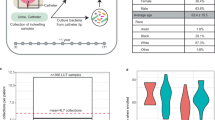
Abstract
Most hospital-acquired urinary tract infections are the result of implanted urinary catheter, with majority of studies focused on a single species colonisation, but recently polymicrobial colonisations are being reported. In this study, indwelling urinary catheters were collected from ICU patients and the colonising microbiome was isolated and identified by the traditional; culturing method and metagenomics. It was observed that majority of catheters were colonised by polymicrobial biofilms, containing both bacterial and fungal isolates making them diverse and complex. However, the metagenomics results were quite surprising showing the presence of multiple organisms of which only 1or 2 showed growth when cultured. Later, in vitro assays were performed by selecting 6 combinations, with each combination containing one Candida spp. – C. albicans or C. tropicalis with one bacteria K. pneumoniae, P. aeruginosa or E. coli. It was observed that polymicrobial biofilms were stronger than mono-microbial biofilms, suggesting their increased surface adhesion. Furthermore, to simulate the dynamic environment in which cells are exposed to a certain level of fluid movement, a flow system was established to imitate the flow generated in colonized urinary catheter. We have observed changes in biofilm architecture, adhesion and thickness under flow conditions compared with static conditions, with a uniformly adhered biofilm with increased thickness of polymicrobial biofilms as compared to mono-species biofilms. The biofilm formed under flow was more viable than the static biofilm with higher number of live cells in flow condition.
Similar content being viewed by others
Introduction
Healthcare associated infections (HAI) are one of the leading causes of morbidity and mortality worldwide, with catheter associated urinary tract infections (CAUTI) being the most common type1. An indwelling urinary catheter is usually used in hospitalised patients for various purposes which leads to increased instances of CAUTIs. Approximately 12–16% of adult hospital inpatients have an indwelling urinary catheter at some time during their hospitalisation2. While most patients with short-term indwelling catheters do not acquire microbial colonisation in their urine, but will eventually demonstrate bacteria in their urine if catheterization continues for a prolonged duration. Each day that an indwelling catheter is maintained, the risk of bacterial colonisation increases by 5–10%3. Common bacterial and fungal colonisers of urethral catheters include Escherichia coli, Enterococcus spp., and Candida spp.4. Despite abiding by all the recommendations, patients develop CAUTI; most being females. CAUTIs constitute about 4–50% of the HAIs in the United States, where approximately five million patients are put on catheters annually5. The main risk factors for CAUTI include patients with comorbidities (diabetes mellitus and renal failure, serum creatinine > 2 mg/dL, at catheterization); females over 50 years of age; cases of catheter insertion after the 6th day of hospitalisation; increased colonisation of the perineum; catheter insertion outside the operating room6. The fundamental starting point of CAUTI is the development of a contagious or infective biofilm on the surface of the indwelling urinary catheter.
Biofilm formation is a key characteristic of these organisms, enhancing resistance against environmental stressors. Biofilm formation is crucial in the development of urinary tract infections(UTIs), as they play a central role in CAUTIs. The interactions among microbes are intricate, involving competition for both space and nutrients. The physiology and function of biofilm communities often undergo changes, regulated by various interactions between different species. Microbial species are structured into different spatial forms based on their types, including interspecific segregation, coaggregation, and stratification. Chronic infections related to biofilms often involve multiple microbial species, leading to increased genetic material exchange, metabolic cooperation, antibiotic resistance development, niche optimization, modulation of the host immune system, and virulence induction7. One major issue with polymicrobial infections is their heightened virulence and increased resistance to antimicrobial agents, which can exceed that of mono-species biofilms. This resistance is attributed to the diverse array of extracellular polymeric substances (EPSs) produced by heterogeneously distributed bacteria, which impede the penetration of drugs8.
Polymicrobial bacteraemia UTI is also associated with an increased mortality rate compared to monomicrobial bacteraemia UTI. Cases of disseminated polymicrobial infection occur most often in compromised persons with underlying risk factors for UTI and frequently include nontraditional microbes or opportunistic pathogens9. The true prevalences of polymicrobial asymptomatic bacteriuria, UTI and CAUTI are therefore likely underreported, and the potential contribution of the urinary microbiota to lower urinary tract symptoms and risk of infection is also just beginning to be explored.
This study was initially started with the curiosity to observe the type of colonisation on urinary catheters in hospitalised patients, which lead to the discovery of polymicrobial colonisation, containing bacteria and Candida being most prominent colonisers on urinary catheters. Upon the culturable method of species identification, whole metagenomics sequencing was also used to identify the non-culturable population in the catheter biofilm microbiome. Hence, this study was then narrowed down to understand the type of biofilm formed in vitro using the collected colonisers, with all forming strongly adherent biofilms. In present study, a flow system was also established using ibidi µ-slide VI0.4 to mimic the flow occurring in a colonised urinary catheter. Candida tropicalis, that is also ranked as a high-priority pathogen as per WHO fungal priority list10, is used in the mixed biofilm model as it is an under-studied pathogen in terms of biofilms in general. The study highlights various morphological changes in the biofilm architecture and thickness amongst the individual biofilms and various combinations used.
Materials and methods
Collection, isolation and identification of culture microbes from indwelling urinary catheters
Indwelling urinary catheters (n = 28) were collected from the intensive care unit (ICU) patients (with more than 2 days of catheter insertion), upon removal in a sterile container from Bhailal Amin General Hospital, Vadodara, Gujarat. A 5 cm piece11,12,13 was cut from the catheter tip and was submerged in 5 ml 0.85% saline, followed by vortexing for 5 min for biofilm extraction. Saline containing resuspended biofilm was serially diluted and each dilution was spread on Luria agar (LA) plates in triplicates and incubated at 37 °C for 24–48 h. Colonies were distinguished as per physical appearance and each colony was maintained as pure culture, followed by storing for further use. Once the pure culture was obtained, standard Gram staining was performed and the isolates were distinguished as gram-negative/positive bacteria or fungi.
As per putative identification from colony characteristics and Gram staining, isolates were processed for rapid DNA extraction followed by PCR for identification (Fig. 1).
Rapid DNA extraction for putative bacterial isolates was done using 1% Triton X100. Briefly, in a sterile microfuge tube 100 µl 1% Triton X100 a few isolated colonies were added and vortexed for 30 s, and was later incubated in a boiling waterbath for 10 min. The mixture was then centrifuged at 7500 rpm for 5 min and the resulting supernatant was used for PCR.
Rapid DNA extraction for putative fungal isolates was done as per Looke et al.14, with a few modifications. Briefly, pellet from an overnight culture or pure yeast colonies were used and to it 100 µl of 0.2 M LiOAc − 1% SDS was added and homogenised. It was then incubated at 70 °C for 5 min. To the supernatant, 300 µl absolute ethanol was added and was centrifuged at 10,000 rpm for 3 min. Later, the pellet was washed with 70% ethanol and the pellet was resuspended in nuclease-free water.
PCR for bacterial isolates was done using the primers 27F (5’ AGAGTTTGATCMTGGCTCAG 3’, M = A or C) and 1525R (5’ AAGGAGGTGWTCCARCC 3’, W = A or T) and for fungal isolates ITS1 (5’ TCCGTAGGTGAACCTGCGG 3’) and ITS4 (5’ TCCTCCGCTTATTGATATGC 3’). The resulting PCR product was then verified on agarose gel, followed by Sanger sequencing. The raw reads from sequencing were analysed using ChromasPro software and species identification was done using NCBI BLAST. The identification was confirmed only with 100% query coverage and greater than 98% percentage identity.
