Abstract
To develop a deep learning model using transfer learning for automatic detection and segmentation of neck lymph nodes (LNs) in computed tomography (CT) images, the study included 11,013 annotated LNs with a short-axis diameter ≥ 3 mm from 626 head and neck cancer patients across four hospitals. The nnUNet model was used as a baseline, pre-trained on a large-scale head and neck dataset, and then fine-tuned with 4,729 LNs from hospital A for detection and segmentation. Validation was conducted on an internal testing cohort (ITC A) and three external testing cohorts (ETCs B, C, and D), with 1684 and 4600 LNs, respectively. Detection was evaluated via sensitivity, positive predictive value (PPV), and false positive rate per case (FP/vol), while segmentation was assessed using the Dice similarity coefficient (DSC) and Hausdorff distance (HD95). For detection, the sensitivity, PPV, and FP/vol in ITC A were 54.6%, 69.0%, and 3.4, respectively. In ETCs, the sensitivity ranged from 45.7% at 3.9 FP/vol to 63.5% at 5.8 FP/vol. Segmentation achieved a mean DSC of 0.72 in ITC A and 0.72 to 0.74 in ETCs, as well as a mean HD95 of 3.78 mm in ITC A and 2.73 mm to 2.85 mm in ETCs. No significant sensitivity difference was found between contrast-enhanced and unenhanced CT images (p = 0.502) or repeated CT images (p = 0.815) during adaptive radiotherapy. The model’s segmentation accuracy was comparable to that of experienced oncologists. The model shows promise in automatically detecting and segmenting neck LNs in CT images, potentially reducing oncologists’ segmentation workload.
Similar content being viewed by others
Introduction
Head and neck cancer (HNC), including nasopharyngeal carcinoma (NPC), is among the most prevalent malignancies worldwide1. Given that neck lymph nodes (LNs) serve as the primary metastasis site for HNC, determining LN involvement plays a pivotal role in treatment decisions and patient outcomes1,2.
Computed tomography (CT) is a primary choice for LN evaluation3,4,5. LNs with a short-axis diameter (SAD) ≥ 10 mm are usually considered as malignant6. However, in HNC, microscopic metastases may also be present in non-enlarged LNs, such as those with an SAD of 5 mm or less, underscoring that small LNs should not be overlooked4,7. Identifying all LNs in a CT scan is important, however, this task is challenging. In practice, LNs ≥ 3 mm may be missed by both inexperienced and experienced radiation oncologists (ROs)8.
Precise LN delineation is critical for HNC radiotherapy. Since manual detection and segmentation of LNs ≥ 3 mm in a CT scan is quite a burdensome task and heavily dependent on an observer’s expertise, a fully automatic LN detection and segmentation system is highly desirable. Recent developments in deep learning (DL) have shown remarkable achievements in various medical tasks, such as lesion detection9,10, organ and tumor segmentation11,12,13,14, and tumor efficacy prediction15,16. In the context of LN detection, some algorithms have been proposed to detect or segment mediastinal, abdominal, pelvic and neck LNs17,18,19,20,21,22,23,24. Zhao et al. introduced an “auto-LNDS” model through transfer learning for LN detection in rectum cancer in magnetic resonance imaging, achieving a sensitivity of 0.8022. Cao et al. proposed a DL system for mediastinal enlarged LNs segmentation using CT images, with the system achieving a patient-level sensitivity of 0.94 in an internal testing dataset23. Additionally, Mehrtash et al. introduced a DL model with weak supervision for mediastinal LN segmentation20, and Xu et al. developed a “Mask R-CNN” model to identify positive neck LNs in oral cancer in CT images24.
Previous studies have yielded promising results, with sensitivity and Dice scores ranging from 50 to 90% and 0.5 to 0.7, respectively. However, due to the considerable individual differences in the ___location and size of neck LNs, the task of LN detection and segmentation remains challenging. Some limitations might exist in previous studies on neck LN segmentation, such as small sample sizes, limited data sources, focusing solely on large LNs, or utilizing outdated segmentation methods8,21,25,26,27. Moreover, contrast-enhanced CT (ceCT) and unenhanced CT (ueCT) are both routinely used in clinical practice. The appearance and margin of LNs differ between these two types of CT, which may impact the accuracy of DL models when segmenting LNs on both modalities. However, this issue has been rarely addressed in previous studies. In conclusion, the robustness, reproducibility and detection ability of DL models for LNs, particularly small LNs, might require further examination.
To address the potential limitations, we retrospectively collected a dataset involving 626 subjects with 11,013 LNs with an SAD ≥ 3 mm from four hospitals, which was used to train and validate a fully automated LN detection and segmentation model. In summary, this work has three main contributions as follows: (1) the study presents a novel application of deep transfer learning through the nnUNet model specifically designed for automatic detection and segmentation of neck LNs in CT images. This innovative approach significantly enhances accuracy, particularly for small LNs that are frequently missed by existing methods. (2) By leveraging a substantial multi-center dataset, this research greatly improves the model’s robustness and generalizability. This diverse dataset addresses limitations seen in prior studies, providing a more comprehensive foundation for effective LN assessment. (3) The model’s segmentation accuracy is shown to be comparable to that of experienced ROs, highlighting its potential as a valuable clinical tool. By assisting clinicians in the segmentation process, the model aims to reduce the manual workload and enhance patient care outcomes.
