Abstract
Chronic rhinosinusitis with nasal polyps (CRSwNP) is a prevalent inflammatory disease where immunomodulation plays a pivotal role. However, immuno-transcriptomic characteristics and its clinical relevance remains largely known. We analyzed transcriptome data of 48 patients with CRSwNP and 34 healthy control subjects from different cohorts and investigated the immuno-transcriptomic characteristics. Differential immune-related genes (DIRGs) were identified and subjected to enrichment analysis. Protein–protein interaction (PPI) networks were constructed to identify hub genes. The least absolute shrinkage and selection operator (LASSO) regression model and multivariate support vector machine recursive feature elimination (mSVM-RFE) were used to identify potential biomarkers, which were validated using the real time quantitative polymerase chain reaction (RT-PCR) and immunohistochemistry (IHC). Infiltration abundance of immune cells in the microenvironment were estimated using CIBERSORT algorithm. Our study identified a total of 660 differentially expressed genes (DEGs) and 81 differentially immune-related genes (DIRGs) in CRSwNP compared to controls. Functional enrichment analysis revealed that the DIRGs were primarily associated with cell chemotaxis and leukocyte migration, and cytokine-cytokine receptor interaction. Through machine learning, we further identified five candidate genes, CXCR1, CCL13, CCR3, PPBP, and MMP9. These five potential CRSwNP biomarkers were experimentally verified in our in-house cohort. Analysis of immune cell infiltration landscape revealed significant variations in the abundance of macrophages and mast cells between CRSwNP and healthy control. Our findings illuminate the significance of immune characteristics in CRSwNP pathogenesis. Future studies focusing on these candidate genes can help elucidate the underlying mechanisms and identify potential therapeutic targets for CRSwNP.
Similar content being viewed by others
Introduction
Chronic rhinosinusitis (CRS) ranks among the most prevalent chronic inflammatory conditions, impacting around 10% of people globally, leading to significant economic challenges1,2. The primary symptoms of CRS include nasal congestion, mucopurulent nasal discharge, facial pain/pressure/fullness, and a reduced sense of smell3. The current staging of CRS is complex and most often divided into two phenotypes: CRS with nasal polyps (CRSwNP) and CRS without nasal polyps (CRSsNP)1,4. However, the cytokine combinations of inflammatory types in patients with CRSwNP vary considerably from region to region, and there are geographic variations even within a single country5. The aim of endophenotyping chronic uncontrolled moderate-to-severe airway disease (e.g., CRS) is to optimize and individualize treatment and to reduce the risk to the patient due to incorrect treatment6.
Nasal polyps encompass various mechanisms, such as the transition of epithelial to mesenchymal cells, the development of fibrosis, and localized immune responses involving lymphocytes. These immune responses include the synthesis of antibodies like IgA, IgM, IgE, and IgG, in addition to the activation of cells such as neutrophils, eosinophils, and mast cells7,8. In addition, defective sinus epithelial cell barriers can lead to increased exposure to pathogenic bacteria, while colonizing bacteria are believed to have a significant impact on disease development9. Chronic rhinosinusitis with nasal polyps (CRSwNP) is commonly associated with asthma and allergic rhinitis, and the inflammatory response caused by a variety of cells and cytokines aggravates the progression of CRSwNP, and further resolution of the inflammatory pathways may hold new promise for the treatment of CRSwNP. Immunomodulation plays a crucial role in the management of CRSwNP. Several studies have shown that a variety of immune cells such as eosinophils, neutrophils, macrophages and immune-related genes play important roles in the pathogenesis of CRSwNP. Further screening of immune-related genes is important for understanding the pathogenesis of the disease and discovering new therapeutic targets1,10,11.
Currently, machine learning is increasingly used in bioinformatics to mine potential mechanisms, prospective biomarkers and therapeutic targets for various diseases12. In this study, we further identified different immune-related genes in CRSwNP through machine learning. In addition, we explored the potential relationship between immune cells and CRSwNP.
Methods
Data collection
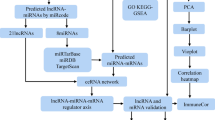
The dataset used in this study was obtained from the GEO database. The search terms were “(Chronic [All Fields] AND Rhinosinusitis[All Fields] AND (‘nasal polyps’[MeSH Terms] OR Nasal Polyps[All] Fields])) AND “Homo sapiens”[porgn]”. The GSE136825 and GSE36830 datasets were retrieved and collected. GSE136825 dataset consists of 28 inferior turbinate tissues collected from healthy controls and 42 nasal polyp tissues from patients with CRSwNP. The GSE36830 dataset consists of 6 nasal polyp samples and 6 normal control samples. The GSE136825 is the training dataset, and the GSE36830 is the validation dataset. We acquired data pertaining to immune-related genes (IRGs) from the ImmPort database, which is accessible at https://www.immport.org/shared/. From this resource, we were able to gather a total of 1509 IRGs for our research. The flow diagram analyzed in this study is shown in Fig. 1.