Metagenomics workflow
Metagenomics sequencing
Whole genome metagenomics was done for two catheter samples, BU13 and BU23. After plating the samples, the remaining saline was spun-down to obtain the total cell pellets, which was the stored at -20 °C until metagenomic sequencing. DNA extraction, library preparation and sequencing using Illumina NovaSeq6000 (2 × 150 bp chemistry) was done at Eurofins Genomics India Pvt. Ltd. (Bengaluru, India). Upon receipt of the total pellet, total DNA was extracted from pellet was extracted using DNA extraction kit from NucleoSpin DNA extraction kit. The quality of extracted DNA was quantified using NanoDrop - A260/A280 ratio and concentration, followed by agarose gel electrophoresis to verify the integrity and QC passed DNA samples were proceeded for library preparation. Pair-ended library preparation was done using Illumina TruSeq Nano DNA library preparation kit and quality assessment of prepared library was done using Agilent 4200 Tape Station. The QC qualified library was then proceeded for sequencing using Illumina NovaSeq6000 platform.
Data analysis
Upon sequencing, bioinformatic analysis was proceeded using Galaxy Europe (https://usegalaxy.eu/). The quality of raw reads was accessed using FastQC (Galaxy Version 0.74 + galaxy1) and trimming was carried out using Trimmomatic (Galaxy Version 0.39 + galaxy2). Upon obtaining high quality pair-ended reads, de novo assembly was prepared using metaSPAdes (Galaxy Version 3.15.5 + galaxy2), using the default parameters. The assembly was then used for taxonomical characterisation using Kaiju (kaiju.binf.ku.dk), accessed in December 202315.
Quantification of biofilms
Biofilm formation was quantified and isolates were categorised using crystal violet assay for all isolates (n = 41), followed by selection of 5 isolates - C. albicans (Ca)- U23.2, C. tropicalis (Ct)- U06.1 and 3 bacteria (K. pneumoniae (Kp) – U02.2, P. aeruginosa (Pa) – U02.3, E. coli (Ec) – U17.2 for further study. Further crystal violet assay was performed for the combinations where, each combination contained one Candida spp. each with one bacteria, hence 6 combinations in total- Ca + Kp, Ca + Pa, Ca + Ec, Ct + Kp, Ct + Pa, Ct + Ec. Crystal violet assay was performed as per the method by Stepanovic et al.16 with a few modifications. The assay was performed in triplicates with sterile BHI media as the negative control. Briefly, 25 µl of fresh culture with an OD600 nm − 0.1 for bacterial isolates and OD600 nm − 0.2 for Candida spp. for individual isolates and for combinations, a 1:1 (v/v) mixture of bacterial and Candidal culture with respective OD600nm was prepared and added to 225 µl sterile BHI media and for and inoculated in sterile 96-well flat bottom microtiter plate (Laxbro Bio-Medical Aids Pvt. Ltd., Pune, India) and was incubated at 37 °C for 24 h. After 24 h, the non-adherent cells were washed twice with sterile 0.85% saline and the adhered cells were fixed with 250 µl absolute methanol for 15 min. Later, the plates were allowed to air dry to remove residual methanol and then the adhered biofilm was stained with 250 µl of 0.1% crystal violet for 15 min. Excess stain was removed by washing the wells twice with 0.85% saline and bound stain was resolubilized with 250 µl of 33% acetic acid and its OD was measured at 590 nm using a microtiter plate reader (Multiskan Go, ThermoFisher Scientific, Waltham, MA, USA). The isolates were categorised into 3 categories- strong, moderate or weak biofilm producers based on the cut-off OD (ODc)16. ODc is defined as three standard deviations above the mean OD590nm of negative control. The categorisation was done as follows: OD ≤ ODc - no biofilm producer, ODc < OD ≤ 2ODc - weak biofilm producer, 2ODc < OD ≤ 4ODc - moderate biofilm producer, 4ODc < OD - strong biofilm producer. The statistical analysis and graphs were plotted using GraphPad Prism 10.
Comparison of biofilm formed under static and flow condition
Biofilm formation was visualised and quantified under static and flow condition. The set-up for static and flow condition has been mentioned below:
Static set-up
Sterile glass cover-slips (22 mm in diameter) were placed in each well of a sterile 6-well plate (Laxbro Bio-Medical Aids Pvt. Ltd., Pune, India). To each well containing 1800 µl sterile BHI media, 200 µl of fresh culture with OD600 nm − 0.1 for bacterial isolates and OD600 nm − 0.2 for Candida spp. for individual isolates and for combinations, a 1:1 (v/v) mixture of bacterial and Candidal culture with respective OD600nm was added and incubated for 72 h at 37 °C. Each plate contained a negative control well containing a sterile coverslip and 2 ml BHI media. After the incubation, the coverslip was picked from the well and was rinsed with sterile 0.85% saline to remove non-adherent biofilm, the coverslip was then proceeded with staining.
Flow set-up
Adhesion assay
Before starting the flow, it was essential to verify microbial attachment to ibidi µ-slide VI0.4, hence adhesion assay was performed for all samples – 2 Candida spp., 3 bacteria and 6 combinations. Briefly, 50 µl of fresh cultures with an OD600 nm − 0.1 for bacterial isolates and OD600 nm − 0.2 for Candida spp. for individual isolates and for combinations, a 1:1 (v/v) mixture of bacterial and Candidal culture with respective OD600nm, were seeded into the slide and were allowed to adhere for 4 h at room temperature. After that, unbound cells were washed with sterile 0.85% saline and cells adhered were subjected to staining by safranine. Brightfield microscopy (Olympus CX43, Tokyo, Japan) was used to observe those adhered cells and conclusions were made.
The flow system consisted of a slide, for biofilm formation- ibidi µ-slide VI0.4 (ibidi, Gräfelfing, Germany) containing 6 channels, each channel with a length of 17 mm, width of 3.8 mm, height of 0.4 mm and volume capacity of 30 µl. Each channel had 2 luer adapters that would assist in connections to inlet and outlet flasks, a pumping system – volumetric infusion pump (Nareena Life sciences, Delhi, India), sterile intravenous infusion sets (IV set, ATPL nano infusion set, Gujarat, India) for the unidirectional flow of sterile BHI media from the inlet flask through the slide to the outlet flask.
For carrying out the flow experiment, initially the cells were allowed to adhere for 4 h, after which each channel was washed with sterile 0.85% saline to remove the non-adherent cells. The system was set as mentioned above in sterile condition in a biosafety cabinet and continuous flow with a flow rate of 200 µl/min was maintained for 72 h. Figure 2 shows the flow system established in the biosafety cabinet. Additionally, supplementary Fig. 1 shows a closer look of the IV set passing through the volumetric infusion pump and maintaining the required flow rate (a), connection of the IV sets to the slide (b) and all IV sets connected to the slide (c).
After 72 h, the slide was removed from the system and the channels were washed with 0.85% saline to remove non-adherent biofilm, followed by staining of the slide.
Flow system: The flow system was set up in a biosafety cabinet to maintain sterility of the experiment. The actual experimental system containing the inlet flasks containing sterile BHI media, the IV sets used for flowing media to the channel of the ibidi µ-slide VI 0.4, and the outlet flasks to collect the out flown media.
