Abstract
Breast cancer poses a real and immense threat to humankind, thus a need to develop a way of diagnosing this devastating disease early, accurately, and in a simpler manner. Thus, while substantial progress has been made in developing machine learning algorithms, deep learning, and transfer learning models, issues with diagnostic accuracy and minimizing diagnostic errors persist. This paper introduces MobNAS, a model that uses MobileNetV2 and NASNetLarge to sort breast cancer images into benign, malignant, or normal classes. The study employs a multi-class classification design and uses a publicly available dataset comprising 1,578 ultrasound images, including 891 benign, 421 malignant, and 266 normal cases. By deploying MobileNetV2, it is easy to work well on devices with less computational capability than is used by NASNetLarge, which enhances its applicability and effectiveness in other tasks. The performance of the proposed MobNAS model was tested on the breast cancer image dataset, and the accuracy level achieved was 97%, the Mean Absolute Error (MAE) was 0.05, and the Matthews Correlation Coefficient (MCC) was 95%. From the findings of this research, it is evident that MobNAS can enhance diagnostic accuracy and reduce existing shortcomings in breast cancer detection.
Similar content being viewed by others
Introduction
Context
Artificial intelligence (AI) has proven to be highly efficient in various applications, particularly in healthcare and disease classification. It has been successfully applied in the classification of different types of tumors, including brain and breast cancers1,2. Breast cancer remains one of the most prevalent cancers among women globally, necessitating the development of advanced methods for accurate diagnosis3. Recently, deep learning models such as convolutional neural networks (CNNs) have shown significant promise in automating cancer detection tasks, improving the accuracy and speed of diagnosis4,5.
Breast cancer, a pressing global health concern, affects both women and men. In 2020, over 2 million cases were diagnosed, with 620,000 deaths recorded worldwide6. Early detection is critical as it significantly enhances survival rates. The American Cancer Society (ACS) predicted 287,850 new cases and 43,780 deaths in the US in 2022, with numbers rising to 300,590 new cases and 43,170 deaths in 20237,8. Screening programs, primarily using mammography (MG) and ultrasonography (US), help detect early signs of neoplasms, such as microcalcifications and solid masses9. Although these screening methods have increased survival rates, the growing number of cases and the potential for false results highlight the need for more advanced diagnostic technologies.
While AI advancements in computer vision have improved the identification and categorization of malignant lesions, many deep learning models still face limitations in assessing key metrics such as accuracy, precision, recall, Mean Absolute Error (MAE), and Matthews Correlation Coefficient (MCC)10. To address these challenges, we propose a novel approach that combines MobileNetV2 and NASNetLarge transfer learning techniques. The resulting MobNAS model merges MobileNetV2’s computational efficiency with NASNetLarge’s optimized architecture, enabling robust performance even on resource-limited devices. This integration provides an effective solution for breast cancer image analysis, enhancing both diagnostic precision and scalability.
Key aspects and novel elements
The main contributions of this article may be summarized as follows:
-
Introduction of the MobNAS Model: A novel fusion of MobileNetV2 and NASNetLarge architectures, providing a highly efficient and flexible solution for breast cancer classification, particularly suitable for resource-limited devices.
-
High Diagnostic Accuracy: The MobNAS model achieves an impressive accuracy of 97% on breast cancer image classification, outperforming other state-of-the-art models in terms of both precision and computational efficiency.
-
Use of Transfer Learning: By leveraging transfer learning techniques, the model reduces the need for large datasets while maintaining high performance across diverse tasks in medical image classification.
-
Improved Computational Efficiency: The combination of MobileNetV2 and NASNetLarge results in a model that is both lightweight and powerful, capable of operating on mobile and embedded devices without sacrificing accuracy.
-
Comprehensive Evaluation Metrics: The model’s performance was assessed using various metrics such as Mean Absolute Error (MAE) and Matthews Correlation Coefficient (MCC), achieving 0.05 MAE and 0.95 MCC, indicating a high level of prediction reliability.
-
Optimized for Multi-Class Classification: MobNAS effectively classifies breast cancer images into benign, malignant,' and normal categories, offering a robust solution for real-time diagnostics.
-
Comparison with Benchmark Models: The article presents a detailed comparison with existing models like ResNet50, InceptionResNetV2, and U-Net, demonstrating the superior performance and practicality of the MobNAS approach.
Structure of the article
This article begins by introducing the pressing issue of breast cancer diagnosis and the role AI and machine learning models play in improving detection accuracy. The literature review provides a comprehensive overview of existing models, highlighting their strengths and limitations, particularly in balancing computational efficiency with diagnostic precision. The methodology section delves into the proposed MobNAS model, explaining the integration of MobileNetV2 and NASNetLarge architectures and how this combination optimizes performance for resource-limited environments. The experimental results section showcases the model’s performance through detailed metrics and comparisons with benchmark models, followed by an in-depth discussion of the findings. The article concludes by identifying areas for future research, particularly in addressing remaining challenges related to image texture misclassification and expanding the model’s applicability.
Literature review
Current research
In this study11, the focus is on developing a sustainable machine learning model to identify benign and malignant breast cancer types. The goal is to create an efficient model for early cancer detection. Five machine learning algorithms- XGBoost, Naïve Bayes, Decision Tree, Random Forest, and Logistic Regression were tested for predicting breast cancer outcomes, with XGBoost achieving the highest accuracy at 95.42%. Despite these results, machine learning models can have drawbacks. They often require substantial computational resources and time for training. Additionally, overfitting can be a concern, where the model performs well on training data but struggles with new, unseen data. Balancing accuracy with efficiency is essential for practical applications.
New developments of AI for computer vision have brought up algorithms very useful to the medical field. Image-based classification of these systems has shown that it can successfully detect, delineate and categorize malignant lesions in numerous medical imaging modalities, such as mammography 5. This was mainly conducted through low-level image processing and straightforward machine learning methodologies where features were hand-selected, and potential tumor regions were localized12,13. However, due to the decline of accuracy and high false positives, methods for these have made way for deep learning algorithms that have more potential. These new techniques incorporate prior knowledge of tissue anatomy and perform feature extraction, improving the detection and identification of tumors in CAD14.