Materials and methods
Data
This study included HNC patients treated in four tertiary hospitals between January 2017 and December 2020. Eligible patients were histologically confirmed as HNC who underwent ceCT or ueCT imaging of the head and neck regions. Patients were excluded if they had prior neck surgery or poor CT image quality. Distribution of sex, age and scanning parameters are presented in Table 1.
All patients in this study underwent radiotherapy-dedicated CT examinations (simulated CT). Firstly, they were immobilized in a supine position using a thermoplastic head, neck and shoulder mask. CT examinations were then performed, covering a scanning range from 3 cm above the suprasellar cistern to 2 cm below the sternal end of the clavicle. The CT images from hospital A and hospital D were acquired using a Brilliance CT Big Bore system from Philips Healthcare (Philips Healthcare, Best, the Netherlands), with the following scanning conditions: bulb voltage at 120 kV, current ranging from 275 to 375 mA, scan thickness of 3.0 mm, and a resolution of 512 × 512. An injected contrast agent, iohexol, was used during the ceCT examination. CT images from hospital B and hospital C were acquired using a Somatom Definition AS 40 system from Siemens Healthcare (Siemens Healthcare, Forcheim, Germany), with the similar scanning conditions as described above.
Hospital A contained 650 CT scans from 325 patients. Each case underwent an ueCT scan followed by a ceCT scan in the same body position within several minutes. This was randomly split into 257 cases for training and 68 cases for internal testing (internal testing cohort, ITC A) (Table 1). Patients from hospitals B, C and D were used to externally validate the performance of the model, which were sequentially recorded as external testing cohort B (ETC B), ETC C and ETC D. The flow chart of this study is illustrated in Fig. 1.
The study flow diagram. The top right sub-figure shows the proposed method, consisting of pretraining and fine-tuning stages. In the pretraining stage, we collected a large-scale head and neck organ-at-risk (OAR) segmentation dataset to pre-train the parameters of the nnUNet. Then, in the finetuning stage, we applied the trained nnUNet weights as initial parameters and then fine-tuned the pre-trained nnUNet on a lymph node segmentation dataset. This method can transfer the learned knowledge from head and neck OAR segmentation task to the HNC segmentation task and boost the models’ performance and robustness. HNC = head and neck cancer, RT = radiotherapy, CT = computed tomography, pts = patients, LN = lymph node, SAD = short-axis diameter, ITC = internal testing cohort, ETC = external testing cohort.
This retrospective study was approved by the Ethics Committee on Biomedical Research of these hospitals (SCCHEC-02-2023-005). Due to the retrospective nature of the study, the Ethics Committee for Medical Research of Sichuan Cancer Hospital waived the need of obtaining informed consent. Furthermore, we confirm that all data collection and analysis methods adhere to the Declaration of Helsinki.
Ground truth segmentation
All CT images in this study were dedicated for radiotherapy. Original CT images and LN contours were extracted from Treatment Planning Systems. One RO with over 20 years of experience in head and neck radiotherapy, who was also the main author of three papers28,29,30 involved in LN distribution study in HNC, was invited to check and delineate LNs according to our requirements. All neck LNs (including retropharyngeal regions, level Ib, and levels II-V) with an SAD ≥ 3 mm were required to be segmented. LNs outside these areas were excluded. For patients with both ueCT and ceCT, two sets of CT images were registered in the MIM 7.07 Software (Microsoft, Corp)31,32, then manual segmentation was performed.
In training cohort, 4729 LNs were manually annotated (Fig. 1). In ITC A and three ETCs, 1,684 LNs and 4,600 LNs were manually annotated, respectively, resulting 6,284 LNs for model testing (Table 1). Distribution of LNs in these cohorts is presented in Fig. 2.
Distribution of lymph nodes short-diameters in the training cohort, internal testing cohort A and three external testing cohorts. Note, In internal testing cohort A and external testing cohort C, we only calculated the number of lymph nodes in contrast enhanced computed tomography images. TC = training cohort, ITC = internal testing cohort, ETC = external testing cohort, LN = lymph node.