Confocal laser scanning microscopy of the biofilms
Live-dead assay was performed to analyse the biofilm formed under static and flow conditions.
LIVE/DEAD® Viability Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to carry-out live dead assay. The microbial cells in mono- and mixed-species biofilm were stained with SYTO9 and propidium iodide (PI) as per the manufacturer’s instructions. Excess stain was rinsed with 0.85% saline and the coverslip was mounted for confocal laser scanning microscopy (CLSM) and the slide was directly used. Carl Zeiss CLSM 780 microscope was used for visualising the biofilms. The following parameters were used for imaging: SYTO9- green fluorescence (488 nm excitation, emission spectra 492–525 22 nm) and PI- red fluorescence (561 nm excitation, emission spectra 563–652 nm). To visualise the 3D structure Z- stack mode of CLSM was used. Cell count (using cell count plug-in) and biofilm thickness (as described by Biswas et al.17) were analysed for each isolate and combination in static and flow condition using ImageJ software.
Results
Sample collection and characterisation of colonised microflora using culture-based method
For present study, a total of 28 urinary catheters were collected from ICU patients upon their removal by hospital staff as per the physicians’ advice. The days of catheterisation varied from 3 to 16 days as per the patient’s records. The criteria for catheter collection were not predefined, and we were only provided with age and gender of the patients along with days of catheterisation, in order to ensure patient privacy. (details mentioned in Supplementary Table 1). All the catheters were silicone-coated latex catheters, pointing towards its higher usage as compared to latex and silicone catheters in hospital set-ups. Among the 28 catheters, 14 catheters showed polymicrobial colonisations, whereas 10 showed monomicrobial colonisation, it was observed that these catheters had catheterisation ranging from 5 to 16 days (Fig. 3). Only 4 catheters did not show any microbial colonisation and it was noted that all of them were inserted in the patient only for 3 days.
Among the polymicrobial colonisations, 10 catheters showed the presence of mixed-species biofilms indicating the presence of one/two bacterial species along with a yeast in a mixed biofilm consortium. Among the catheters with polybacterial colonisation, 2 catheters (BU17 and BU26) were colonised with 3 bacterial species. Along with polymicrobial colonisation, 10 catheters were observed to have monomicrobial colonisation, of which 5 catheters had bacterial and 5 had fungal colonisation.
Summary of colonisation on urinary catheters: A total of 28 catheters were collected from which 14 catheters had polymicrobial colonisation, 10 had monomicrobial colonisation whereas 4 catheters did not show any microbial colonisation. Among polymicrobial colonisation, mixed species biofilm was found to be the most commonly encountered.
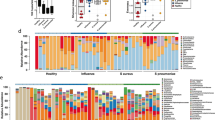
Upon successful isolation from all catheters, a total of 41 isolates were obtained. Among the collected isolates, genus Candida was most common with the presence of various species including C. albicans, C. kefyr, C. tropicalis, C. parapsilosis and C. auris. Among the Candida spp., C. albicans was most prevalent (n = 5), and it was obtained in mixed species biofilms along with various bacterial species like E. coli, P. aeruginosa, K. pneumoniae and with Trichosporon asahii in a polyfungal biofilm (Fig. 4). Among the bacterial species, the prevalence of Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae and Enterococcus faecium were most prevalent and were obtained in different types of biofilms. Details regarding organisms colonising the catheters and the type of biofilm obtained is mentioned in Supplementary Table 1. It was also observed that P. aeruginosa was one of the commonly encountered strain in a polymicrobial colonisation containing three different organisms in both mixed species biofilm and polybacterial biofilms. It was observed that all these organisms were majorly present in a mixed species biofilm rather than a mono-species biofilm, which shows the complexity of colonisation on urinary catheters. Apart from these, other fungal and bacterial isolates like Candida kefyr, Staphylococcus haemolyticus, Trichosporon asahii, Candida auris, Candida parapsilosis, Stenotrophomonas maltophila, Providencia vermicola and others were also present but the frequency of occurance was low.
The data concludes a higher prevalence of polymicrobial colonisation on urinary cathers with maximum instances of mixed species biofilms of Candida with bacteria.
Disribution of bacterial and fungal isolates from urinary catheters: The most common fungal isolate obtained were C. albicans(n = 5), and C. tropicalis which were majorly present in mixed species biofilm. The most common bacterial isolates were E. coli, E. faecium, K. pneumoniae and P. aeruginosa which again were more prominently present in mixed species biofilms. The inner pie shows the names and number of microbial species, whereas the outer pie indicates the type and number of times it which it was obtained.
Metagenomics analysis
Metagenomic sequencing was done to analyse the population of unculturable microbes inhabiting the urinary catheter. It was performed for 2 catheter samples, BU13 and BU23. When colonisation on catheters BU13 and BU23 were cultured, 2 distinct microbial colonies were obtained in case of each catheter. Upon culture-based identification, Staphylococcus haemolyticus and Candida parapsilosis were obtained from BU13 and Klebsiella pneumoniae and Candida albicans were obtained from BU23, however various species were obtained upon metagenomics.
Metagenomic analysis of colonisation on BU13 showed that ‘Bacterial Kingdom’ was the most abundant (78.38%) kingdom followed by Eukaryota, 19.87% (Fig. 5a). As per metagenomics among the identified species, C. parapsilosis was the most abundant organism which was also obtained upon culturing. However, the second most abundant organism as per metagenomics was found to be Acinetobacter baumannii followed by E. coli, B. thuringiensis, Staphylococcus haemolyticus and P. viridiflava (Fig. 5b). Similar results were obtained in case of catheter BU23, with the of Bacterial kingdom being most prevalent with 75.76% abundance, followed by Eukaryota with 23.19% abundance (Fig. 5c). Among the identified isolates C. albicans was the most prevalent organism followed by Acinetobacter baumannii, E. coli, B. thuringiensis, P. viridiflava and K. pneumoniae (Fig. 5d). An interesting observation was made, even though both the catheters were isolated from different patients, the abundant species in both the catheters are quite common with A. baumannii being the second abundant organism, which did not show and microbial growth in either of the case. The most abundant organisms in both the catheters were Candida spp., which showed microbial growth when cultured. Among the identified species using metagenomics, we found a few organisms that were more abundant (green highlight in Fig. 5b and d) than S. haemolyticus and K. pneumoniae (yellow highlight in Fig. 5b and d) on catheters BU13 and BU23 respectively, but they did not show any microbial growth upon culturing. An interactive Krona chart showing the completely identified microbiome using metagenomics was prepared and is represented in supplementary files 2 and 3 for BU13 and BU23 respectively.
Biofilm quantification and categorization
In case of all 41 isolates, majority of isolates formed strong biofilms (n = 24) indicating higher adhesion (Fig. 6a). A few isolates formed moderate biofilms (n = 10) and weak biofilms (n = 7) as well. It was observed that all Candida strains formed strong biofilms except for two C. parapsilosis strains. Among the bacterial species, strains of Enterococcus faecium either form a moderate or weak biofilm where as majority of K. pneumoniae, E. coli and P. aeruginosa isolates formed strong biofilms. Hence, the future study was carried out on C. albicans- Ca, C. tropicalis- Ct, K. pneumoniae- Kp, P. aeruginosa- Pa and E. coli- Ec, all of them forming strong biofilms.
