet to generate more precise feature maps by combining shallow and deep features for efficient detection16. Ma et al. combined YOLOv3 with DMRF-CNN to obtain deep edge and texture features and used multi-scale detection to identify nodules of different sizes17. Zhang et al. improved YOLOv3 by replacing the original backbone network with a high-resolution network (HRNet) and optimizing anchor boxes using a clustering algorithm to improve the accuracy of nodule localization and identification18. Yang et al. introduced involution, deformable convolution, and an attention mechanism into YOLOX, significantly improving thyroid nodules’ detection, especially early nodules19.
Early detection is crucial for preventing complications associated with thyroid cancer, so developing models that can accurately diagnose early-stage nodules promptly is of great significance for the clinical treatment of thyroid nodules. This study addresses the challenges of insufficient detection accuracy and difficulty in identifying the morphology of early nodules. By optimizing the algorithm structure, the model’s ability to characterize nodule features is improved. Using YOLOv8-L as the baseline model, we propose a detection framework specifically for thyroid nodule ultrasound images and systematically evaluate its application in ultrasound detection. The structure of the research process is shown in Fig. 2.
Method
The overall architecture of this study is shown in Fig. 3, which includes the backbone (feature extraction network), neck (feature fusion network), and head (detection head). In the backbone, the traditional convolution in the C2f. module of S2 is replaced with the Deformable Convolutional Networks (DCN)21, and the Channel Prior Convolutional Attention (CPCA)22 is introduced to weight the offset learned by DCN so that it pays more attention to the target region. DCN can extract multi-scale features at different levels, more comprehensively describe the diversity and complexity of nodules, and has strong adaptability, effectively capturing the rich local information in shallow features.
Before feature fusion, we use CPCA to perform weighted operations on the four feature maps extracted from the backbone network to reduce the impact of background interference in thyroid ultrasound images. To address the problem of small individual nodules and insignificant features in early nodules, we use a feature fusion network based on the GD mechanism23, which achieves a balance between latency and accuracy and significantly enhances the ability to fuse multi-scale features. With the self-attention mechanism, nodule features are better captured and enhanced, improving the model’s ability to detect early nodules.
In the first detection step, we use a decoupled head, which includes three branches: cls, reg, and projection Conv. Label Assign adopts the AnchorFree format and uses TAL dynamic matching to select high-quality anchors as positive samples from the perspective of task alignment and incorporate them into the loss design. Calculating the loss with the aid of a border further enhances the model’s generalization ability20.
Backbone
This study divides the backbone feature extraction network into five stages (as shown in Fig. 4, S1–S5). In stage S1, the height and width information of the ultrasound image is first converted into channel features using the first layer of the base structure. Then, a second channel is used to compress the feature map. Further, the number of channels increases, and the feature map is observed for down sampling. Then, a feature map with a rich gradient flow is extracted through the C2f. module.
In stages S2 to S5, the feature map’s width and height are further compressed, increasing the number of channels. The spatial pyramid pooling module (SPPF) converts the feature map of variable size to a uniform size to achieve multi-pixel feature fusion. Stages S2 to S5 generate four feature maps as input to the neck network.
We replaced the traditional convolution in stage S2 during the optimization process with the DCN structure. DCN automatically adjusts the receptive field by adaptively learning the offset of the feature map, and at the same time, through attention weighting, it has more flexible sampling ability and adaptability than traditional convolution. DCN can better adapt to nodules with different morphological features and enhance the model’s ability to capture shallow features. To address the problem of background interference in ultrasound image data, CPCA is introduced after the SPPF module to perform attention weighting on the input feature map, reducing the impact of background information on detection performance and improving the representational power and robustness of the model.
Deformable convolution
The superficial features of thyroid nodules are rich in local information, and the efficient extraction of superficial features is crucial to the nodule detection process. However, due to their fixed geometry, traditional convolution units cause the receptive field size of the same CNN to be the same when processing different regions of the image and cannot adapt to features that change in position.