Model development
Model pretraining with self-supervised learning has recently shown compelling and promising results in many image analysis tasks33,34,35. Inspired by those existing works, this work attempts to develop a LN detection and segmentation model via pretraining and finetuning. The overview of this framework is shown in Fig. 1, which consists of model pre-trained and fine-tuning stages. Differently from previous self-supervised and contrastive learning methods33,34,35, this work applies a fully supervised manner to pre-train a foundational segmentation model for good initial parameter weights, where the head and neck organs at risk (OARs) segmentation was used as the pretraining task. In this work, we previously collected a head and neck OARs segmentation dataset to train the segmentation network, which included more than 5000 CT volumes (ueCT and ceCT) with 45 OARs (like SegRap202336) from several public datasets and private hospitals. Then, we finetuned the pre-trained segmentation model weights on an annotated LN dataset to segment the LN accurately and robustly, where the last classification layer channel of the pre-trained model was modified to 2. This framework has three advantages, (1) Compared with self-supervised learning and contrastive learning, the fully supervised manner can better capture the context and anatomical information; (2) the pre-trained dataset consists of multi-center heterogeneous CT volumes, which can increase the data diversity and boost the generalization of the pre-trained model; (3) fine-tuning can transfer the learned knowledge efficiently and reduce the LN annotation cost. Moreover, the finetuned LN segmentation model can segment unseen center patients without any additional retraining and finetuning.
During the training stage, we applied the random cropped patch as input with a size of 64 × 128 × 224, and the batch size was 2. The model is a general nnUNet37 with five downsampling and upsampling blocks, all with a kernel size of 3 × 3 × 3 except the first block with a kernel size of 1 × 3 × 3. The optimization loss function is a weighted average combination of cross-entropy and dice losses. During the inference stage, the patient-level prediction was generated via a sliding window with a window size of 32 × 64 × 112, and the prediction size was the same as the raw CT scan.
Preprocessing images
In pre-training and finetuning stages, we used a body segmentation network to locate the region of interest and remove the impact of bed and other non-relevant regions. Given the heterogeneous CT volumes from different institutions, which could have distribution differences, we applied several pre-processing strategies to reduce the data difference: (1) we converted all CT volumes into a fixed orientation RAI, where the human pose is right-to-left, anterior-to-posterior, and inferior-to-superior; (2) we adjusted the voxel-spacing to the median value of the whole dataset of 1.0 × 1.0 × 3.0, which is also a common spacing in radiation-related CT images; (3) to reduce the intensity distribution difference across different patients, we removed the intensity outlier via the intensity histogram (just considered the inside distribution between 0.5 and 99.5 and normalized each patient intensity to zero mean and unit variance. In the inference stage, we removed predictions in which short-axis diameters were smaller than 3 mm and employed morphologic operations to separate the overlapping LNs to produce the final node-level predictions.
Implementation details
This framework was developed on an Ubuntu 22.04 desktop with two NVIDIA 3090 GPUs, PyTorch 2.0, and Python 3.10. We used the full-resolution nnUNet37 as our segmentation baseline, most parameters were self-configured based on the pre-trained datasets. There were several modifications: (1) in the pretraining stage, we increased the training epochs from 100 to 3000 to converge in the large-scale dataset; (2) in the fine-tuning stage, we modified the channel number of the last layer to 2 for LN segmentation and employed a larger positive sampling ratio of 0.66 to randomly crop the input patches to further improve the model segmentation and detection sensitivity. Additionally, the initialization weights of the LN segmentation model are copied from the pre-trained model except for the last layer as the channel number modification, where the last layer weights are initiated randomly. Note that in this work the pre-trained model was just used to provide a good initialization for the LN segmentation model development, it can be used to segment OARs if needed but it is out scope of this work.
Evaluation metrics
Performance evaluation included detection and segmentation. A true positive (TP) means that a model correctly detects an area where LNs are present as LNs, while a false negative (FN) occurs when a model fails to detect LNs that are present. A detection is considered to be false positive (FP) if a model incorrectly detects an area without LNs as LNs38. The sensitivity and positive predictive value (PPV) were utilized to evaluate the detection performance. The higher values of the both, the better performance of a model.
Sensitivity is the proportion of the true LNs detected by our model to total true LNs, defined as:
PPV is the proportion of the true LNs detected by our model to all LNs detected by our model, defined as:
The false positive per volume (FP/vol) is a metric that quantifies the mean number of false positives per case, defined as:
Additionally, the Dice similarity coefficient (DSC) and 95th percentile Hausdorff distance (HD95) were used to evaluating segmentation accuracy. The DSC measures the volumetric overlap between the model-generated contour (A) and the ground truth contour (G) (Eq. 1)39. The HD (mm) measures the boundary distance between two contours. In Eqs. 2 and 3, where Sa and Sb represent the set of surface points of one organ segmented by a DL model and the ground truth, respectively. HD95 (mm) is similar to HD, and it uses the 95-th percentile instead of the maximal value in Eq. 340. In general, a superior model should generate a higher value for DSC (maximum of 1) and a lower value for HD95 (minimum of 0).
Statistical analysis
Statistical analysis was performed using SPSS software package (Version 22.0, IBM SPSS Inc). Numeric variables were represented as mean ± standard deviation and compared using the t-test (two groups) or variance analysis (≥ three groups). A two-tailed p-value < 0.05 was considered significant.
Results
Detection performance
To assess the detection accuracy of our model, metrics of sensitivity, PPV and PF/vol were calculated for each testing cohort, as shown in Table 2. In the ITC A, the sensitivity, PPV, and PF/vol were 54.6%, 69.0% and 3.4, respectively. Across three ETCs, the performance ranged from 45.7% sensitivity at 3.9 FP/vol to 63.5% sensitivity at 5.8 FP/vol, with the ETC D achieving the highest sensitivity and the ETC C achieving the highest PPV.