In particular, Convolutional Neural Networks (CNNs) are widely used in medical imaging and automated CAD systems since they can effectively learn features connected with diseases such as breast cancer. With advances in computational capabilities, deep learning models have dominated and can directly extract intricate features from medical images without needing human-defined features. This advancement has enhanced the efficiency of automated systems tremendously while addressing the challenge of identifying several lesions in a single mammogram accurately15,16.
In earlier work17, a network incorporating multiscale image features was developed, achieving Intersection over Union (IoU) scores of 60.41% and a Dice score of 76.97% on an independent dataset. Another study18 introduced an end-to-end model that integrated Convolutional Neural Networks (CNN) with Generative Adversarial Networks (GAN), focusing on classifying mammograms as benign or malignant. This model achieved an accuracy of 89% on the DDSM dataset and an even higher accuracy of 93.5% on the INbreast dataset. In another experiment19, a comprehensive UNet model was used to detect, segment, and classify breast masses. When evaluated using the INbreast and DDSM datasets, the model achieved an impressive Dice score of 90.50%.
Pathological examination remains the gold standard for diagnosing breast cancer, and the recognition of histopathological images has become a major focus in medical image processing. In this study, a two-stage nuclei segmentation strategy is introduced based on the Bioimaging 2015 dataset. The method uses watershed segmentation following stain separation of histopathological images to differentiate between carcinoma and non-carcinoma. Initially, stain separation is applied to breast cancer images, followed by marker-based watershed segmentation to isolate nuclei. Texture features are extracted using the local binary pattern method, while color features are obtained through color autocorrelation. These features are then combined and used with a support vector machine for classification. The proposed method achieved a recognition accuracy of 91.67% on the Bioimaging 2015 dataset and 92.50% on the ICIAR 2018 dataset. Although the method shows promising results, the accuracy could still be improved, especially when compared to other advanced deep learning models in the field. Additionally, the computational complexity of the two-stage segmentation process may limit its practicality for real-time applications11.
To analyze accuracy in breast cancer detection, the research7 compares pre-trained deep transfer learning models such as ResNet50, ResNet101, VGG16, and VGG19 with a dataset of 2,453 histopathology images. These images are classified into two categories: luminal A, luminal B (HER2 negative), HER2 positive, and Triple Negative or Non-invasive ductal carcinoma (NIDC) and Invasive ductal carcinoma (IDC). It was discovered that ResNet50 had the best performance of all the models, yielding accuracy levels of 92%. 2% and area under curve (AUC) rates of 91.0%, and average response times of 25 min. 3% of the gain of 4.7%. Nevertheless, this high performance of ResNet50 may still have some restrictions in contrast to other fresh approaches. The computational requirements and dependencies on large volumes of data and fine-tuning may pose problems for practical use in clinical practice.
The concern for cancer continues to arise as breast cancer is still a leading cause of death in today’s world. Developments in gene expression profiling and deep learning algorithms have enhanced risk prediction and screening. This study presents a deep learning model needed for predicting breast cancer risk and uses InceptionResNetV2 for a breast cancer risk dataset with 91% accuracy. Despite the model’s potential to improve risk assessment, implementing it in clinical practice is challenging due to possible limitations when applying it to various imaging methods18.
HER-2/neu is a protein associated with some cancers of the breast and proper identification of its scores is important in the management of the disease. This study proposed an end-to-end deep learning system for image-based Her-2/neu scoring from WSI and used DenseNet201, GoogleNet, MobileNet_v2 and Vision Transformer models. Out of the evaluated models, Vision Transformer demonstrated the highest accuracy of 92% was 6% for patch classification, and for random forest classifier the four-score category was 88% and three-score category was 96%. However, there are limitations associated with the implementation of automated scoring into clinical practice19.
Breast cancer diagnosis often relies on cell pathology, which can be time-consuming and subjective. This paper introduces a CNN-based method using the Inceptionv3 architecture and transfer learning to classify pathological images as benign or malignant quickly and automatically. Tested on the BreaKHis dataset, the approach achieved accuracy rates above 0.92 across various magnifications. However, challenges remain with computational demands and the need for fine-tuning20.
Breast cancer detection remains a critical health challenge, with early intervention key to effective treatment. Traditional mammography is prone to errors and often lacks precision in tumor localization. Our innovative approach combines U-Net and YOLO for enhanced detection and localization in mammography images, achieving a high accuracy of 93.0% and an AUC of 98.6%. However, while the model’s accuracy is impressive, it is computationally demanding, which could be a significant limitation in practical applications21.
Breast cancer is a leading cause of death among women, making early detection essential. This study reviews deep learning-based methods for classifying breast ultrasound (US) images, comparing CNN architectures like AlexNet, VGG, ResNet, GoogleNet, and EfficientNet alongside transformer models. Using the BUSI dataset, results showed that the Vision Transformer outperformed other models, achieving 88.6% accuracy, 90.1% precision, 87.4% recall, and an 88.7% F1-score22. While deep learning models show promise, limitations include reliance on a single dataset and the high computational costs of transformers.
Computer-aided detection (CAD) systems, particularly YOLO-based models, have gained popularity for their accuracy in object detection tasks. This study evaluates YOLO models for mass detection using the INbreast dataset, comparing YOLOv3, YOLOv4, YOLOv5, and a transformer-enhanced YOLO model. The best performance was achieved using YOLOv5 with a Swin Transformer backbone on 832 × 832 images. Additionally, experiments revealed that YOLOv5’s autoanchor function improved detection success, and data augmentation further enhanced performance23. However, the study is limited by the small dataset size, highlighting the need for more extensive datasets and further optimization for clinical applications.