When the individual isolates were compared with the combinations, it was observed that Ct formed the strongest biofilms of all and as compared to individual isolates (Fig. 6b), combinations formed a significantly stronger biofilm. In case of combinations of Ca and Ct, with Kp and Ec, the combinations, Ct + Kp and Ct + Ec formed significantly stronger biofilm than the combination of Ca with Kp and Ec. However, there was no significant difference between the biofilms formed by Ca + Pa and Ct + Pa.
Quantification and categorisation of biofilms: (a) Categorisation of biofilm formed by all collected samples (n = 41) where majority of samples form strong biofilm, (b) Quantification of biofilm formed by selected isolates (n = 5) and combinations (n = 6), the combinations form significantly stronger and adherent biofilm as compared to individual isolates and combinations containing Ct form stronger biofilm as compared to combinations containing Ca, except the combinations containing Pa as the partner. The statistical analysis and graphs were prepared using GraphPad Prism 10, where * - p < 0.05; ** - p < 0.01, *** - p < 0.001, **** - p < 0.0001, and ns indicates p > 0.05.
Adhesion assay
To study the adhesion capacity onto the ibidi µ-slide VI 0.4 slide, the isolates were subjected to cell adhesion assay followed by light microscopy to verify cell adhesion before starting the flow. When present individually, both Ca and Ct uniformly adhered to the slide’s surface (Fig. 7a, b) and initialisation of hyphal formation was seen in Ca. In case of individually adhered bacterial species, Kp was seen to adhere uniformly on the surface (Fig. 7c) whereas compared to Kp and Ec, fewer Pa cells were attached, the adhesion was seen on different focal planes and a few microcolonies were observed (Fig. 7d). Similar to Pa, the adhesion of Ec was observed on different focal planes, but increased microcolony formation was observed (Fig. 7e) upon adhesion of 4 h.
Adhesion assay for individual isolates: (a) uniform attachment to the surface was observed in Ca along with initialisation of hyphal formation whereas (b) in Ct, a uniform attachment was seen; (c) Kp was uniformly attached to the surface whereas (d) fewer adherent cells were observed in case of Pa along with a few microcolony formation; (e) higher extent of cell adhesion along with many microcolony formation was seen in Ec. The microscopy was done at 40x magnification and the scale is 22 μm.
Upon the adhesion of isolates in combination, distinct results were observed as compared to their individual adhesion. In the mixed biofilm of Ca + Kp, microcolony formation is observed on different focal planes and Kp have started aggregating around yeast cells (Fig. 8a). In case of Ca + Pa, both the isolates were seen to independently adhere in different focal planes (Fig. 8b). Similar to Ca + Kp, microcolony formation is seen in case of Ca + Ec in different focal planes (Fig. 8c). On contrary to the combination Ca + Kp, in Ct + Kp both the isolates are independently adhered on a similar focal plane with no microcolony formation and bacterial aggregation near the yeast cells (Fig. 8d). A similarity in adhesion pattern was seen in case of Ct + Pa with Ca + Pa, where both the isolates have adhered independently in different focal planes (Fig. 8e). Some differences were observed in Ct + Ec as compared to Ca + Ec, where adhered cells have reduced in number and no microcolony formation, but the adhesion is in different focal planes (Fig. 8f).
Adhesion assay for combinations: (a) In combination, Ca + Kp, cell adhesion is seen in different focal planes with microcolony formation and bacterial cell aggregation around yeast cells, whereas, in Ct + Kp (d) independent adhesion of both the isolates is seen. A similarity in adhesion pattern is seen in Ca + Pa (b) and Ct + Pa (e), with both the isolates adhering independently to the surface. In the combination, Ca + Ec (c), higher cell adhesion is seen with microcolony formation in different planes, whereas in Ct + Ec (f), the number of adhered cells is low and no microcolony formation observed. The microscopy was done at 40x magnification and the scale is 22 μm.
Comparison of biofilm formed under static and flow condition
To visualise and characterize the biofilm architecture, confocal laser scanning microscopy (CLSM) was performed for all the isolates individually as well as in combination where, each Candida spp. was studied with respective bacteria. Live and dead cells embedded in the biofilm matrix to evaluate the cell death along with variation in biofilm thickness was quantified. Distinct differences in the biofilm structure and thickness were observed among biofilms formed in static and in flow conditions.
In case of Ca, under static condition (Fig. 9a) only a few yeast cells were present and the population was higher as compared to live ones, whereas under the flow condition (Fig. 9b) denser cell population was observed and the present cells were live in the biofilm. The 72-hour biofilm of Ct appeared a little different than Ca. In Ct static biofilm, scattered population of yeast cells is seen (Fig. 9d) whereas in flow condition densely packed live cells embedded in the biofilm matrix (Fig. 9e). Biofilm formed by Kp, was observed to be densely packed in the matrix in both the conditions, however a mixed population of live and dead cells was observed in static condition (Fig. 9g), whereas only live cell population was observed in flow condition (Fig. 9h). Pa was observed to have high exopolymeric substratum that would bind the cells in biofilm, hence even in case of lesser number of cells biofilm appeared to be strong and adherent. Surprisingly, under static condition a high population of adherent cells were seen but the cells were observed to be dead (Fig. 9j), whereas in flow condition, the population was found to be live but very few bacterial cells were embedded in the biofilm matrix (Fig. 9k). Ec had strongly adhered biofilm in both conditions, but similar to Kp, Ec biofilm also had a mixed cell population in static condition (Fig. 9m) whereas majority of live cells were observed in flow condition (Fig. 9n) and interestingly the Ec cells in the biofilm under flow were observed to have an increase in their size. The graphical representation of cell count has been shown for each isolate in Fig. 9c, f, i, l, o, and the graphs were created using GraphPad Prism10.
Biofilm for individual isolates under static and flow condition: The biofilm of Ca and Ct were strongly adhered in case of flow condition (b, e respectively), whereas the static biofilms were scarcely adhered with a mixed population of live and dead cells (a, d respectively). In case of Kp and Ec, a mixed population of live and dead cells was observed in static condition (g,m respectively) whereas a densely packed biofilm with majority of live cells was observed in floe condition (h, n respectively). Extremely high cell death was observed in Pa biofilm under static condition (j) whereas very few live cells embedded in exopolymeric substrate was observed in flow condition (k). The graphical representation of cell count has been shown for each isolate in (c,f,i,l,o). The scale on the image is 22 μm.
In mixed biofilm of Ca + Kp under static condition (Fig. 10a), a densely adhered biofilm was observed with majority of dead Ca cells and only a few live Kp cells, with a higher population of dead Kp cells. However, under the flow condition (Fig. 10b), an adherent biofilm embedded in biofilm matrix was observed with live cells, with reduction in cell density and cell number- specifically Kp, as compared to static. An interesting observation was observed in Ca + Pa biofilm, under static condition Ca cells were found to be absent and all Pa cells were dead (Fig. 10d), whereas biofilm under flow condition had a few Ca cells and a mixed live and dead population of Pa cells (Fig. 10e). In Ca + Ec biofilm under static condition, a mixed live and dead population of both Ca and Pa cells is seen (Fig. 10g) whereas interestingly under flow condition, high population of live Ca cells was observed with only a few Ec cells (Fig. 10h). The graphical representation of cell count has been shown for each isolate in Fig. 10c, f, i, and the graphs were created using GraphPad Prism10.