Segmentation performance
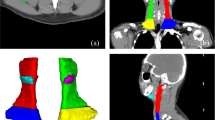
Similarly, to evaluate the segmentation accuracy of our model, we calculated the metrics of DSC and HD95 for each testing cohort. In patient level, the mean DSC (mDSC) and mean HD95 (mHD95) were 0.69 and 20.37 mm, respectively in the ITC A (Table 2). The mDSC for three ETCs ranged from 0.61 to 0.66, and the mHD95 ranged from 22.28 mm to 26.13 mm. When only calculating TP LNs detected by our model, the mDSC in the ITC A increased to 0.72 and the mHD95 decreased to 3.78 mm. Similarly, among three ETCs, a significant increase in mDSC and a considerable decrease in mHD95 were observed, ranging from 0.72 to 0.74, and 2.73 mm to 2.85 mm, respectively (Table 2). Visualization of the predicted segmentations in comparison with the manual segmentations is depicted in Fig. 3.
The DSC and HD95 distribution of LNs with different SAD in the ITC A and ETCs is presented in Fig. 4. Both the mDSC for varying LN sizes in the ITC A and ETCs exceeded 0.70 and 0.72, respectively (Fig. 4A). Regarding the HD95, there was a tendency for the mHD95 to increase as an LN SAD increased (Fig. 4C and D). In general, the mHD95 was lower than 5 mm for LNs with an SAD less than 10 mm.
DSC and HD95 distribution of lymph node with different short-axis diameters in internal testing cohort A and three external testing cohorts. In ITC A and ETC C, we only calculated the accuracy of lymph nodes in contrast-enhanced computed tomography images. DSC = Dice similarity coefficient, HD95 = 95% Hausdorff distance; ITC = internal testing cohort; ETC = external testing cohort.
Performance comparison in subgroups
To assess whether different CT modalities affect the segmentation performance of our model, we compared the detection and segmentation accuracy across different CT modalities. In terms of detection accuracy, there was no statistical difference in sensitivity (ceCT = 54.6%, ueCT = 51.9%; p = 0.502), or in PPV(ceCT = 69.0%, ueCT = 64.1%; p = 0.147) between ceCT and ueCT groups in the ITC A (Fig. 5A and B). Consistent results were observed in the ETC C, where the sensitivity and PPV in ceCT and ueCT groups were 45.7% vs 44.6% (p = 0.746), and 64.6% vs 64.1% (p = 0.889), respectively (Fig. 5C and D). Regarding segmentation accuracy, both in the ITC A and ETC C, the DSC in ceCT group was statistically higher than that in ueCT group (ITC A, 0.72 vs 0.67, p < 0.001, ETC C, 0.73 vs 0.71, p = 0.023) (Fig. 5E and G. However, in the two cohorts, the HD95 in ceCT group did not differ statistically from that in ueCT group (ITC A, 3.78 mm vs 4.23 mm, p = 0.073, ETC C, 2.83 mm vs 3.04 mm, p = 0.411) (Fig. 5F and H).
Accuracy comparison between contrast-enhanced and unenhanced CT images in internal testing cohort A and external testing cohort C. ceCT = contrasted-enhanced CT, ueCT = unenhanced CT, PPV = positive predictive value, DSC = Dice similarity coefficient, HD95 = 95% Hausdorff distance; ITC = internal testing cohort; ETC = external testing cohort. Data were denoted as mean with standard deviation, and p values were obtained by unpaired t-test.
To evaluate whether treatment-related CT affects the segmentation performance of our model, a similar comparison was conducted. There was no statistical difference in sensitivity (69.1% vs 64.4%, p = 0.809), or in PPV (58.8% vs 50.8%, p = 0.182) between CT images obtained before chemotherapy and post chemotherapy (Fig. 6A and B). No statistical difference was found in sensitivity among the three radiation-related CT groups (CTRA vs CTRB vs CTRC: 66.4% vs 75.0% vs 60.7%; p = 0.815) (Fig. 6C). However, the PPV in CTRC was statistically lower (CTRA vs CTRB vs CTRC: 52.3% vs 47.8% vs 37.1%; p = 0.035) (Fig. 6D). In terms of segmentation accuracy, the DSC in CT images obtained before chemotherapy and post chemotherapy was similar (0.75 vs 0.76, p = 0.723) (Fig. 6E), and this was also true for HD95 (2.33 mm vs 2.42 mm, p = 0.681) (Fig. 6F). The DSC and HD95 metrics revealed no statistically significant differences among the three radiation-related CT groups (see Fig. 6G and H).