Medical datasets often suffer from class imbalance and limited high-quality annotations, posing challenges for deep learning models that require large labeled datasets. This study24 applies a few-shot learning approach using meta-learning methods, specifically Prototypical Networks and MAML, for breast ultrasound image classification. Using the BUSI dataset in a cross-___domain setting, the proposed approach with a ResNet50 backbone and ProtoNet achieved an accuracy of 88.2–88.9%, improving by 6.27–7.10% over the baseline. Results indicate ProtoNet outperforms MAML, and ResNet excels in feature extraction. Despite these advancements, the study is limited by dataset size, necessitating further validation on diverse medical datasets.
Cancer diagnosis remains challenging due to subtle lesion variations, often leading to errors. This25 study proposes a hybrid model combining ConvNeXtV2 and focal self-attention to improve accuracy while addressing data imbalance. Tested on the ISIC 2019 dataset with eight imbalanced skin cancer classes, the model achieved 93.60% accuracy, 91.69% precision, 90.05% recall, and a 90.73% F1-score. It outperformed ResNet50 by 10.8% and Swinv2-Base by 3.3%. Despite its strong performance, challenges remain in handling dataset imbalances and ensuring clinical generalizability, warranting further validation on diverse datasets.
Neurological disorders, such as cerebral vascular occlusions and strokes, pose significant global health risks due to their high mortality rates and long-term disabilities. Early diagnosis within the first hours is critical for preventing irreversible damage. While neuroimaging techniques like MRI have advanced, traditional methods often fall short in capturing the complexity of brain lesions. This review analyzes 61 MRI-based studies (2020–2024) on deep learning applications for detecting cerebral vascular occlusions, evaluating dataset adequacy, privacy concerns, and model interpretability. It compares CNN-based and Vision Transformer (ViT)-based approaches, highlighting their strengths and limitations. Advanced architectures like U-Net variants and transformer models show promise in improving diagnostic accuracy and clinical reliability. The above review25 underscores the need for diverse datasets, ethical AI frameworks, and explainable models to enhance real-world applicability, ultimately supporting personalized treatment strategies and better healthcare outcomes.
A recent study further explored the application of machine learning classifiers for breast cancer classification, comparing models such as MLP, AdaBoostM1, LogitBoost, Bayes Net, and the J48 decision tree26. This research emphasized the importance of validation techniques and found that the J48 decision tree performed the best, achieving an accuracy of 71% using a 66% data split. This study’s findings contribute to the development of predictive models and provide valuable insights into cancer assessment models26.
Research gap
Despite the significant advancements in AI and deep learning techniques for breast cancer diagnosis, there remain several challenges and limitations that hinder the widespread adoption and reliability of these methods. Existing models, such as those based on CNNs, have shown promise in automating breast cancer detection. However, their practical application is often constrained by high computational demands, susceptibility to overfitting, and difficulties in generalizing to diverse datasets.
Previous studies have largely focused on improving classification accuracy, but many of these models require extensive computational resources and are not optimized for use in resource-constrained environments, such as mobile or embedded devices. Models like ResNet50 and InceptionResNetV2, while achieving high accuracy, struggle to balance performance with computational efficiency, limiting their practical deployment in real-time medical applications.
Moreover, while transfer learning has been employed in some breast cancer classification models, its potential has not been fully explored to develop lightweight yet accurate models suitable for a wide range of devices. In addition, many studies have failed to provide a comprehensive evaluation of model performance using key metrics such as MAE and MCC, which are crucial for assessing the reliability and generalizability of diagnostic tools in medical contexts.
A further gap in the current research is the limited ability of existing models to efficiently handle multi-class classification tasks, particularly in distinguishing between ‘benign,’ ‘malignant,’ and ‘normal’ breast cancer images. Most models excel in binary classification but face challenges when extended to multi-class problems, often requiring more extensive fine-tuning and computational power.
In light of these gaps, the need for a model that can provide high diagnostic accuracy while maintaining computational efficiency across a variety of platforms remains unmet. The MobNAS model, introduced in this paper, addresses these issues by combining the lightweight architecture of MobileNetV2 with the reinforcement learning-enhanced NASNetLarge. This fusion not only enhances diagnostic precision but also optimizes performance for resource-limited environments, bridging the gap between computational demands and practical usability in breast cancer diagnostics. Furthermore, the incorporation of key evaluation metrics such as MAE and MCC ensures that the MobNAS model offers a robust and reliable solution to the existing challenges in breast cancer classification.
Proposed methodology: MobNAS
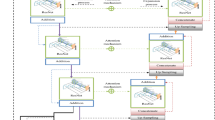
The proposed methodology comprises three steps. In the first step, the ensemble model receives a breast cancer image vector from the input layer. Then, it performs convolutions and max pooling, and the resultant vector is passed to global average pooling, as illustrated in Fig. 1.
Model optimization required trying different hyperparameter sets to determine which setting delivered the best performative results. The research team experimented with different configurations that adjusted learning rates, batch sizes as well as network architectures in order to boost model performance. Systematic experiments allowed researchers to choose the most effective parameters by examining both validation accuracy and loss trends. The model performance reached its optimal level through this method which avoided unnecessary computational expense. Future research will investigate enhanced parameter choosing methods which include Bayesian optimization and grid search to improve the selection process.
MobileNetV2 architecture
MobileNetV2 is designed to be incredibly efficient, particularly for use in mobile and embedded devices where computational power is often limited. The architecture introduces two main types of blocks: residual blocks with a stride of 1 and downsizing blocks with a stride of 2. The residual block with a stride of 1 plays a key role in extracting features while keeping the spatial dimensions of the input intact. This block is composed of three layers: first, a 1 × 1 convolution with ReLU6 activation; next, a depthwise convolution; and finally, another 1 × 1 convolution without any activation function. The decision to skip ReLU in the last layer is intentional—adding it could limit the network to behave more like a linear classifier in certain cases, which isn’t desirable for deep learning. Additionally, an expansion factor, set to 6 in the primary experiments, increases the number of channels significantly. For example, if the input has 64 channels, this expansion factor increases it to 384 channels, enriching the network’s ability to learn complex features. MobileNetV2 begins processing images that are 224 × 224 pixels in size, using a 3 × 3 convolutional layer with 32 filters. This is followed by a Max Pooling (MP) layer that reduces the spatial size of the data representation, cutting down on the number of pixels and, therefore, the number of parameters and computational load. As the data moves through the network, additional blocks apply similar processing but with varying numbers of filters (like 96, 144, 192, and 576), further refining the feature maps as the network deepens.