Biofilm of Ca in combination with bacterial species under static and flow condition: The biofilm formed by Ca + Kp under static condition had a few dead Ca cells whereas a mixed population of live and dead Kp cells were seen (a), on the contrary, under flow condition, the cell number was lesser than static, but a few live Ca and Kp cells were seen (b). In case of static biofilm of Ca + Pa, Ca cells were found to be absent and the population of Pa was dead (d), in flow condition, a mixed population of live and dead Ca and Pa cells were seen (e). In mixed biofilm of Ca + Ec, under static condition, a mixed population of live and dead Ca and Ec cells were seen (g) whereas under the flow condition few Ec cells were seen (h). The graphical representation of cell count has been shown for each isolate in (c,f,i). The scale on the image is 22 μm.
In mixed biofilms of Ct + Kp, the population of Kp has reduced drastically in presence of Ct with only a few Kp cells seen forming small clusters or were present on the surface on Ct cells (Fig. 11a), a mixed live and dead population of Ct was observed. Similar to the static condition, a reduction in population of Kp cells was seen with only a few dead Kp cells aggregated near the corners (Fig. 11b), a reduction in cell number of Ct was also observed but majority of population was live. In case of Ct + Pa, a mixed live and dead population of both Ct and Pa was seen with many Pa cells clustering the Ct population (Fig. 11d), on the contrary in case of flow condition a drastic reduction in cell population was seen with more live cells and a uniformly spread biofilm (Fig. 11e). Interestingly in mixed biofilm of Ct + Ec, Ec cells were found to be absent with a highly dense dead population of Ct in both static and flow condition, the only difference being higher cell death of Ct cells in static (Fig. 11g) as compared to flow (Fig. 11h). The graphical representation of cell count has been shown for each isolate in Fig. 11c, f, i, and the graphs were created using GraphPad Prism10.
Biofilm of Ct in combination with bacterial species under static and flow condition: The population of Kp and Ec was found to be absent in presence of Ct under both static (a, g respectively) and flow (b, h respectively) condition. In case of Ct + Pa, a mixed population of live and dead Ct and Pa cells was seen clustered around Ct cells in static (d) whereas a uniformly spread biofilm was observed inn flow condition (h). The graphical representation of cell count has been shown for each isolate in c, f, i. The scale on the image is 22 μm.
Biofilm thickness
From the Z-stacks, along with live-dead cell assay, biofilm thickness was also calculated. It was observed that Pa biofilm under the flow condition had maximum thickness of all, 63 μm. Even though, a lot of variations were observed in the biofilm morphology and in cell number of both live and dead cells, a significant variation in thickness was observed only in a isolates and combinations. It was a common observation that in all isolates, the biofilm formed under flow condition was thicker as compared to the static condition (Fig. 12). A significant variation have been observed in case of biofilms by Pa, Ca + Kp, Ct + Pa and Ct + Ec.
Biofilm thickness: An increase in biofilm thickness under flow condition was observed for all isolates, but a statistical significance was observed only in Pa, Ca + Kp, Ct + Pa and Ct + Ec. The statistical analysis and graphs were prepared using GraphPad Prism 10, where * - p < 0.05; ** - p < 0.01, *** - p < 0.001, **** - p < 0.0001, and ns indicates p > 0.05.
Discussion
Catheters are the most commonly used indwelling medical device used in hospital set-ups specifically in ICUs, which also leads to their frequent colonisation leading to various other nosocomial infections, which can be proven life threatening. The primary aim of this study was to identify the microflora inhabiting the indwelling urinary catheters. There are multiple studies highlighting the occurrence of microbial colonisation on urinary catheters and urine. Various studies have used culture-based method for identification like, Abu-Shoura, 202418; Xu et al., 201211 have reported E. coli, K. pneumoniae, E. facecium and P. aeruginosa as the commonly obtained colonisers of the urinary catheter, which is similar to our findings in case of bacterial species. Xu et al., 201211 observed these colonisers would lead to development of CAUTI as they would ascend the bladder. They also obtained that the tip of the catheter had more microbial colonisation than urine. Sahai and Kumar, 201819 have reported the presence of non-albicans and C. albicans using the culture-based method, they concluded that about 23% of symptomatic CAUTI were caused by Candida spp., including C. albicans and non-albicans Candida. In present study we have obtained majority of urinary catheters that have polymicrobial colonisation (n = 14, Fig. 3), either mixed species (n = 10), containing colonisation of bacteria with Candida; or polybacterial (n = 3) or polyfungal (n = 1); which is relatively under reported than single species colonisation. We later studied polymicrobial colonisation based on this finding.
Zhang et al.., 201520, detected the presence of > 10 microbial phyla and > 136 diverse microbial genera, separated into 12,224 operational taxonomic units (OTUs) on intravascular catheters. We also found similar results wherein many bacteria were observed in the metagenome data but the culture-based method allowed growth of only few of them. However, upon availability of nutrients the state can be revived21. Further studies are warranted to show the importance and implications of such diverse genera found on catheters. An important concordance between the culture-based method and metagenomic analysis is the occurrence of most prevalent organism – Candida, as per metagenomics showed growth when cultured. Few studies report the presence of many bacterial genera on individual catheters22 while as per our knowledge it is the first report stating the co-occurrence of Candida- bacteria on urinary catheter using both culture-based and metagenomics. Also, in the present metagenomic analysis Acinetobacter baumannii was found to be second most abundant species in both catheter samples. A. baumannii is known to cause UTI with an ability to form biofilms23, and is also listed among the critical pathogens in WHO priority pathogen list 202424. The study by Nye et al.22, has shown the presence of Acinetobacter in urinary catheter as well as in urine. Further, though Acinetobacter was reported as a causative agent of CAUTIs since 1995 25 to the best of our knowledge recent reports on CAUTI’s due to Acinetobacter are scarce in metagenomic studies as well. A. baumannii has an impeccable ability to undergo the VBNC state as a survival strategy and hence it could not be cultured, in spite its abundance in metagenomics data. These results warrant future studies on the importance of A. baumannii in polymicrobial biofilm formation on catheters and the possibility of infection in presence of A. baumannii. To further validate the findings by metagenomics, more samples could be used, we have only used 2 samples as the other selected samples did not clear quality check after DNA extraction. We would like to put an important disclaimer that we were not informed if any of the patients, from whom catheters were collected had CAUTI or UTI. Hence, all organisms found using culture-based and non-culture-based identification are colonizers. It is evident that polymicrobial biofilms can be formed by colonizers and they need not always cause CAUTI. Here, we would like to mention a few limitations of this study; collection of more catheters along with patient data and treatment plan, could have helped in a more comprehensive study and would have would have help in corelating the colonisation with a certain disease/antibiotic. However, it would be interesting to study their behaviour in the biofilm as they may cause infection in due course. Polymicrobial bacteraemia UTIs are associated with increased mortality relative to bacteraemia UTIs caused by a single uropathogen26.
Next, we wanted to observe the effect of flow on mixed species biofilms. Flow cells with glass surfaces have been developed to address this need. These flow cell methods support the growth of mature biofilms while removing planktonic cells through continuous flow, providing a more realistic environment for biofilm studies. Hence in present study, ibidi slides were used due to the ease in biofilm visualisation under flow conditions27,28.