Accuracy comparison between before and post induction chemotherapy CT images and accuracy comparison among repeated CT images during adaptive radiotherapy in external testing cohort D. In C, D, G, and H, each patient experienced three repeated CT scans during adaptive radiotherapy, such as at the completion of 0, 15–20, and 25–30 fractions, which was tagged as CTRA, CTRB, and CTRC, respectively. IC = induction chemotherapy, PPV = positive predictive value, DSC = Dice similarity coefficient, HD95 = 95% Hausdorff distance. In A to D, data were denoted as median with interquartile range. And p values were obtained by Mann–Whitney test (A and B), and Kruskal–Wallis test (C and D), due to the data did not conform to the normal distribution. In E to H, data were denoted as mean with standard deviation. And p values were obtained by t-test (E and F) and ordinary one-way ANOVA test (G and H).
Performance comparison between ROs and our model
To compare the performance of our model with that of experienced ROs, we randomly selected 21 patients from all testing cohorts and assigned them to three ROs with 5–8 years of radiotherapy experience who have not participated in ground truth generation. All LNs with an SAD ≥ 3 mm were annotated. These segmentations were each compared with the ground truth. As observed, the sensitivity and PPV for the three ROs varied from 36.8% to 40.5%, and 74.8% to 90.7%, respectively, higher than the model (32.9%) (Table 3). And the sensitivity and PPV of the RO1 were statistically higher than those of the model (p = 0.022, p < 0.001, respectively). However, in terms of patient-level DSC, node-level DSC and node-level HD95, the model outperforms these ROs or achieves comparable results, except for the patient-level HD95 of the RO1 (10.04 mm vs 21.35 mm, p = 0.019). The patient-level DSC and node-level HD95 of RO1 to RO3 were in the range of 0.60 to 0.62 and 2.66 mm to 3.78 mm, respectively, while the model results were 0.63 and 2.48 mm, respectively.
Discussion
In this study, we proposed a DL model to LN detection and segmentation in heterogenous CT scans for HNC. The main findings can be summarized as follows: (1) the model achieved a promising detection performance in the ITC, with a sensitivity of 54.6% at 3.4 FP/vol; (2) a good segmentation accuracy was observed for LNs with an SAD over 3 mm in the ITC, with a mDSC and a mHD95 of 0.72 and 3.78 mm, respectively; (3) a reasonable generalizability was demonstrated in detection and segmentation performance in ETCs; (4) the performance of the model in detection and segmentation was found to be less influenced by the CT scan phases and treatment conditions; (5) the segmentation accuracy was comparable to that of experienced ROs, with similar mDSC and mHD95.
To develop a LN detection and segmentation model, we first introduced a fully supervised pre-training method to produce good initial parameter weights for LN segmentation model fine-tuning. Thanks to the pre-training task of head and neck OARs segmentation, the pre-trained model is context and anatomical aware which can benefit to the fine-tuning task. In the fine-tuning stage, we employed a larger positive sampling ratio to improve the network sensitivity for LN segmentation and detection. In the inference stage, we employed morphologic operations to separate the overlapping LN predictions and remove small-size predictions. This framework shows promising performance and generalization for LN detection and segmentation in heterogeneous CT images, suggesting its effectiveness and robustness.
In internal testing cohort, the model achieved a promising sensitivity (54.6%), a high PPV (69.0%), a low FP/vol (3.4), and a good DSC (0.70). Barbu et al. acquired good results for pelvic and abdomen LNs, with a PPV of 72.6% and a FP/vol of 3.219. However, their research concentrated on LNs with an SAD over 10 mm, meaning that metastatic LNs with an SAD lower than that might be missed. Dornheim et al. introduced a 3D mass-spring model to segment neck LNs, with good results in HD9526. However, this algorithm requires some manual initialization, thus increasing extra burden for ROs. Recently, Rinneburger et al. proposed a 3D foveal neural network to detect neck LNs with an SAD over 5 mm in ceCT, with a localization rate of 81.1% and a FP/vol of 9.321. Although our sensitivity was lower than that study, our FP was considerably lower. And, we obtained a higher and more stable DSC for various LN sizes, comparing with their model showing a reduced DSC in smaller LNs than larger ones. Ariji et al. used the “DetectNet” to detection LNs for oral squamous cell cancer8. Although they achieved a high ___location rate of 73% for metastatic LNs, the rate for non-metastatic LNs (small LNs) was reduced to 53%, suggesting that detection of small LNs remains a challenging task.
Upon analysis of the model’s performance in external testing cohorts, a decline in sensitivity was observed, particularly in ETC B and ETC C, indicating that the robustness of the model was affected by heterogeneous data distributions. This is a common phenomenon in medical image segmentation tasks, as many studies have also reported a decrease in detection and segmentation performance when deploying of a trained model from a seen data to an unseen data22,41. Nevertheless, the model achieved an average sensitivity of 52.6% for the whole ETCs, which was slightly lower to that obtained in the ITC. Additionally, the segmentation accuracy for each ETC was similar to that acquired in the OARITC. Overall, a reasonable generalizability in detection and segmentation performance across ETCs was achieved.
In addition, we observed that the metric of HD95 was more influenced by an LN diameter than the DSC. The possible reason might be that the HD95 is more sensitive to outliers13. As the SAD of an LN increasing, the LN tend to have a larger volume with more irregular shape and margin. Nevertheless, the HD95 for most LNs with an SAD < 10 mm was lower than 5 mm. Taken together, the model obtained a good segmentation accuracy in heterogenous CT scans.