To solve this problem, we replace the traditional convolution in the C2f. module with DCN21 in the S2 stage of the backbone network. DCN adapts the offset of the feature map through an additional convolution layer and applies it to the sampling position of the standard convolution, enabling the network to respond flexibly to deformation. In addition, to further correct and refine the range of new sampling points, CPCA is incorporated into the deformable convolution structure to improve the model’s ability to resist spatial transformations. Figure 5 shows the structure of the deformable convolution, and Fig. 6 compares the sampling positions of the standard convolution and the deformable convolution.
In the deformable convolution, based on the input image, the traditional convolution kernel is first used to obtain the feature map \({F}_{in}\), and then the obtained feature map is used as input. A convolution is applied, followed by CPCA for weighting to obtain the feature map \({F}_{0}\). The width and height of \({F}_{0}\) correspond to the input feature map, with each pixel representing \(\text{K}\times \text{K}\) sampling points, with an offset \(\Delta \text{p}(\Delta \text{x},\Delta \text{y})\). The learning of these offsets is conducted through interpolation and backpropagation. The weighted values of the offsets are then applied to the corresponding sampling areas (green grid). Finally, the convolution operation is performed in the adjusted sampling area (blue grid), achieving the effect of deformable convolution. This process can be mathematically represented as follows.
First, a set of pixel points R is sampled from the input Feature Map, as shown in Eq. (1). R defines the receptive field. Taking a 3 × 3 convolution as an example, the elements in R represent the relative positions of the sampling points.
Then, based on R, the sampled results are calculated using a convolution operation, as shown in Eq. (2).
Finally, an offset \(\Delta {p}_{n}\) is added to each sampling point \({p}_{0}\)+\({p}_{n}\), where \(\{\Delta {p}_{n}|n=\text{1,2},3,...,N\}\), and the output pixel value \(\text{y}({p}_{0})\) at point \({p}_{0}\) in the Feature Map is obtained through convolution operations. The sampling positions in the Feature Map are usually integers, but the offset \(\Delta {p}_{n}\) is typically a small fractional value. Therefore, bilinear interpolation is used to retrieve values in the input Feature Map. As shown in Eq. (3).
For a floating-point sampling point p,first,the actual pixel points \(\text{x}(\text{q})\) in its four neighboring regions are retrieved.The function G(.) is used to compute the weights for each \(\text{x}(\text{q})\). Finally, the weighted values of \(\text{x}(\text{q})\) are summed and used as the pixel value \(\text{x}(\text{p})\) at position in the input feature map. As shown in Eq. (4).
Attention mechanism
In the thyroid nodule detection method proposed in this study, the backbone network is used to initially extract the nodule features from ultrasound images and output four feature maps for the neck network to use. To reduce the interference of background information on the model, we introduced CPCA22 between the backbone and neck networks. Specifically, before the feature maps extracted by the backbone network are input into the neck network, the attention mechanism is applied to suppress noise and reduce background interference.
The structure of CPCA, as shown in Fig. 6, consists of channel attention and spatial attention sequentially. It dynamically allocates weights across both channel and spatial dimensions, allowing it to capture spatial relationships while retaining channel priorities. First, the spatial information of the feature map is aggregated through average pooling and max pooling operations, and a channel attention map is generated via a shared Multi-Layer Perceptron (MLP). The input features are then multiplied by the channel attention map to produce channel-refined features, which are fed into a deep convolution block to generate a spatial attention map. After generating the spatial attention map, the convolution block continues to receive and perform channel-spatial fusion operations, enhancing feature representation. Finally, the result of channel-spatial fusion is multiplied with the channel-refined features to obtain the optimized feature output. The mathematical expression of this process is shown below:
In this context, \(\text{CA}(\text{F})\) represents the channel attention map, \({F}_{c}\) denotes the channel prior, \(\text{SA}(\text{F})\) stands for the spatial attention map, and \(\widehat{F}\) refers to the refined features output.