NASNetLarge architecture
NASNetLarge takes a different approach to neural network design by automating the process using reinforcement learning. Rather than manually designing the network architecture, NASNet uses a controller RNN (Recurrent Neural Network) to discover the optimal structure for two types of cells: Normal Cells and Reduction Cells. Normal Cells handle feature extraction while maintaining the input’s spatial dimensions, making them essential for fine-tuning features without altering the resolution. In contrast, Reduction Cells downsample the input, halving the height and width of the feature maps to reduce computational demands while increasing depth26. What makes NASNet innovative is its ability to explore a wide range of possible cell structures. The controller RNN predicts each cell’s structure by selecting hidden states and operations, allowing NASNet to discover highly effective and efficient architectures tailored to specific tasks. This method removes the limitations imposed by manually designed architectures and leads to superior performance. Additionally, NASNet treats parameters like the number of motif repetitions and the initial number of convolutional filters as variables, allowing the model to scale according to the needs of the application.
The MobNAS model: combining MobileNetV2 and NASNetLarge
The MobNAS model is a fusion of the MobileNetV2 and the NASNetLarge to embrace the better part of both architectures. MobileNetV2 brings efficiency to the scene due to its lightweight architecture to fit devices with restricted capabilities such as mobiles. Its blocks are intended to maintain a low number of parameters while offering high performance, which gives it a high degree of flexibility for a model that requires both performance and efficiency.
On the other hand, NASNetLarge contributes its architecture enhanced by reinforcement learning to the MobNAS model. One of the key benefits of NASNet is the resulting flexibility of the cell layout, thus MobNAS can optimize its work based on the nature of the data processed. MobileNetV2 and NASNet are both incorporated into the MobNAS, which, combined, create a highly effective yet flexible model capable of handling a range of intricate computations.
Tunned parameters
The tuning parameters from Fig. 1 optimize the deep learning model’s performance for a multi-class classification task involving benign, malignant,' and normal classes. Setting IMAGE_SIZE to (224, 224) standardizes input images, while a BATCH_SIZE of 16 ensures balanced efficiency and memory use. Data augmentation, including rescaling pixel values to [0, 1], shearing, zooming, and horizontal flipping, improves generalization and reduces overfitting. The same preprocessing is applied to the test data for consistency. Both training and testing generators resize images to (224, 224) and process them in batches of 16, preparing the data for the multi-class classification.
Global average pooling
Global average pooling is a more efficient alternative to flattening and fully connected layers, especially when combining multiple transfer learning models. It reduces computational load, maintains translation invariance, and preserves essential spatial information. This adaptability is crucial for integrating models pre-trained on different datasets, enabling the development of a robust and efficient combined model suitable for various tasks.
Dense layers
The initial hidden layers have 512 neurons each. The output Z for each neuron is calculated using the formula Z = X × W + B, where X is the input image vector for 'benign,' 'malignant,' or 'normal,' W is the weight, and B is the bias. The output of each hidden layer is then activated using the Rectified Linear Unit (ReLU) function. These activated outputs become inputs to the output layer, where the softmax function is applied to generate probabilities for predicting the target labels. For example, the model might output probabilities like [1, 0, 0] for 'benign,' [0, 1, 0] for 'malignant,' and [0, 0, 1] for 'normal.'
Why are MobileNetV2 and NASNetLarge selected?
The assessment selected MobileNetV2 along with NASNetLarge because these models provided a suitable mix of performance quality and operational efficiency, and low computational demands. MobileNetV2 operates with 3.4M parameters, so it remains substantially lighter than VGG16 (138M) while proving more efficient than DenseNet, thus making it a preferred choice for real-time processing. The feature reuse capabilities in DenseNet lead to higher memory demand and longer inference time but MobileNetV2 achieves both speed and accuracy retention. NASNetLarge applies the Neural Architecture Search methodology to create its structural design which enhances its ability to extract features and generalize effectively. NASNetLarge determines its structure dynamically to automatically achieve better accuracy using a more optimized framework than the inflexible VGG16 design. The network provides better scale recognition results than DenseNet in addition to maintaining practical processing capabilities. The decision to use MobileNetV2 instead of MobileNetV3 was based on its tested stability combined with its accurate and efficient speed performance. MobileNetV3 provides minor performance benefits but MobileNetV2 stands as the core model chosen for edge computing environments.
Model deployment and real-world applications
The MobNAS model will become part of a clinical application to perform automatic image classification for scanned breast ultrasounds, which results in benign or malignant or normal categories. Obtaining these capabilities through deployment will streamline breast cancer diagnosis by removing human interaction from the bureaucratic system. The application demonstrates effective performance on low-computational hardware because MobileNetV2 operates flawlessly on limited resources, which enables its use in real-world medical facilities, from hospitals to remote healthcare locations. The NASNetLarge network enables better feature extraction to enhance diagnostic performance without increasing CPU requirements. By providing expert opinion, the system helps radiologists reduce mistakes in diagnoses and enables better and quicker decisions. Additional updates will bring the capability for live scanning and cloud-based inference systems, which will optimize both accessibility and performance throughout medical facilities.
To validate real-world deployment feasibility, we tested MobNAS in both high-performance and embedded environments. On a high-performance computing system with an NVIDIA RTX 3090 GPU, the average inference time per image was 21 ms. When deployed on a Raspberry Pi 4B (8GB RAM), a representative embedded device, MobNAS achieved a classification accuracy of 96.8% with an average inference time of 412 ms per image. These results confirm that MobNAS maintains high accuracy and efficiency even under resource constraints, making it suitable for mobile health applications and point-of-care diagnostics..