Previous studies have seldomly investigated the impact of CT scanning phase and post treatment images on the performance of a DL model. In one study, it was observed that a DL model exhibited inferior performance in the segmentation of LN levels in patients who underwent neck surgery compared with those who did not42. In this study, the model exhibits similar detection performance in ueCT and ceCT images, and consistent results were also found in treatment-naïve and post treatment CT images. In segmentation performance, the model tends to generate more accurate segmentations in ceCT compared to ueCT images, as indicated by a statistically higher mDSC. However, the HD95 was not significantly different. These results indicated that the model has an ability to adapt to CT intensity and changes in anatomy during treatment.
Compared with the performance of three ROs, our results show that the DL-based model can achieve a promising accuracy in terms of patient-level DSC and node-level DSC and HD95, especially the DSC outperforms three ROs consistently, suggesting that the DL-based segmentation model can assist the RO in accelerating the LN delineation and reducing the delineation burden. With the model assist, ROs need to check and refine the mis-segmentation and there is no need for manual delineation slice-by-slice. Although the model achieved some improvement in segmentation, it performs inferiorly compared to ROs in sensitivity and PPV. It conforms to common sense as the ROs tend to label all potential LNs rather than delineating the perfect contours but the model pays more attention to generating segmentations to be like the expert annotations. Moreover, we also observed an interobserver variability among the ROs, with some LNs being missed by them (the sensitivity was lower than 50%). It was similar to a study of LN detection in rectum cancer, with sensitivity ranging from 39.2% to 45.6% for radiologists with two to nine years experience, indicating that detecting all LNs is still a difficult task even for experienced experts22. Overall, our results suggest that the model still can not perform like an experienced RO in the LN detection and segmentation tasks but it has a potential to play an assistant role to shorten the detection process, accelerate the delineation workflow and reduce ROs’ burden.
We acknowledge limitations to our research. First, despite the experienced RO in treatment of HNC was invited to annotate LNs, these segmentations might still suffer from subjective variations or incomplete. In the future, inviting more reputable ROs and radiologists to participate in ground truth segmentation might aid in acquiring more representative results. Second, LNs in level_Ia were not included in this study, which might limit the model’s broader applicability. However, this does not diminish the importance of studying LNs in HNC, and future research will address this limitation by including LNs in this region. Third, although we obtained a lower FP/vol compared with most studies and a good segmentation accuracy, there is still much room for further improve the sensitivity for detecting LNs. Fourth, considering the segmentation burden for experts, performance comparison between ROs and our model was performed on a small portion of the patients, and evaluation biases might exist.
Conclusions
In conclusion, this work investigated automatic LN detection and segmentation in heterogeneous CT images using transferring learning. The comprehensive evaluation demonstrated that the proposed framework can achieve an encouraging performance in different centers showing the potential to reduce the oncologist’s segmentation burden. We will release the carefully built dataset with labels and trained models as the first large-scale head and neck LN segmentation benchmark to advance the cut-edge methods and tools development and application. In the future, we plan to conduct clinical trials to validate the performance and clinical value of our model. We also intend to explore our method on other segmentation tasks, such as different locations and other types of tumors.
Data availability
The code for neural network training can be obtained at https://github.com/Luoxd1996/. Additionally, we will release the carefully-annotated dataset lymph node annotations to boost this task research. Other data generated and analyzed during this study can be obtained by contacting the corresponding author with reasonable requirements.
Abbreviations
- CT:
-
Computed tomography
- ceCT:
-
Contrast-enhanced computed tomography
- DL:
-
Deep learning
- DSC:
-
Dice similarity coefficient
- ETC:
-
External testing cohort
- FP:
-
False positive
- FP/vol:
-
False positive rate per case
- FN:
-
False negative
- HNC:
-
Head and neck cancer
- HD95:
-
95th percentile Hausdorff distance
- ITC:
-
Internal testing cohort
- LN:
-
Lymph node
- NPC:
-
Nasopharyngeal carcinoma
- OARs:
-
Organs at risk
- PPV:
-
Positive predictive value
- RO:
-
Radiation oncologist
- RAI:
-
Right-to-left, anterior-to-posterior, and inferior-to-superior
- SAD:
-
Short-axis diameter
- TP:
-
True positive
- ueCT:
-
Unenhanced computed tomography
References
Mody, M. D., Rocco, J. W., Yom, S. S. & Haddad, R. I. Saba NFJTL: Head and neck cancer. Lancet 398(10318), 2289–2299 (2021).
De Bree, R. et al. Advances in diagnostic modalities to detect occult lymph node metastases in head and neck squamous cell carcinoma. Head Neck. 37(12), 1829–1839 (2015).
Som, P. M. & Curtin, H. D. Mancuso AAJAJoR: Imaging-based nodal classification for evaluation of neck metastatic adenopathy. AJR Am. J. Roentgenol. 174(3), 837–844 (2000).