Neck
This study introduces the GD mechanism23 as the Neck structure in the YOLO series to improve the integration of cross-layer information. This mechanism solves the difficulties and losses in the information interaction and fusion process of the traditional FPN structure. Through convolution and self-attention mechanisms, the GD mechanism can efficiently fuse multi-scale features on a global scale and inject global information into features at each layer, overcoming the shortcomings of FPN, improving the efficiency of information transmission, and ultimately enhancing the model’s ability to detect early thyroid nodules.
The GD mechanism includes a Low- Gather and Distribute (Low-GD) and a High-Gathering and Distribute (High-GD), as shown in Fig. 7a, b. The aggregation and distribution process consists of three parts: the Feature Alignment Module (FAM), the Information Fusion Module (IFM), and the Injection module (Inject). FAM collects and aligns features at each level, IFM is responsible for fusing the aligned features to generate global information, and Inject is accountable for distributing the fused global information to the feature maps at each level and injecting it into the features at each level through a self-attention mechanism.
The input to the Neck is derived from the four effective feature layers, B2, B3, B4, and B5, extracted from the backbone network, where \({B}_{i}\in {R}^{N\times {C}_{{B}_{i}}\times {R}_{{B}_{i}}}\), with batch size represented by N, channels by C, and spatial dimensions by R(where \(\text{R}=\text{HXW}\)). Additionally, the dimensions of \({R}_{B2}\),\({R}_{B3}\),\({R}_{B4}\),\({R}_{B5}\) are \(R\),\(\frac{1}{2}R\),\(\frac{1}{4}R\),\(\frac{1}{8}R\), respectively.
Low-GD
Low-GD includes LOW-FAM, LOW-IFM, and the Injection module, which perform fusion on B2, B3, B4, and B5 to capture high-resolution features of small target information.
In LOW-FAM, AvgPool is used to down sampling the input features, thereby unifying the feature sizes. LOW-IFM, on the other hand, consists of multiple layers of \(RepBlock\) and \(Split\). The \(RepBlock\) aligns the input features \({F}_{align}\) (where \((\text{channel}=\text{sum}({\text{C}}_{\text{B}2},{\text{C}}_{\text{B}3},{\text{C}}_{\text{B}4},{\text{C}}_{\text{B}5},)\)) to generate \({F}_{fuse}(\text{channel}=({\text{C}}_{\text{B}4}+{\text{C}}_{\text{B}5})\). The intermediate channels are adjusted according to the size of the target model. The features generated by \(RepBlock\) are split into two dimensions, denoted as \({F}_{inj\_P3}\) and \({F}_{inj\_P4}\), which correspond to different levels of feature fusion. The Injection module divides the input features \({F}_{align}\), as well as the globally injected information \({F}_{inj}\), for fusion. Through Conv calculation, the local features \({F}_{\text{act}}\) and the global features \({F}_{\text{global}\_\text{embed}}\) are obtained. The local features \({F}_{\text{local}\_\text{embed}}\) are then fused through Conv and attention mechanisms to generate the final output features \({F}_{out}\). Since there is a size difference between the local and global features, average pooling and bilinear interpolation are employed to adjust the sizes of \({F}_{inj}\), \({F}_{\text{global}\_\text{embed}}\) and \({F}_{\text{act}}\), ensuring proper alignment. To more effectively inject global information into different layers, a self-attention mechanism is used to control the fusion, along with adding \(RepBlock\) to further extract and fuse information. Among these,\({F}_{local}\) is equivalent to \(\text{Bi}\). The mathematical representation of the above process is shown below:
High-GD
High-GD comprises High-FAM, High-IFM, and the Injection module, responsible for further fusing the outputs of Low-GD from {P3, P4, P5}.