Adhesion-related studies have also been reported for monomicrobial as well as for polymicrobial species to elucidate adherence of the cells onto the surface of static and flow cell systems. Earlier reports show that a group had studied adhesion between polymicrobial interactions of C. albicans and Staphylococcus aureus on a static platform where they found the initial adhesion after 24 h was significantly stronger and firmer29. Another group performed adhesion to conclude that the polymicrobial biofilm had a complex structure, with C. albicans serving as a scaffold where S. aureus adheres, preferentially to the hyphal form of the fungus30. Study related to variation in adhesion of various Candidal species was evaluated and it was observed that non-albicans Candida formed stronger biofilms than C. albicans31. However, reports encompassing the combinations used in this study (Candida with Kp or Ec or Pa) are very scarce and dispersed. Regarding adhesion in flow systems or under flowing conditions, it is indispensable to understand the flow rate at which the fluid, in other words, urine will flow inside the urinary catheter. Different literature surveys displayed varied use of flow rates for their studies32,33,34,35,36, therefore for conducting our adhesion related studies, we have used the flow of 200 ul/min.
Negri and his group studied biofilm quantification and adhesion using an instrumentation mechanism almost similar to the one executed in our study37. They found out that the isolate adhered to the urinary catheters were significantly greater in flow as opposed to the static conditions and also displayed the capacity to colonize distal sites. In a study conducted by Klayman and his team, they found that bacterial cells notably E. coli and P. aeruginosa significantly increased their cell population when they were kept under flow conditions38. Nielsen and his group also showed the increased biofilm architecture between Pseudomonas aeruginosa and Saccharomyces cerevisiae using a similar flow setup used in this study39. Notably, apart from these, there are other findings with mono-microbial and polymicrobial species too that highlight the significance of using flow cell systems over static conditions.
In our study, all the mono species as well as the mixed species isolates present in flow conditions showed a remarkable increase in the overall live cell population as compared to the static conditions. The mixed species biofilms were observed on different planes and they displayed greater aggregation of cells showing enhanced biofilm architecture and adhesion. Noteworthy to say, increased live cell population led to more stronger biofilm adherence and a firm biofilm architecture. Additionally, biofilm thickness was also measured in static as well in flow that displayed the significant increase in the overall length of the produced polymicrobial biofilm under flow conditions as compared to the static condition.
Generally, studies regarding Candida biofilms have used static models. In a study, mature biofilms of C. tropicalis consisted of a dense network of yeast cells and filamentous forms40. In another study from C. tropicalis isolated from hospital patients, it was observed that the isolate adhered more to the epithelial lining rather than silicone following hyphae and pseudohyphae in serum expressing total hemolytic activity37. Although some studies used in vitro flow models to attempt to mimic Candida biofilm development in vivo either to assess Candida-bacteria polymicrobial interactions in drug response, analyse the biofilm architectures or even to understand the flow dynamics and shear stresses that the isolates undergo under flow conditions41,42,43. Similarly, the study of mono-bacterial or polybacterial combinations using in vitro static and flow models have also been reported. In a study, wild-type P. aeruginosa was grown and subjected to laminar and turbulent flows to investigate relative contributions of hydrodynamics and cell signalling for biofilm formations44. Other cases of how K. pneumoniae biofilms are formed and maintained demonstrating resistance to drugs have also been reported45. Vejborg and his team demonstrated the diversity E.coli can achieve when colonized inside urinary catheters under flow showing different structural phenotype, where they observed formation of chains46, which was not observed in our study, however we have observed cell elongation in case of individual E. coli biofilms. Though, information is scarce regarding the flow system and conditions used in this study.
The interaction between Candida and bacteria can influence the expression of specific virulence factors and may influence the outcome of co-infections47. Many of these co-infections are more pathogenic than single species infections48. There are many reports citing the presence of K. pneumoniae49, P. aeruginosa and E. coli50,51, C. albicans and C. tropicalis52,53,54,55 in mono as well as mixed species biofilms but the combinations used in this study are highly under reported. There are reports that demonstrate synergistic effects of polymicrobial biofilms hence strengthening of the overall biofilm architecture56,57,58,59 and on the other hand, antagonistic effects of certain bacteria and yeast species that lead to inhibition of one species and survival of the other47,60,61,62,63.
C. albicans producing moderate to strong biofilms and Klebsiella pneumoniae producing strong biofilms are well reported64,65. But mixed species biofilms of C. albicans and K. pneumoniae show very strong biofilm formation corroborating the idea that there are significant changes in adherence and biofilm formation66,67 that aligns with our finding as well. Previous reports show that there was a difference in OD between P. aeruginosa and the OD of P. aeruginosa with exposure to C. albicans, where there was an increase in OD which indicated a synergistic interaction68. C. albicans produces ethanol due to the induction by phenazine from P. aeruginosa and the ethanol which stimulates adhesion and biofilm formation of P. aeruginosa. There is a positive feedback mechanism where C. albicans ethanol production can increase 5-methyl-phenazine-1-carboxylic acid (5-MPCA) production in P. aeruginosa and increase biofilm formation. Then, 5-MPCA stimulates ethanol production in C. albicans69. Similar findings were also found in a study conducted in 2021 by Kasetty et al.., which showed that P. aeruginosa biofilms containing C. albicans had higher OD levels than P. aeruginosa biofilms alone70. It also bore similarities to Phuengmaung’s research from 2022, which discovered that prominent interkingdom biofilms with more severe infections were induced by Pseudomonas and Candida in the lungs of patients suffering from cystic fibrosis and ventilation-associated pneumonia (VAP)71. In this study as well, we found synergistic relationship between P. aeruginosa and C. albicans and it can be hypothesised that the synergy could be because of the aforementioned mechanisms. The biofilm produced were relatively much stronger, had greater adherence and were present on different planes. Again, in this study we found that in the presence of E. coli, there was an increase in the adhesion and attachment of candidal cells with E. coli being present on the surface of yeast. Previously, it has been documented that E. coli cells formed more biofilm when they were growing simultaneously with C. albicans72. Bandara et al.. reported that the secretory products from the E. coli biofilm modulate Candida biofilm formation73.
Another important finding of present study is the negative effect of C. tropicalis on K. pneumoniae and E coli. However, to the best of our knowledge this is the first study on biofilms formed in flow conditions containing the combinations of C. tropicalis with either of bacteria: K. pneumoniae or P. aeruginosa or E. coli. C. tropicalis is shown to form stronger biofilms than other species under study. We have observed decrease in population of K. pneumoniae and E coli cells in combination with C. tropicalis. Hence studies on individual and mixed biofilms of C. tropicalis in depth are recommended for future.
Some of the original experiments in this study were on studying the polymicrobial biofilm in flow. Some combinations (for e.g. Ct + Kp, Ct + Pa, Ct + Ec) are studied for the first time. Further, the selection of isolates and combinations was based on the frequently found mixed species together on catheters. This to our knowledge is extremely important considering the growing prevalence of uropathogens and CAUTI’s. We conclude that, C. albicans shows synergy by keeping bacteria alive while C. tropicalis shows antagonism by killing bacteria, particularly Kp and Ec.
The present study contributes to a better understanding towards the composition of polymicrobial communities and the behaviour of mixed species biofilms in flow. These will help in executing targeted treatments that disrupt the community structure, making individual uropathogens more susceptible to treatment.
Data availability
The raw datasets generated during the current study are available on NCBI, in the BioProject (ID-PRJNA1188362) and the raw read files can be accessed with the accession number - SRX26798577 and SRX26798578.
References
Werneburg, G. T. Catheter-Associated Urinary Tract Infections: Current Challenges and Future Prospects. RRU 14, 109–133 (2022).
Eckert, L. et al. Reducing the risk of indwelling Catheter–Associated urinary tract infection in female patients by implementing an alternative female external urinary collection device: A quality improvement project. J. Wound Ostomy Cont. Nurs. 47, 50–53 (2020).