Yonetsu, K., Sumi, M., Izumi, M., Ohki, M. & Eida, S. Nakamura TJAjon: Contribution of Doppler sonography blood flow information to the diagnosis of metastatic cervical nodes in patients with head and neck cancer: Assessment in relation to anatomic levels of the neck. AJNR Am. J. Neuroradiol. 22(1), 163–169 (2001).
Roh, J.-L. et al. 18F fluorodeoxyglucose PET/CT in head and neck squamous cell carcinoma with negative neck palpation findings: A prospective study. Radiology 271(1), 153–161 (2014).
Schwartz, L. et al. Eisenhauer EJEjoc: Evaluation of lymph nodes with RECIST 1.1. Eur. J. Cancer 45(2), 261–267 (2009).
Chen, A., Deeley, M. A., Niermann, K. J. & Moretti, L. Dawant BMJMp: Combining registration and active shape models for the automatic segmentation of the lymph node regions in head and neck CT images. Med. Phys. 37(12), 6338–6346 (2010).
Ariji, Y. et al. Automatic detection of cervical lymph nodes in patients with oral squamous cell carcinoma using a deep learning technique: A preliminary study. Oral Radiol. 37, 290–296 (2021).
Nam, J. G. et al. Development and validation of deep learning–based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology 290(1), 218–228 (2019).
Ying, H. et al. A multicenter clinical AI system study for detection and diagnosis of focal liver lesions. Nat. Commun. 15(1), 1131 (2024).
Chen, X. et al. A deep learning-based auto-segmentation system for organs-at-risk on whole-body computed tomography images for radiation therapy. Radiother. Oncol. 160, 175–184 (2021).
Liao, W. et al. Comprehensive evaluation of a deep learning model for automatic organs at risk segmentation on heterogeneous computed tomography images for abdominal radiation therapy. Int. J. Radiat. Oncol. Biol. Phy. 15, 994–1006 (2023).
Tang, H., Chen, X., Liu, Y. & Lu, Z. Xie XJNMI: Clinically applicable deep learning framework for organs at risk delineation in CT images. Nat. Mach Intell. 1(10), 480–491 (2019).
Oh, Y. et al. Ye JCJNC: LLM-driven multimodal target volume contouring in radiation oncology. Nat. Commun. 15(1), 9186 (2024).
Li, Z. et al. Multiregional dynamic contrast-enhanced MRI-based integrated system for predicting pathological complete response of axillary lymph node to neoadjuvant chemotherapy in breast cancer: Multicentre study. EBioMedicine 107, 105311 (2024).
Ramtohul, T. et al. Use of pretreatment perfusion MRI–based intratumoral heterogeneity to predict pathologic response of triple-negative breast cancer to neoadjuvant chemoimmunotherapy. Radiology 312(3), e240575 (2024).
Roth, H. R., Lu, L., Seff, A., Cherry, K. M., Hoffman, J., Wang, S., Liu, J. & Turkbey, E. Summers RM: A new 2.5 D representation for lymph node detection using random sets of deep convolutional neural network observations. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2014: 17th International Conference, Boston, MA, USA, September 14–18, 2014, Proceedings, Part I 17: 2014: Springer; 2014: 520–527.
Kitasaka, T., Tsujimura, Y., Nakamura, Y., Mori, K., Suenaga, Y., Ito, M. & Nawano S: Automated extraction of lymph nodes from 3-D abdominal CT images using 3-D minimum directional difference filter. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2007: 10th International Conference, Brisbane, Australia, October 29-November 2, 2007, Proceedings, Part II 10: 2007: Springer; 2007: 336–343.
Barbu, A., Suehling, M., Xu, X., Liu, D. & Zhou, S. K. Comaniciu DJIToMI: Automatic detection and segmentation of lymph nodes from CT data. IEEE Trans. Med. Imaging 31(2), 240–250 (2011).
Mehrtash, A. et al. Evaluation of mediastinal lymph node segmentation of heterogeneous CT data with full and weak supervision. Comput. Med. Imaging Graph. 111, 102312 (2024).
Rinneburger, M. et al. Laqua FCJERE: Automated localization and segmentation of cervical lymph nodes on contrast-enhanced CT using a 3D foveal fully convolutional neural network. Eur. Radiol. Exp. 7(1), 45 (2023).
Zhao, X. et al. Deep learning–based fully automated detection and segmentation of lymph nodes on multiparametric-mri for rectal cancer: A multicentre study. EBioMedicine 56, 102780 (2020).
Cao, Y. et al. Mao LJLrm: LNAS: A clinically applicable deep-learning system for mediastinal enlarged lymph nodes segmentation and station mapping without regard to the pathogenesis using unenhanced CT images. Radiol. Med. 129(2), 229–238 (2024).
Xu, X. et al. Deep learning assisted contrast-enhanced CT-based diagnosis of cervical lymph node metastasis of oral cancer: A retrospective study of 1466 cases. Eur. Radiol. 33(6), 4303–4312 (2023).