In High-FAM, the input feature dimensions are reduced and unified into a consistent size. High-IFM includes a Transformer module and \(Split\) process, which operates in three steps: (1) The Transformer merges \({F}_{align}\) to obtain \({F}_{fuse}\)0. (2)The Conv layer reduces \({F}_{fuse}\) to the sum of \(({\text{C}}_{\text{P}4},{\text{C}}_{\text{P}5})\)0. (3) The \(Split\) process divides \({F}_{fuse}\) along the channel dimension into \({F}_{inj\_N4}\) and \({F}_{inj\_N5}\), which are then fused with the corresponding features from the previous layers.The Injection process is identical to that in Low-GD. In this context, \({F}_{local}\) is equivalent to \(\text{Pi}\). The mathematical representation of the above process is as follows:
Head
In the detection head step, a decoupled head structure24 and AnchorFree25 strategies are used, as shown in Fig. 8. This structure separates the classification branch and the regression branch to correspond to different task requirements, where Varifocal Loss (VFL Loss)26 is the classification loss and Distribution Focal Loss (DFL Loss)27 + CIOU Loss is the bounding box regression loss. In addition, the Anchor-Free strategy is used, where the target is represented by multiple key points or centroid points and corresponding boundary information without preset anchors so that target detection can be performed directly. This avoids the complexity of manually designing and adjusting the prior box hyperparameters25.
Experimentation and results
Experimental data
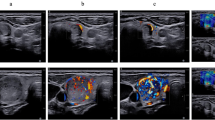
A partner hospital provided the thyroid ultrasound image data used in this study. Under the premise of fully protecting patient privacy, 3,055 ultrasound images of 205 patients aged 10 to 77 years were collected (2019 to 2023). Each image contains at least one thyroid nodule region for 3088 nodules, including 1752 benign and 1336 malignant nodules, divided into training and test sets in a 7:3 ratio. In addition, we have also included the currently publicly accessible DDTI public database28 as part of the test set. All data were labeled by professional physicians in the ultrasound department to ensure the authenticity and reliability of the data. An example of the ultrasound image data is shown in Fig. 9.
Data processing
Data quality is affected by equipment acquisition, doctor’s operation, and patient cooperation. Deep learning relies on high-quality data to ensure the accuracy and reliability of the model. Therefore, this study processed the thyroid ultrasound image data by sharpening and other means to enhance the image contrast and highlight the nodules and their boundary information. The processed ultrasound images (as shown in Fig. 10) have higher contrast, more explicit nodule boundaries, and more prominent features.
Experimental environment and training strategies
The system utilized in this research is equipped with an RTX 4070 GPU (32 GB memory), an Intel Core i9-13980HX processor, and is configured with CUDA 11.1 and cuDNN 8.0.5 for accelerated GPU computing. The development platform is PyCharm, with the project built on PyTorch 2.0.During model training, a transfer learning strategy was employed to optimize performance. Initial training was conducted using the VOC dataset to obtain pre-trained weights, followed by fine-tuning. The optimizer used is Adaptive Moment Estimation (Adam), with a momentum of 0.95 and an initial learning rate of 0.01. Training was conducted for a total of 200 epochs with a batch size of 16.Fig. 11 illustrates the change in loss and mAP@50 as the number of training epochs increases. By the 185th epoch, both loss and mAP stabilized. From the 190th epoch onwards, the loss decreased significantly and quickly plateaued. To prevent overfitting, Mosaic data augmentation was disabled during the final 10 epochs. Each experiment was repeated three times to ensure data reliability, and the average values were taken to minimize the influence of random error.
Comparative experiments
Experimental index assessment
This study employs three key metrics to comprehensively evaluate the performance of the thyroid nodule detection model: Precision (P), Recall (R), and mAP@50. Precision reflects the proportion of predicted positive samples that are truly positive, measuring the model’s ability to minimize false positives, thereby reducing unnecessary examinations and treatments. Recall evaluates the proportion of actual positive samples correctly identified by the model, which is critical for reducing missed diagnoses a particularly important aspect in medical diagnosis.
To balance precision and recall, we introduce mAP@50 as a comprehensive performance evaluation metric. mAP@50 considers not only the overlap between the predicted bounding box and the ground truth but also measures accuracy across different thresholds, fully reflecting the model’s localization and classification capabilities. A higher mAP@50 indicates better classification accuracy and bounding box localization in nodule detection.