Sedor, J., Mulholland, S. G., Hospital-acquired urinary tract infections associated with the indwelling catheter. Urol. Clin. North Am. 26, 821–828 (1999).
Hooton, T. M. et al. Diagnosis, prevention, and treatment of Catheter-Associated urinary tract infection in adults: 2009 international clinical practice guidelines from the infectious diseases society of America. Clin. Infect. Dis. 50, 625–663 (2010).
Mukhit Kazi, M. Catheter associated urinary tract infections (CAUTI) and antibiotic sensitivity pattern from confirmed cases of CAUTI in a tertiary care hospital: A prospective study. Clin. Microbiol. 04, (2015).
Chenoweth, C. E. & Saint, S. Urinary tract infections. Infect. Dis. Clin. N. Am. 30, 869–885 (2016).
Geredew Kifelew, L., Mitchell, J. G. & Speck, P. Mini-review: efficacy of lytic bacteriophages on multispecies biofilms. Biofouling 35, 472–481 (2019).
Topka-Bielecka, G. et al. Bacteriophage-Derived depolymerases against bacterial biofilm. Antibiotics 10, 175 (2021).
Fourcade, C., Canini, L., Lavigne, J. P. & Sotto, A. A comparison of monomicrobial versus polymicrobial Enterococcus faecalis bacteriuria in a French university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1667–1673 (2015).
WHO Fungal Priority Pathogens List To Guide Research, Development and Public Health Action. (World Health Organization, 2022).
Xu, Y. et al. Culture-Dependent and -Independent investigations of microbial diversity on urinary catheters. J. Clin. Microbiol. 50, 3901–3908 (2012).
Farhana, A., Basher, H., Alim, S. & Khan, S. Detection of catheter related blood stream infections in an ICU of a tertiary care center in Northern India. IJMR 8, 102–107 (2021).
Sousa, M. F. et al. Microbiological and microstructural analysis of indwelling bladder catheters and urinary tract infection prevention. Rev. Esc Enferm USP. 56, e20210552 (2022).
Lõoke, M., Kristjuhan, K. & Kristjuhan, A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques 50, 325–328 (2011).
Menzel, P., Ng, K. L. & Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 7, 11257 (2016).
Stepanović, S., Vuković, D., Dakić, I. & Savić, B. Švabić-Vlahović, M. A modified microtiter-plate test for quantification of Staphylococcal biofilm formation. J. Microbiol. Methods. 40, 175–179 (2000).
Biswas, B. et al. A novel robust method mimicking human substratum to dissect the heterogeneity of Candida auris biofilm formation. Microbiol. Spectr. 11, e00892–e00823 (2023).
Abu-Shoura, E. J. I., Hudu, S. A. & Al- Quadan, T. F. Exploring genetic and phenotypic factors contributing to urethral catheter biofilm formation in hospitalised patients in Jordan. Biomed. Pharmacol. J. 17, 1125–1134 (2024).
Sahai, S. & Kumar, A. Role of Candida in catheter associated urinary tract infection. IJCRR 10, 15–19 (2018).
Zhang, L. et al. Microbial diversity on intravascular catheters from paediatric patients. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2463–2470 (2015).
Zhang, X. H., Ahmad, W., Zhu, X. Y., Chen, J. & Austin, B. Viable but nonculturable bacteria and their resuscitation: implications for cultivating uncultured marine microorganisms. Mar. Life Sci. Technol. 3, 189–203 (2021).
Nye, T. M. et al. Microbial co-occurrences on catheters from long-term catheterized patients. Nat. Commun. 15, 61 (2024).
Gedefie, A. et al. Acinetobacter baumannii biofilm formation and its role in disease pathogenesis: A review. IDR Volume. 14, 3711–3719 (2021).
WHO & Bacterial Priority Pathogens List 2024: Bacterial Pathogens of Public Health Importance, To Guide Research, Development, and Strategies To Prevent and Control Antimicrobial Resistance (World Health Organization, 2024).
Seifert, H. Acinetobacter species as a cause of Catheter-related infections. Zentralblatt Für Bakteriologie. 283, 161–168 (1995).
Siegman-Igra, Y., Kulka, T., Schwartz, D. & Konforti, N. Polymicrobial and monomicrobial bacteraemic urinary tract infection. J. Hosp. Infect. 28, 49–56 (1994).
Raas, M. W. D. et al. Whole slide imaging is a high-throughput method to assess Candida biofilm formation. Microbiol. Res. 250, 126806 (2021).
Olsen, N. M. C. et al. Priority of early colonizers but no effect on cohabitants in a synergistic biofilm community. Front. Microbiol. 10, 1949 (2019).
Harriott, M. M. & Noverr, M. C. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob. Agents Chemother. 53, 3914–3922 (2009).
Hernandez-Cuellar, E. et al. Characterization of Candida albicans and Staphylococcus aureus polymicrobial biofilm on different surfaces. Revista Iberoamericana De Micología. 39, 36–43 (2022).
Silva, S. et al. Silicone colonization by non-Candida albicans Candida species in the presence of urine. J. Med. Microbiol. 59, 747–754 (2010).
Jones, C. J. et al. ChIP-Seq and RNA-Seq reveal an AmrZ-Mediated mechanism for Cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 10, e1003984 (2014).
Moreira, J. M. R. et al. Influence of flow rate variation on the development of Escherichia coli biofilms. Bioprocess. Biosyst Eng. 36, 1787–1796 (2013).
Radonicic, V. et al. Single-Cell optical nanomotion of Candida albicans in microwells for rapid antifungal susceptibility testing. Fermentation 9, 365 (2023).
Sharma, K. et al. Dynamic persistence of UPEC intracellular bacterial communities in a human bladder-chip model of urinary tract infection. eLife 10, e66481 (2021).
Teodósio, J. S., Simões, M., Melo, L. F. & Mergulhão, F. J. Flow cell hydrodynamics and their effects on E. coli biofilm formation under different nutrient conditions and turbulent flow. Biofouling 27, 1–11 (2011).
Negri, M. et al. Candida tropicalis biofilms: artificial urine, urinary catheters and flow model. Med. Mycol. 1–9. https://doi.org/10.3109/13693786.2011.560619 (2011).
Klayman, B. J., Volden, P. A., Stewart, P. S. & Camper, A. K. Escherichia coli O157:H7 requires colonizing partner to adhere and persist in a capillary flow cell. Environ. Sci. Technol. 43, 2105–2111 (2009).
Weiss Nielsen, M., Sternberg, C., Molin, S. & Regenberg, B. Pseudomonas aeruginosa and Saccharomyces cerevisiae biofilm in flow cells. JoVE 2383 https://doi.org/10.3791/2383 (2011).
Bizerra, F. C. et al. Characteristics of biofilm formation by Candida tropicalis and antifungal resistance: C. tropicalis biofilm formation and antifungal resistance. FEMS Yeast Res. 8, 442–450 (2008).
Sellam, A. et al. A Candida albicans early stage biofilm detachment event in rich medium. BMC Microbiol. 9, 25 (2009).
Uppuluri, P., Chaturvedi, A. K. & Lopez-Ribot, J. L. Design of a simple model of Candida albicans biofilms formed under conditions of flow: development, architecture, and drug resistance. Mycopathologia 168, 101–109 (2009).
Uppuluri, P. et al. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6, e1000828 (2010).