Taku, N. et al. Auto-detection and segmentation of involved lymph nodes in HPV-associated oropharyngeal cancer using a convolutional deep learning neural network. Clin. Transl. Radiat. Oncol. 36, 47–55 (2022).
Dornheim, J., Seim, H., Preim, B., Hertel, I. & Strauss, G. Segmentation of neck lymph nodes in CT datasets with stable 3D mass-spring models. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2006: 9th International Conference, Copenhagen, Denmark, October 1–6, 2006 Proceedings, Part II 9: 2006: Springer; 2006: 904–911.
Sjöberg, C. et al. Clinical evaluation of multi-atlas based segmentation of lymph node regions in head and neck and prostate cancer patients. Radiat. Oncol. 8, 1–7 (2013).
Zhang, J. et al. Level IIb CTV delineation based on cervical fascia anatomy in nasopharyngeal cancer. Radiother. Oncol. 115(1), 46–49 (2015).
Zhao, Y. et al. Level Ib CTV delineation in nasopharyngeal carcinoma based on lymph node distribution and topographic anatomy. Radiother. Oncol. 172, 10–17 (2022).
Zhong, Z. et al. Lymph drainage and cervical fascia anatomy-oriented differential nodal CTV delineation at the supraclavicular region for esophageal cancer and nasopharyngeal cancer. Radiother. Oncol. 177, 113–120 (2022).
Nakajima, Y. et al. Evaluation of the effect of user-guided deformable image registration of thoracic images on registration accuracy among users. Med. Dosim. 45(3), 206–212 (2020).
Pukala, J. et al. Meeks SLJJoacmp: Benchmarking of five commercial deformable image registration algorithms for head and neck patients. Appl. Clin. Med. Phys. 17(3), 25–40 (2016).
Huang, S.-C., Pareek, A., Jensen, M., Lungren, M. P. & Yeung, S. Chaudhari ASJNDM: Self-supervised learning for medical image classification: A systematic review and implementation guidelines. NPJ. Digit. Med. 6(1), 74 (2023).
Azizi, S. et al. Strachan PJNBE: Robust and data-efficient generalization of self-supervised machine learning for diagnostic imaging. Nat. Biomed. Eng. 7(6), 756–779 (2023).
Wasserthal, J. et al. Yang SJRAI: Totalsegmentator: Robust segmentation of 104 anatomic structures in CT images. Radiol. Artif. Intell. https://doi.org/10.1148/ryai.230024 (2023).
Luo, X., Fu, J., Zhong, Y., Liu, S., Han, B., Astaraki, M., Bendazzoli, S., Toma-Dasu, I., Ye, Y. & Chen, Z. J. Segrap2023: A benchmark of organs-at-risk and gross tumor volume segmentation for radiotherapy planning of nasopharyngeal carcinoma (2023).
Isensee, F., Jaeger, P. F., Kohl, S. A. & Petersen, J. Maier-Hein KHJNm: nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021).
Stapleford, L. J. et al. Fox TJIJoROBP: Evaluation of automatic atlas-based lymph node segmentation for head-and-neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 77(3), 959–966 (2010).
Sherer, M. V. et al. Oncology: Metrics to evaluate the performance of auto-segmentation for radiation treatment planning: A critical review. Radiother. Oncol. 160, 185–191 (2021).
Zhao, Q. et al. Wang GJIToMI: Efficient multi-organ segmentation from 3D abdominal CT images with lightweight network and knowledge distillation. IEEE Trans. Med. Imaging 42(9), 2513–2523 (2023).
Zhang, L. et al. Xu DJItomi: Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE Trans. Med. Imaging 39(7), 2531–2540 (2020).
Cardenas, C. E. et al. Generating high-quality lymph node clinical target volumes for head and neck cancer radiation therapy using a fully automated deep learning-based approach. Int. J. Radiat. Oncol. Biol. Phys. 109(3), 801–812 (2021).
Funding
This work was supported by the National Natural Science Foundation of China under Grant 82203197, and the Sichuan Provincial Cadre Health Research Project (Grant/award number: 2023-803).
Author information
Authors and Affiliations
Contributions
Wenjun Liao: Writing-original draft, review and editing, Data curation, Formal analysis, Funding acquisition; Xiangde Luo: Writing-review and editing, Formal analysis, Methodology, Software, Visualization; Lu Li: Data curation, Resources, and Validation; Jinfeng Xu: Data curation, Resources, and Validation; Yuan He: Data curation and Resources; Hui Huang: Data curation and Resources. Shichuan Zhang: Conceptualization, Data curation, Writing-review and editing, Project administration, and Supervision.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests.
Informed consent
Due to the retrospective nature of the study, the Ethics Committee for Medical Research of Sichuan Cancer Hospital waived the need of obtaining informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liao, W., Luo, X., Li, L. et al. Automatic cervical lymph nodes detection and segmentation in heterogeneous computed tomography images using deep transfer learning. Sci Rep 15, 4250 (2025). https://doi.org/10.1038/s41598-024-84804-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-84804-3