In summary, these three indicators Precision, Recall, and mAP@50 offer a comprehensive evaluation of the model’s performance in thyroid nodule detection, ensuring its accuracy, sensitivity, and reliability in practical application.
Comparative experiments on different models
YOLOv8 demonstrated outstanding detection performance in thyroid ultrasound diagnosis. Based on comparative experiments with different model weights, and considering both inference efficiency and detection accuracy, YOLOv8-L showed the best overall performance, as illustrated in Table 1. Therefore, YOLOv8-L was selected as the baseline model for this study.
To meet the characteristics of ultrasound imaging and clinical diagnostic needs, we optimized the baseline algorithm to enhance its feature representation capabilities and improve the detection of small nodules. Experimental results indicate that the mAP@50 of the improved model increased by 6.6% compared to the baseline algorithm, with all performance metrics surpassing the baseline model, particularly in the detection of malignant nodules.
To verify the feasibility and effectiveness of our model, we compared it with several single- and two-stage models as well as their baseline algorithms. All experiments were conducted using the same thyroid ultrasound dataset, training strategies, and evaluation metrics. The comparison results are shown in Table 2. Zheng et al.’s Cascade Mask-RCNN model demonstrated better performance in detecting benign nodules, improving mAP@50 by 4.7% over its baseline. Although Yang et al.'s Improved-YOLOX model accounted for the diversity of nodule morphology and the needs of early diagnosis, it was limited by the randomness of deformable convolutional offset sampling and the FPN architecture, achieving only a 5.2% increase in mAP@50, which still falls short compared to our model.
Comparison of attention mechanisms
In this study, four attention mechanisms were integrated into the network to reduce the impact of redundant information in ultrasound images on model performance: Coordinate Attention (CA)29, Spatial Transformer Networks (STN)30, Convolutional Block Attention Module (CBAM)31, and CPCA. The comparison results of these four attention mechanisms are presented in Table 3. Experimental results show that CPCA significantly improved the detection performance of thyroid nodules, with an mAP@50 of 91.5%. In comparison, CA and STN only improved mAP@50 by 0.2% and 0.4%, respectively, over the baseline model. As a hybrid attention mechanism that combines spatial and channel information, CBAM achieved an mAP@50 of 90.8% when integrated into the baseline model but still fell short of CPCA. This is because CBAM enforces a uniform spatial attention distribution across the channel dimension, which limits its ability to dynamically adjust attention weights between channel and spatial dimensions.
Ablation experiments
To verify the effectiveness of the improvements made to the baseline model in this study, ablation experiments were conducted on individual modules (DCN, CPCA, and GD), and the results are presented in Table 4.
DCN module: This replaces the traditional convolution in the C2f. module at stage S2, enhancing the model’s ability to capture shallow features. Experimental results show that the DCN module increased the mAP@50 for thyroid nodules to 89.8%, which is a 0.9% improvement over the baseline model. The DCN’s ability to flexibly adjust the sampling area according to different nodule shapes strengthens the network’s representational and detection capabilities.
Enhanced DCN (with CPCA): While DCN enables flexible sampling by learning offsets, it further improves the model’s robustness to spatial transformations through attention-weighted mechanisms. The combined use of DCN and CPCA resulted in a 2.8% increase in mAP@50.
CPCA module: CPCA applies attention weighting to the four feature maps output by the backbone network to mitigate the interference of background information. The results show that the introduction of CPCA improved the mAP@50 by 1.5%.
GD module: The GD mechanism efficiently integrates and exchanges information through convolution and self-attention, enhancing the model’s ability to fuse multi-scale features. Results indicate that the GD module increased the mAP@50 by 1.4%.
Results
To provide a clear comparison of the detection performance of our proposed model against advanced algorithms in thyroid ultrasound imaging, we conducted comparative experiments using the test set as the sample. The results, as shown in Fig. 12, indicate that our model significantly outperforms others in terms of precision, recall, and mAP@50. This demonstrates the model’s effectiveness in accurately locating and identifying nodules in thyroid ultrasound images.