Purevdorj, B., Costerton, J. W. & Stoodley, P. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 68, 4457–4464 (2002).
Ribeiro, S. M., Cardoso, M. H., Cândido, E. D. S. & Franco, O. L. Understanding, preventing and eradicating Klebsiella Pneumoniae biofilms. Future Microbiol. 11, 527–538 (2016).
Vejborg, R. M. & Klemm, P. Cellular chain formation in Escherichia coli biofilms. Microbiology 155, 1407–1417 (2009).
Allison, D. L. et al. Candida -Bacteria interactions: their impact on human disease. in Virulence Mechanisms of Bacterial Pathogens (ed Kudva, I. T.) 103–136 (ASM, 2016). https://doi.org/10.1128/9781555819286.ch5.
Pohl, C. H. Recent advances and opportunities in the study of Candida albicans polymicrobial biofilms. Front. Cell. Infect. Microbiol. 12, 836379 (2022).
Lee, K. W. K. et al. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 8, 894–907 (2014).
Kaur, S., Chhibber, S. & Bansal, S. Disrupting the mixed-species biofilm of Klebsiella pneumoniae B5055 and Pseudomonas aeruginosa PAO using bacteriophages alone or in combination with xylitol. Microbiology 161, 1369–1377 (2015).
Mironova, A. V., Karimova, A. V., Bogachev, M. I., Kayumov, A. R. & Trizna, E. Y. Alterations in antibiotic susceptibility of Staphylococcus aureus and Klebsiella pneumoniae in dual species biofilms. IJMS 24, 8475 (2023).
Lin, Y. J. et al. Interactions between Candida albicans and Staphylococcus aureus within mixed species biofilms. BIOS 84, 30–39 (2013).
Ponde, N. O., Lortal, L., Ramage, G., Naglik, J. R. & Richardson, J. P. Candida albicans biofilms and polymicrobial interactions. Crit. Rev. Microbiol. 47, 91–111 (2021).
Thein, Z. M., Seneviratne, C. J. & Samaranayake, Y. H. Samaranayake, L. P. Community lifestyle of Candida in mixed biofilms: a mini review. Mycoses 52, 467–475 (2009).
Trejo-Hernández, A., Andrade-Domínguez, A., Hernández, M. & Encarnación, S. Interspecies competition triggers virulence and mutability in Candida albicans – Pseudomonas aeruginosa mixed biofilms. ISME J. 8, 1974–1988 (2014).
Ferrer-Espada, R., Liu, X., Goh, X. S. & Dai, T. Antimicrobial blue light inactivation of polymicrobial biofilms. Front. Microbiol. 10, 721 (2019).
Garsin, D. A. & Lorenz, M. C. Candida albicans and Enterococcus faecalis in the Gut: synergy in commensalism? Gut Microbes. 4, 409–415 (2013).
Jin, P., Wang, L., Chen, D. & Chen, Y. Unveiling the complexity of early childhood caries: Candida albicans and Streptococcus mutans cooperative strategies in carbohydrate metabolism and virulence. J. Oral Microbiol. 16, 2339161 (2024).
Zhu, Y. et al. Porphyromonas gingivalis and Treponema denticola synergistic polymicrobial biofilm development. PLoS ONE. 8, e71727 (2013).
Förster, T. M. et al. Enemies and brothers in arms: Candida albicans and gram-positive bacteria: Candida albicans and gram-positive bacteria. Cell. Microbiol. 18, 1709–1715 (2016).
Fourie, R. & Pohl, C. H. Beyond antagonism: the interaction between Candida species and Pseudomonas aeruginosa. JoF 5, 34 (2019).
Pérez, J. C. The interplay between gut bacteria and the yeast Candida albicans. Gut Microbes. 13, 1979877 (2021).
Shirtliff, M. E., Peters, B. M. & Jabra-Rizk, M. A. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 299, 1–8 (2009).
Galdiero, E. et al. Allium ursinum and Allium oschaninii against Klebsiella pneumoniae and Candida albicans Mono- and polymicrobic biofilms in in vitro static and dynamic models. Microorganisms 8, 336 (2020).
Desai, S., Sanghrajka, K. & Gajjar, D. High adhesion and increased cell death contribute to strong biofilm formation in Klebsiella pneumoniae. Pathogens 8, 277 (2019).
Bowen, W. H., Burne, R. A., Wu, H. & Koo, H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242 (2018).
James, G. A. et al. Bacterial adhesion and biofilm formation on textured breast implant shell materials. Aesth Plast. Surg. 43, 490–497 (2019).
Andriana, Y., Widodo, A. D. W. & Arfijanto, M. V. Synergistic interactions between Pseudomonas aeruginosa and Candida albicans, Candida glabrata, Candida Krusei, Candida parapsilosis as well as Candida tropicalis in the formation of polymicrobial biofilms. J. Pure Appl. Microbiol. 18, 219–228 (2024).
Chen, A. I. et al. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-Controlled biofilm formation as part of a Cyclic relationship involving phenazines. PLoS Pathog. 10, e1004480 (2014).
Kasetty, S., Mould, D. L., Hogan, D. A. & Nadell, C. D. Both Pseudomonas aeruginosa and Candida albicans Accumulate Greater Biomass in Dual-Species Biofilms under Flow. mSphere 6, e00416-21 (2021).
Phuengmaung, P. et al. Rapid synergistic biofilm production of Pseudomonas and Candida on the pulmonary cell surface and in mice, a possible cause of chronic mixed organismal lung lesions. IJMS 23, 9202 (2022).
Peters, B. M. et al. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology 158, 2975–2986 (2012).
Bandara, H. M. H. N., Yau, J. Y. Y., Watt, R. M., Jin, L. J. & Samaranayake, L. P. Escherichia coli and its lipopolysaccharide modulate in vitro Candida biofilm formation. J. Med. Microbiol. 58, 1623–1631 (2009).
Acknowledgements
We sincerely thank the staff of Bhailal Amin General Hospital, Vadodara for providing the indwelling urinary catheters. We acknowledge Central Facility at VSI-CMB Department, The M. S. University of Baroda for Confocal Laser Scanning Microscopy (Carl Zeiss LSM 710). We are grateful to DBT India for funding the Phase II of M. Sc. Biotechnology Teaching program. PJ acknowledges SHODH fellowship.
Author information
Authors and Affiliations
Contributions
PJ : Writing original draft, reviewing and editing, designed and carried out experiments, prepared figures and data analysis; RB: Writing original draft and editing, carried out experiments (flow system), image data analysis; MS: Writing original draft, carried out experiments (static); DG: Reviewing and editing manuscript, conceptualization, supervision, fund acquisition.The authors declare no competing interests.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical statement
This study was approved by the Institutional Biosafety Committee (IBSC) of The Maharaja Sayajirao University of Baroda and conducted in accordance with the principles outlined in the Declaration of Helsinki. As this was a retrospective study involving urinary catheters removed by healthcare workers based on physicians’ advice, with no direct or indirect interaction between the researchers and patients, the IBSC granted a waiver of informed consent. Demographic data, including gender, age, and duration of catheterization, were collected and analyzed in an anonymized format to ensure participants’ privacy.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Joshi, P., Bhattacharjee, R., Sahu, M. et al. Insights into urinary catheter colonisation and polymicrobial biofilms of Candida- bacteria under flow condition. Sci Rep 15, 15375 (2025). https://doi.org/10.1038/s41598-025-00457-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-00457-w