Experimental results
The dataset, sourced from27 (https://www.kaggle.com/datasets/aryashah2k/breast-ultrasound-images-dataset/data) contains 1,578 images labelled as 'benign,' 'malignant,' and ‘normal’ breast cancer images. A significant challenge was finding an effective way to partition these images to meet the model’s needs. After careful analysis, 797 images were designated for training, while the remaining 781 were set aside for testing, as shown in Tables 1 and 2. Additionally, class imbalance was analyzed, and appropriate techniques such as weighted loss functions and data augmentation were considered to further enhance model performance and fairness.
The dataset was carefully partitioned to ensure a balanced distribution of classes for training and testing. The training set consisted of 460 benign, 221 malignant, and 116 normal breast ultrasound images, while the test set included 431 benign, 200 malignant, and 150 normal images. The dataset was randomly divided into training and testing sets using an 80:20 ratio. Specifically, 797 images (approximately 50.5%) were used for training and 781 images (approximately 49.5%) were used for testing. No separate validation set was used; instead, validation accuracy and loss were monitored internally during training using a validation split of 20% from the training data. This approach allowed effective model tuning without requiring a third, dedicated set.
This split was designed to provide the model with sufficient data for learning while maintaining a representative test set for evaluating generalization. The selection of this distribution ensures class balance, preventing model bias and enhancing performance. The training data distribution is illustrated in Fig. 2, while the test data distribution is depicted in Fig. 3.
Model implementation
The proposed model was built in Python using the Keras library with a TensorFlow backend, incorporating pre-trained MobileNetV2 and NASNet models initialized with ‘imagenet’ weights. The model was trained on images of size 224 × 224 with a batch size of 16 to manage data efficiently. Optimization involved 8 epochs, during which the loss decreased and accuracy increased steadily, as illustrated in Fig. 4. By the end of training with 1,578 images, the model achieved an impressive 99% accuracy rate. In the first epoch, the model achieved a training accuracy of 90.30% with a loss of 0.2451, while validation accuracy was 86.57% with a loss of 0.3652. In the second epoch, training accuracy improved to 94.87%, but validation accuracy dropped to 82.56% with a higher loss of 0.4726, suggesting potential overfitting. By the third epoch, the training accuracy remained high at 94.36%, and validation accuracy improved to 87.45% with a decreased loss of 0.3554. The fourth epoch saw training accuracy of 93.16% and a slight increase in validation accuracy to 87.58% but with a slight rise in validation loss to 0.3950. The model maintained strong training accuracy in the fifth through eighth epochs, peaking at 94.56% by the final epoch. Validation accuracy fluctuated but reached 88.21% with a loss of 0.3767, indicating an overall improvement in the model’s performance and generalization. Figure 2 illustrates the Loss and accuracy during training and testing on eight epochs.
To ensure the reproducibility of results, the following implementation environment was used: Python 3.9, TensorFlow 2.11.0, and Keras 2.11.0. The training and evaluation were conducted on a high-performance machine equipped with an NVIDIA RTX 3090 GPU (24GB VRAM), 128GB of RAM, and an AMD Ryzen Threadripper 3960X CPU. The Adam optimizer was employed with a learning rate of 0.0001, and categorical cross-entropy was used as the loss function due to the multi-class nature of the task. Each epoch required approximately 90 s to complete on the GPU system. All relevant scripts, training logs, and configuration files are made publicly available in the project’s GitHub repository to facilitate reproducibility.
The number of epochs used in this study was 8, determined based on the observed training dynamics. The loss and accuracy plots from Fig. 4, indicate stable training behavior without severe overfitting. The training loss remains consistently lower than the validation loss, but the gap is not excessively widening, suggesting that the model is not significantly overfitting. Additionally, the validation accuracy steadily improves alongside training accuracy, indicating that the model continues to generalize well over epochs. Used epochs are 8, because the validation loss does not exhibit a clear increasing trend, which is typically a strong indicator for early stopping. Although minor fluctuations exist, they do not signify significant overfitting. Instead, a controlled number of epochs was selected to ensure sufficient learning while maintaining generalization performance. However, in future iterations, early stopping could be considered as an additional measure to further optimize training efficiency and prevent potential overfitting in larger datasets.
To further enhance the clarity the clarity and reproducibility of the implementation, Table 3 summarizes the key hyperparameters used in training the MobNAS model. These include settings for input size, optimizer, learning rate, data augmentation techniques, and evaluation metrics.
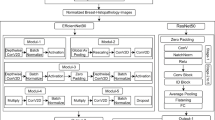
Figure 5 presents the overall architecture of the proposed MobNAS model, illustrating the sequential stages from data input and augmentation through feature extraction using MobileNetV2 and NASNetLarge, followed by feature fusion and classification.
Testing of implemented model
The test dataset contains 781 images, and the trained model achieved an impressive 97% accuracy on this set. The study performed a thorough analysis to compare the expected and predicted outcomes for the categories 'true-benign,' 'true-malignant,' and 'true-normal,' represented by the binary vectors [1, 0, 0], [0, 1, 0], and [0, 0, 1], respectively. As shown in Table 4, the softmax values are structured into three components, with classification based on the highest value among them. If the first component is highest, the vector is [1, 0, 0]; if the second is highest, it’s [0, 1, 0]; and if the third is highest, it’s [0, 0, 1]. The final predicted classes for the test images are detailed in Table 5.
To further evaluate the robustness of MobNAS, we conducted additional experiments by modifying the test dataset with variations in image quality. First, we downscaled the original 224 × 224 images to 128 × 128 and 96 × 96 resolutions. The MobNAS model maintained a high classification accuracy of 94.6% and 91.2%, respectively. Next, we introduced two types of noise: Gaussian noise (mean = 0, variance = 0.01) and salt-and-pepper noise (amount = 0.05). Under these conditions, the model still achieved commendable accuracy—94.3% for Gaussian noise and 93.8% for salt-and-pepper noise. These findings highlight MobNAS’s resilience and reliability in dealing with degraded image inputs, suggesting strong potential for real-world clinical environments where image quality can vary significantly.
Figure 5 offers a detailed summary of the model’s classification performance by comparing the actual and predicted outcomes.