Additionally, we conducted follow-up evaluations with users, and the results suggest that the proposed method surpasses clinical doctors’ visual diagnosis in overall performance while requiring minimal time for the entire diagnostic process. Therefore, this method demonstrates notable accuracy and efficiency in the diagnosis and treatment of thyroid nodules in ultrasound images, indicating its vast potential for clinical application.
Conclusion and discussion
Conclusion
The objective of this study is to enhance the detection accuracy of nodules in thyroid ultrasound images, particularly for early-stage nodules. By utilizing the YOLOv8-L model as a baseline, we propose a novel framework that integrates advanced feature extraction techniques and model optimization strategies, achieving precise localization and classification of nodular lesions. Experimental results demonstrate that the optimized model performs exceptionally well in detecting early small nodules, indicating its promising clinical application potential.
In this study, statistical uncertainty is also a crucial consideration. Although our method shows favorable performance in improving detection sensitivity and accuracy, there are inherent statistical uncertainties introduced by factors such as sample selection, data noise, and model parameter estimation. This uncertainty may affect the stability and reliability of the detection results. Therefore, we recommend that future research employ larger and more diverse datasets to mitigate the impact of sample variation on the results. Additionally, incorporating statistical methods such as confidence intervals and error analysis can provide a more comprehensive evaluation of model performance, thereby enhancing the credibility of the results.
By improving detection sensitivity and accuracy, we anticipate that this method can serve as an effective auxiliary tool for radiologists, subsequently increasing the efficiency of early screening for thyroid diseases and ultimately improving patient diagnosis and prognosis. Furthermore, our optimization approach is not limited to YOLO-based models but is applicable to various mature object detection models. This broad applicability suggests that the method has the potential for widespread adoption, particularly in the early screening and diagnosis of thyroid diseases. By promoting the advancement of intelligent medical technology in the field of imaging, we aim to provide more precise and efficient healthcare services to a wider patient population, further enhancing the scientific basis and timeliness of medical decision-making, thereby improving patient prognosis and quality of life.
Discussion
The proposed method in this study primarily focuses on improving nodule detection accuracy and early nodule detection rates, resulting in some sacrifices in efficiency. As such, it does not yet meet the requirements for real-time diagnostics. However, in clinical settings, radiologists typically perform better when analyzing ultrasound videos, indicating significant potential for real-time diagnostic development in video imaging. Therefore, we hope that future researchers can build on the results of this study to achieve lightweight optimizations, developing more efficient deep learning solutions that maintain detection accuracy while enhancing the system’s real-time performance. This would further promote the application of intelligent medical technology.
Currently, the use of deep learning technologies for intelligent diagnosis of thyroid ultrasound images has become a research hotspot, yielding significant progress. However, this field still faces challenges such as the limited availability of publicly accessible datasets and the lack of standardized evaluation metrics, which need to be addressed in future work. As a data-driven discipline, the quality of data directly influences the final performance of the model. The inherent presence of speckle noise and low contrast obscures important information in visual diagnosis, reducing the quality and reliability of ultrasound images. Therefore, future work should focus on the development of denoising and filtering algorithms for thyroid ultrasound images to further improve the visual interpretability of ultrasound data and ensure data quality.
Data availability
The data that support the findings of this study are available from The Affiliated Hospital to Changchun University of Traditional Chinese Medicine but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the corresponding author upon reasonable request and with permission of The Affiliated Hospital to Changchun University of Traditional Chinese Medicine.
References
Das, D., Iyengar, M. S., Majdi, M. S., Rodriguez, J. J. & Alsayed, M. Deep learning for thyroid nodule examination: A technical review. Artif. Intell. Rev. 57(3), 47. https://doi.org/10.1007/s10462-023-10635-9 (2024).
‘4 Image Quality Assessment Parameters for Despeckling Filters.pdf’.