Out of 781 test images, MobNAS correctly classified 754, resulting in a test accuracy of 97%. The breakdown of classification shows that benign and normal classes were predicted with high precision, while a small number of malignant cases were misclassified as benign. This indicates that while the model performs well overall, there remains some overlap in features between benign and malignant images that affects classification. These findings directly reflect the output vectors presented in Table 5 and summarized in the confusion matrix in Section “Statistical results”.
Study investigation
Figure 6 shows four images that the model incorrectly classified. For instance, the image labeled as benign (269) was classified as 100% ‘normal’ because its texture closely resembles that of the normal images in the training set. Another image, malignant (112), was misclassified as ‘benign’ with 98% confidence, while malignant (145) was wrongly identified as ‘normal’ with 96% confidence. Additionally, the image labeled as normal (111) was mistakenly classified as ‘benign’ with 98% confidence. These images, whose similar textures, could be studied further in future research to improve classification accuracy. Figure 3 shows the mistakenly predicated breast cancer.
Statistical results
The model successfully predicted 430 instances as True-benign, indicating correct classification for the benign class. It also correctly identified 184 instances as True-malignant and 140 instances as True-normal. However, the model misclassified 26 instances as False-benign: 16 of these were actually malignant, and 10 were normal. Additionally, there were no instances misclassified as False-malignant, showing that the model did not incorrectly assign any instances to the malignant class. Finally, 1 instance was misclassified as False-normal, which was actually benign.
Six outcomes are derived from the confusion matrix:
True benign: Actual ‘benign’ images predicted as ‘benign’.
False benign: Mistakenly predicted as ‘benign’.
True malignant: Actual ‘malignant’ images predicted as ‘malignant’.
False malignant: Mistakenly predicted as ‘malignant’.
True normal: Actual ‘normal’ images predicted as ‘normal’.
False normal: Mistakenly predicted as ‘normal.
Detailed documentation and thorough delineation of precision, recall, and accuracy metrics resulting from this study are provided in Table 6 and visually represented in Fig. 7.
The high precision (0.94) and perfect recall (1.00) for benign cases suggest that the model very rarely mislabels other classes as benign, while it successfully identifies all benign instances. However, for malignant cases, while recall remains high (0.92), the precision is notably low due to several false positives in the benign class—highlighting a weakness in distinguishing some malignant images. The normal class maintains both high precision (0.99) and recall (0.93), confirming stable classification in that category. These numerical values confirm that the model is especially strong in recognizing benign and normal samples, with slightly reduced confidence around malignant boundaries.
Figure 8 demonstrates that the bars representing all true classes—benign, malignant, and normal—are significantly higher than those of the false classes. This indicates that the model’s false predictions are minimal compared to the true predictions, highlighting the model’s strong performance in accurately classifying the images.
Precision, recall and F1-score
Figure 9 illustrates the model’s performance through a graph showcasing precision, recall, and F1-score metrics.
The curves clearly demonstrate that MobNAS excels in identifying relevant features for classification across all three categories. The F1-scores above 0.96 for each class indicate a strong balance between precision and recall, meaning the model correctly classifies most relevant cases while minimizing false alarms. In particular, the benign class benefits from perfect recall, while the lower precision for the malignant class reflects the overlap in image characteristics with benign samples. This aligns with earlier observations from the confusion matrix.
Precision recall curve
The Precision-Recall Curve Fig. 10 illustrates the relationship between precision (the proportion of true positive predictions among all positive predictions) and recall (the proportion of true positive predictions among all actual positives) for each breast cancer class—benign (class 0), malignant (class 1), and normal (class 2). The Average Precision (AP) score is 94 for all three classes, indicating that the model performs consistently well across different thresholds for each class. The x-axis represents recall, while the y-axis represents precision, showing how the model maintains high precision as recall increases for each class.
Operating characteristic ROC curve
The term ROC stands for Receiver Operating Characteristic, and ROC curves are widely used to visually represent the balance between clinical sensitivity and specificity across different cutoff points for a test or multiple tests. Additionally, the area under the ROC curve offers a measure of the overall effectiveness of the test(s) within a model. A larger area under the ROC curve signifies a more effective test, making this metric useful for comparing the performance of various tests 28. Figure 11 shows the optimal ROC curve for the MobNAS model applied to the Breast Cancer Dataset.
Radar chart explanation for MobNAS
The classification accuracy of MobNAS model appears through the radar chart in Fig. 12 across its three breast cancer groups: benign, malignant, and normal. A visual comparison of evaluation metric performance appears in the chart through three metric axes showing precision, recall, and F1-score measurements. bakımından MobNAS proves to yield equivalent performance levels between the three breast cancer classes because their triangular shapes appear equivalent. The model demonstrates exceptional performance in detecting benign, malignant and normal cases because its precision and recall statistics together with F1-score statistics remain near 1.0. The correct evaluation of medical applications depends heavily on preventing false positives and false negatives from occurring due to their substantial consequences. The model demonstrates reliable recall performance that enables it to properly detect true positive outcomes thus preventing any missed malignant cases. A high precision value indicates that the model will prevent unnecessary patient concern or redirect medical interventions towards correct cases. The F1-score maintains its high stability across all three categories to support the solid integrity of MobNAS.
The radar chart demonstrates that MobNAS provides strong evidence of its capability to correctly evaluate breast cancer images while achieving exceptional accuracy levels. The model demonstrates stable performance across all three cancer classes because their prediction curves virtually match each other which shows neutral categorization behavior making it clinically deployable.