Yadav, N. Deep learning-based CAD system design for thyroid tumor characterization using ultrasound images. Multimed. Tools Appl. (2024).
Yadav, N. A systematic review of machine learning based thyroid tumor characterisation using ultrasonographic images. J. Ultrasound. (2024).
Yildirim, D. et al. Current radiological approach in thyroid nodules. J. Cancer Ther. 08(05), 423–442. https://doi.org/10.4236/jct.2017.85037 (2017).
Wu, S., Zhu, Q. & Xie, Y. Evaluation of various speckle reduction filters on medical ultrasound images. In 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) 1148–1151 (IEEE, 2013). https://doi.org/10.1109/EMBC.2013.6609709
Yadav, N. Objective assessment of segmentation models for thyroid ultrasound images. J. Ultrasound (2023).
Yadav, N. Despeckling filters applied to thyroid ultrasound images: a comparative analysis. Multimed. Tools Appl. (2022).
Virmani, J. & Agarwal, R. Assessment of despeckle filtering algorithms for segmentation of breast tumours from ultrasound images. Biocybern. Biomed. Eng. (2019).
Bibicu, D., Moraru, L. & Biswas, A. Thyroid nodule recognition based on feature selection and pixel classification methods. J. Digit. Imaging 26(1), 119–128. https://doi.org/10.1007/s10278-012-9475-5 (2013).
Tsantis, S., Dimitropoulos, N., Cavouras, D. & Nikiforidis, G. Morphological and wavelet features towards sonographic thyroid nodules evaluation. Comput. Med. Imaging Graph. 33(2), 91–99. https://doi.org/10.1016/j.compmedimag.2008.10.010 (2009).
Chang, C.-Y., Chen, S.-J. & Tsai, M.-F. Application of support-vector-machine-based method for feature selection and classification of thyroid nodules in ultrasound images. Pattern Recognit. 43(10), 3494–3506. https://doi.org/10.1016/j.patcog.2010.04.023 (2010).
Zheng, Y. et al. Automated detection and recognition of thyroid nodules in ultrasound images using improve cascade mask R-CNN. Multimed. Tools Appl. 81(10), 13253–13273. https://doi.org/10.1007/s11042-021-10939-4 (2022).
Fang, H. et al. Reliable thyroid carcinoma detection with real-time intelligent analysis of ultrasound images. Ultrasound Med. Biol. 47(3), 590–602. https://doi.org/10.1016/j.ultrasmedbio.2020.11.024 (2021).
Song, W. et al. Multitask cascade convolution neural networks for automatic thyroid nodule detection and recognition. IEEE J. Biomed. Health Inform. 23(3), 1215–1224. https://doi.org/10.1109/JBHI.2018.2852718 (2019).
Wang, L. et al. Automatic thyroid nodule recognition and diagnosis in ultrasound imaging with the YOLOv2 neural network. World J. Surg. Oncol. 17(1), 12. https://doi.org/10.1186/s12957-019-1558-z (2019).
Ma, J. et al. Efficient deep learning architecture for detection and recognition of thyroid nodules. Comput. Intell. Neurosci. 2020, 1–15. https://doi.org/10.1155/2020/1242781 (2020).
Zhang, L. et al. Automated ___location of thyroid nodules in ultrasound images with improved YOLOV3 network. J. X-Ray Sci. Technol. 29(1), 75–90. https://doi.org/10.3233/XST-200775 (2021).
Yang, T.-Y., Zhou, L.-Q., Li, D., Han, X.-H. & Piao, J.-C. An improved CNN-based thyroid nodule screening algorithm in ultrasound images. Biomed. Signal Process. Control 87, 105371. https://doi.org/10.1016/j.bspc.2023.105371 (2024).