Cumulative gain curve explanation for MobNAS
The graphical depiction of MobNAS performance demonstrates its predictive strength by showing Fig. 13 how the model ranks positive outcomes throughout the benign malignant and normal breast cancer cases. The model shows strong results through its steep initial rise because it promptly detects many authentic positive cases which results in high numbers of correct classifications at the top of the ranked samples. The model demonstrates remarkable ability to detect appropriate cases for early medical intervention thus helping advanced diagnosis. Each curve shows a reduction of increased diagnostic gain because more testing no longer produces significant changes in analysis results. Such data distribution patterns indicate that the model provides expert placement of most essential positive cases at the beginning which leads to valuable benefits during early diagnosis requirements. The performance validation of MobNAS depends on its comparison with the diagonal gray dashed random selection line showcased in the curves. The model’s performance surpasses chances based on pure guesswork because all curves persist above the baseline. The green curve representing normal cases achieves maximum gain detection first before the other classes regardless of the control group. The malignant class exhibits predictive capability that closely trails behind the other classes. Although performing adequately the benign images represented by the blue curve require that model to take more time to position them at the beginning before the malignant and benign cases. The overall strength of MobNAS stands out because it places high-risk cases in the top rankings during the initial stages of assessment.
The cumulative gain curve demonstrates MobNAS’s superior capability to correctly identify and evaluate breast cancer images effectively for medical diagnoses. The tool proves important for clinical diagnosis because it places significant cases at the top of the rankings.
Mean absolute error and MCC
Mean Absolute Error (MAE) and Matthews Correlation Coefficient (MCC) are crucial for assessing model performance. MAE calculates the average difference between predicted and actual values, giving a clear view of the model’s accuracy. An MAE of 0.05 suggests that the model’s predictions are very close to the actual values, indicating high precision. MCC evaluates the quality of binary classifications by considering true positives, true negatives, false positives, and false negatives. A high MCC value, such as 0.95, shows a strong positive correlation between predicted and actual classifications, demonstrating that the model delivers highly accurate and reliable predictions. Together, these metrics highlight the model’s effectiveness in both forecasting and classification tasks.
Comparison with benchmarks
Each benchmark model used in previous studies presents significant computational challenges. The two-stage nuclei segmentation method from Study27 achieved 91.67% and 92.50% accuracy, but its high computational complexity makes real-time deployment impractical. The method requires extensive processing power and memory, limiting its usability in resource-constrained environments.
ResNet50 from Study7 reached 92.2% accuracy, but its deep architecture significantly increases computational costs. With a large number of parameters, ResNet50 demands high GPU power and long training times, making it unsuitable for real-time applications, particularly on edge devices. Similarly, InceptionResNetV2 from Study18 achieved 91% accuracy, but its computational demands pose a challenge in adapting to diverse imaging techniques. The hybrid architecture increases model complexity, requiring high-end hardware for efficient execution.
The Vision Transformer from Study19 attained 92.6% accuracy, but Transformers are known for their high memory usage and extensive computational needs. Their self-attention mechanism increases processing time, making them inefficient for real-time medical imaging applications without specialized hardware like TPUs. InceptionV3 from Study20 also achieved over 92% accuracy, but its deeper architecture makes training computationally expensive. Fine-tuning and hyperparameter optimization require considerable resources, making it less feasible for real-time deployment.
U-Net and YOLO from Study21 used for tumor detection and localization achieved 93.0% accuracy, but both models demand high computational power. U-Net’s pixel-wise segmentation approach makes it memory-intensive, while YOLO’s object detection mechanism is costly in terms of processing speed and inference time. These limitations restrict their application in real-world clinical settings.
Deep learning-based methods for classifying breast ultrasound (US) images have been extensively explored, with models like AlexNet, VGG, ResNet, GoogleNet, EfficientNet, and Vision Transformers evaluated on the BUSI dataset22. The Vision Transformer outperformed other models, achieving 88.6% accuracy, 90.1% precision, 87.4% recall, and an 88.7% F1-score. However, transformers require significant computational power and extensively labeled data, limiting their real-world applicability. In contrast, MobNAS achieves 8.4% higher accuracy while being lightweight and suitable for deployment on lower-end devices.
Object detection-based models, particularly YOLO architectures, have been widely used for mass detection in mammography images23. A study comparing YOLOv3, YOLOv4, YOLOv5, and a Swin Transformer-enhanced YOLO model found that YOLOv5 with a Swin Transformer backbone on 832 × 832 images performed best, achieving 94% accuracy, 94% precision, 92% recall, and a 93% F1-score. While YOLO excels in detecting masses, it requires more pre-processing and computational resources than classification-based models. MobNAS surpasses YOLOv5 in accuracy by 3% and is optimized specifically for classification, making it more efficient for breast cancer detection.
Few-shot learning approaches have been explored to address the limitations of small medical datasets, leveraging meta-learning methods like Prototypical Networks and MAML24. By using ResNet50 as the backbone, the model achieved 88.2–88.9% accuracy, improving 6.27–7.10% over baseline models. Despite this improvement, few-shot learning struggles with generalization and often requires specialized training strategies for new datasets. In comparison, MobNAS achieves an additional 8–9% improvement in accuracy without requiring extensive dataset adaptation, making it more scalable and robust.
In contrast, MobNAS achieves 97% accuracy with superior computational efficiency as shown in Table 7. By leveraging MobileNetV2 and NASNetLarge, MobNAS maintains high performance while reducing resource consumption, making it the most practical and deployable solution for real-time applications.
Adding further comparisons beyond these would require significant additional experimentation and may exceed the intended scope of this work, which focuses on proposing a novel and lightweight ensemble architecture (MobNAS) specifically optimized for resource-constrained environments. Nevertheless, we remain committed to future work that will include additional SOTA models as part of broader benchmarking across different datasets and tasks.
Conclusion
In this study, we put forward a new model name MobNAS, based on a lightweight and high-accuracy model combination of MobileNetV2 and NASNetLarge to classify multi-class breast cancer. The model was trained on a dataset of 1,578 ultrasound images, yielding an accuracy of 97%, MAE of 0.05, MCC of 0.95. Such results are promising for diagnostic use, particularly in resource-poor settings.
MobNAS obtains higher performance than well-known models such as VGG16, ResNet while being computationally cheap. While this seems promising, more experiments are needed to eliminate misclassification induced by the similar textures of images. Future research efforts should target working on cross the datasets validation, improving fusion methods and considering further pre-processing techniques to enhance robustness.
Data availability
The dataset and code used to support the findings of this study is available at https://github.com/SaqibSheikhSaqi/MobNas/blob/main/BCancer_MOBILENET_NASNET.ipynb.