Feng, C., Zhong, Y., Gao, Y., Scott, M. R. & Huang, W. TOOD: Task-aligned one-stage object detection. arXiv 2021. Accessed Apr. 09, 2024. Available: http://arxiv.org/abs/2108.07755
Dai, J. et al. Deformable Convolutional Networks (2017). arXiv: arXiv:1703.06211. Accessed: Mar. 26, 2024. [Online]. Available: http://arxiv.org/abs/1703.06211
Huang, H., Chen, Z., Zou, Y., Lu, M. & Chen, C. Channel prior convolutional attention for medical image segmentation (2023). arXiv: arXiv:2306.05196. Accessed: Mar. 26, 2024. [Online]. Available: http://arxiv.org/abs/2306.05196
Wang, C. et al. Gold-YOLO: Efficient object detector via gather-and-distribute mechanism (2023). arXiv: arXiv:2309.11331. Accessed: Mar. 26, 2024. [Online]. Available: http://arxiv.org/abs/2309.11331
Wu, Y. et al. Rethinking classification and localization for object detection. In 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA 10183–10192 (IEEE, 2020). https://doi.org/10.1109/CVPR42600.2020.01020
Zhang, X., Wan, F., Liu, C., Ji, R. & Ye, Q. FreeAnchor: Learning to match anchors for visual object detection (2019). arXiv: arXiv:1909.02466. Accessed: Mar. 26, 2024. [Online]. Available: http://arxiv.org/abs/1909.02466
Zhang, H., Wang, Y., Dayoub, F. & Sünderhauf, N. VarifocalNet: An IoU-aware dense object detector (2021). arXiv: arXiv:2008.13367. Accessed: Mar. 26, 2024. [Online]. Available: http://arxiv.org/abs/2008.13367
Li, X. et al. Generalized focal loss: Learning qualified and distributed bounding boxes for dense object detection (2020), arXiv: arXiv:2006.04388. Accessed: Mar. 26, 2024. [Online]. Available: http://arxiv.org/abs/2006.04388
Pedraza, L., Vargas, C., Narváez, F. et al. An open access thyroid ultrasound image database. In: 10th International Symposium on Medical Information Processing and Analysis 9287:92870W1–6 (2015). https://doi.org/10.1117/12.2073532
Hou, Q., Zhou, D. & Feng, J. Coordinate attention for efficient mobile network design (2021). arXiv: arXiv:2103.02907. Accessed: Mar. 26, 2024. [Online]. Available: http://arxiv.org/abs/2103.02907
Jaderberg, M., Simonyan, K. & Zisserman, A. Spatial Transformer Networks.
Woo, S., Park, J., Lee, J.-Y. & Kweon, I. S. CBAM: Convolutional block attention module (2018). arXiv: arXiv:1807.06521. Accessed: Mar. 26, 2024. [Online]. Available: http://arxiv.org/abs/1807.06521
Acknowledgements
This work has received funding support from the Jilin Provincial Department of Science and Technology’s Technical Innovation Guidance for the Development of the Medical and Health Industry Special Project (No. 20220401086YY).
Author information
Authors and Affiliations
Contributions
Xu Yang: Project conceptualization, Method analysis, Experimental analysis, Investigation, and Writing of the original draft. Hongliang Geng: Investigation, Visualization analysis, Data processing, and Writing of the original draft. Xue Wang: Resource docking, Data collection, and Verification. Lingxiao Li: Data management, Project management. Zhibin Cong: Project conceptualization, Supervision and Verification. Xiaofeng An: Supervision, Verification, Project management and Funding acquisition.
Experimental statement
All methods were performed in accordance with the relevant guidelines and regulations. The experimental data acquisition was confirmed to have received informed consent from all subjects and their legal guardians. Additionally, the Ethics Committee of The Affiliated Hospital of Changchun University of Traditional Chinese Medicine approved all experimental protocols involved.
Corresponding authors
Ethics declarations
Competing interests
The author declares no potential conflicts of interest regarding the research, the author’s identity, and the viewpoints presented in this article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, X., Geng, H., Wang, X. et al. Identification of lesion ___location and discrimination between benign and malignant findings in thyroid ultrasound imaging. Sci Rep 14, 32118 (2024). https://doi.org/10.1038/s41598-024-83888-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-83888-1