References
Mohiud din dar, G. et al. A novel framework for classification of different Alzheimer’s disease stages using CNN model. Electronics 12(2), 469 (2023).
Raza, A. et al. A hybrid deep learning-based approach for brain tumor classification. Electronics 11(7), 1146 (2022).
Mehak, S. et al. Automated grading of breast cancer histopathology images using multilayered autoencoder. Comput. Mater. Contin. 71(2), 3407 (2022).
Kaushik, H. et al. Diabetic retinopathy diagnosis from fundus images using stacked generalization of deep models. IEEE Access 9, 108276–108292 (2021).
Guefrachi, S. et al. Automated diabetic retinopathy screening using deep learning. Multimed. Tools Appl. 83(24), 65249–65266 (2024).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249 (2021).
Ray, R. K. et al. Transforming breast cancer identification: An in-depth examination of advanced machine learning models applied to histopathological images. J. Comput. Sci. Technol. Stud. 6(1), 155–161 (2024).
Clancy, E. ACS report shows prostate cancer on the rise, cervical cancer on the decline. Renal & Urology News (2023).
Marmot, M. G. et al. The benefits and harms of breast cancer screening: An independent review. Br. J. Cancer 108(11), 2205–2240 (2013).
AsgariTaghanaki, S. et al. Deep semantic segmentation of natural and medical images: A review. Artif. Intell. Rev. 54, 137–178 (2021).
Khan, R. H. et al. A comparative study of machine learning algorithms for detecting breast cancer. In 2023 IEEE 13th Annual Computing and Communication Workshop and Conference (CCWC) (IEEE, 2023).
Kriti, et al. PCA-PNN and PCA-SVM based CAD systems for breast density classification. In Applications of Intelligent Optimization in Biology and Medicine: Current Trends and Open Problems 159–180 (Springer, 2015).
Li, H. et al. Breast masses in mammography classification with local contour features. Biomed. Eng. Online 16, 1–12 (2017).
Yassin, N. I. et al. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: A systematic review. Comput. Methods Programs Biomed. 156, 25–45 (2018).
Qiu, Y. et al. A new approach to develop computer-aided diagnosis scheme of breast mass classification using deep learning technology. J. Xray Sci. Technol. 25(5), 751–763 (2017).
Suzuki, K. Overview of deep learning in medical imaging. Radiol. Phys. Technol. 10(3), 257–273 (2017).
Hai, J. et al. Fully convolutional densenet with multiscale context for automated breast tumor segmentation. J. Healthc. Eng. 2019(1), 8415485 (2019).
Humayun, M. et al. Framework for detecting breast cancer risk presence using deep learning. Electronics 12(2), 403 (2023).
Saidul, K. et al. The utility of a deep learning-based approach in Her-2/neu assessment in breast cancer (2023).
Xiao, M. et al. Convolutional neural network classification of cancer cytopathology images: Taking breast cancer as an example. In Proceedings of the 2024 7th International Conference on Machine Vision and Applications (2024).
Rahman, M. M. et al. Breast cancer detection and localizing the mass area using deep learning. Big Data Cogn. Comput. 8(7), 80 (2024).
Pacal, İ. Deep learning approaches for classification of breast cancer in ultrasound (US) images. J. Inst. Sci. Technol. 12(4), 1917–1927 (2022).
Coşkun, D. et al. A comparative study of YOLO models and a transformer-based YOLOv5 model for mass detection in mammograms. Turk. J. Electr. Eng. Comput. Sci. 31(7), 1294–1313 (2023).
Işık, G. & Paçal, İ. Few-shot classification of ultrasound breast cancer images using meta-learning algorithms. Neural Comput. Appl. 36(20), 12047–12059 (2024).
Ozdemir, B. & Pacal, I. An innovative Deep learning framework for skin cancer detection employing ConvNeXtV2 and focal self-attention mechanisms. Res. Eng. 25, 103692 (2025).
Haq, I. et al. Exploring machine learning classifiers for breast cancer classification. KSII Trans. Internet Inf. Syst. TIIS 18(4), 860–880 (2024).
Hu, H. et al. Breast cancer histopathological images recognition based on two-stage nuclei segmentation strategy. PLoS ONE 17(4), e0266973 (2022).
Gengtian, S., Bing, B. & Guoyou, Z. Efficientnet-based deep learning approach for breast cancer detection with mammography images. In 2023 8th International Conference on Computer and Communication Systems (ICCCS) (IEEE, 2023).
Nalifabegam, J. et al. Cancer classification revolution: Employing advanced deep CNNs for multi-class detection of breast irregularities. In 2023 Third International Conference on Smart Technologies, Communication and Robotics (STCR) (IEEE, 2023).
Acknowledgements
This work was supported by King Saud University, Riyadh, Saudi Arabia, through Researchers Supporting Project number (RSP2025R184).
Author information
Authors and Affiliations
Contributions
Habib Hamam and Tariq Shahzad perform the Original Writing Part, Software, and Methodology; Sheikh Muhammad Saqib and Tehseen Mazhar perform Rewriting, investigation, design Methodology, and Conceptualization; Habib Hamam, Muhammad Iqbal and Ahmad Almogren perform the related work part and manage results and discussions; Yazeed YasinGhadi, Mamoon M. Saeed and Tehseen Mazhar perform related work part and manage results and discussion; Habib Hamam ,Yazeed YasinGhadi, Mamoon M. Saeed and Tariq Shahzad perform Rewriting, design Methodology, and Visualization; Sheikh Muhammad Saqib and Ahmad Almogren performs Rewriting, design Methodology, and Visualization.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The dataset used in this study is publicly available and fully anonymized, with no personally identifiable information. All data are de-identified in compliance with ethical standards for medical imaging research.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shahzad, T., Saqib, S.M., Mazhar, T. et al. MobNas ensembled model for breast cancer prediction. Sci Rep 15, 18238 (2025). https://doi.org/10.1038/s41598-025-01920-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-01920-4