Differential expression analysis
The differentially expressed genes (DEGs) between the CRSwNP and inferior turbinate tissues were screened using the R package ‘limma’13. Differential expression analysis was performed using the limma package, as previously described in studies of immune-related disorders14.The results were subsequently filtered using the following parameters: P-value less than 0.05. In total, 660 differentially expressed genes (DEGs) and 1509 immune-related genes (IRGs) were subsequently intersected to identify the differentially expressed immune-related genes (DIRGs).
Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses
Genes were analyzed for GO and KEGG pathway enrichment using the R package “Cluster Profiler”15. The enrichment results were visualized using the R package “ggplot2”.
Protein–protein interaction analysis
We obtained all human PPIs from the STRING (https://string-db.org/) database16. The PPI network was then constructed by Cytoscape 3.9.1 (https://cytoscape.org/). We used the cytoHubba plugin to identify the top ten hub genes.
Construction of an DIRG prediction model for CRSwNP
DIRGs with |log2-fold change (FC)|> 2 were selected, and obtained by Spearman correlation analysis of the correlation between these genes. To identify potential diagnostic biomarkers for CRSwNP in differentially expressed genes, we implemented two machine learning algorithms, Least Absolute Shrinkage and Selection Option (LASSO) logistic regression and Multiple Support Vector Machine Recursive Feature Elimination (mSVM-RFE).
LASSO is executed using the R package “glmnet”, which is a regression analysis algorithm that uses regularization for variable selection17. mSVM-RFE stabilizes the feature ordering by using a resampling technique in each iteration and identifies the most relevant features by removing the feature vectors generated by SVM through supervised machine learning techniques18. We performed the mSVM-RFE algorithm using the “e1071” R package to filter out the best variables.Ultimately, the candidate genes identified by the two algorithms were integrated to derive potential prognostic genes for CRSwNP.
Patient recruitment
32 subjects were recruited for the study, including 20 CRSwNP patients and 12 healthy controls. They received either functional endoscopic sinus surgery or inferior turbinoplasty. We collected nasal polyps from CRSwNP patients and inferior turbinates from control patients who underwent septoplasty for deviated septum. The diagnosis of CRSwNP patients was determined according to the European Rhinitis and Nasal Polyps Position Paper 2020. Exclusion criteria included severe systemic disease, immunodeficiency, fungal sinusitis, and pregnancy. Participants had not received systemic or topical corticosteroids 4 weeks prior to study entry. In addition, patients receiving biologics or immunotherapy were explicitly excluded from the study. The subjects in this study signed an informed consent form prior to participation in the study. The Ethics Committee of the Second Affiliated Hospital of Harbin Medical University reviewed and approved the study protocol. We confirmed that all experiments were performed in accordance with relevant guidelines and regulations.The demographic characteristics of the subjects who participated in this study are shown in Table 1.
Cell culture and cell transfection
The human nasal epithelial cell line (HNEpC) was obtained from Procell (Catalogue No. CP-H252) and cultured in Human Nasal Mucosal Epithelial Cell Complete Medium (No. CM-H252), and the cells were incubated in an incubator with 5% CO2 at 37 °C. The cells were then transfected with CXCR1 small interfering RNA or PPBP overexpression plasmid (GenePharma, China). We stimulated the cells by using CXCR1 small interfering RNA or PPBP overexpression plasmid (GenePharma, China) and used them for subsequent experimental analyses 48 h after transfection. After the cell was stabilised, 30 μg/mL LPS was added to stimulate the cells, and the cells were incubated for 24–48 h.
Real time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from nasal polyp tissues and control mucosa using TRIzol (Invitrogen, USA) and reverse transcribed using PrimeScript RT kit (TaKaRa, Dalian, China). Subsequently, RT-qPCR was repeated three times using FastStart Universal SYBR Green Master (ROX) (Roche) with GAPDH as an internal control. The relative mRNA levels were calculated using the 2(-Delta Delta CT) method. The primers used for RT-qPCR are listed in Table 2.
Western blot
Total protein was isolated using RIPA lysates with PMSF. Total protein concentration was determined using a BCA kit (Biorab). Protein samples (20 μg) were electrophoresed in an SDS-PAGE gel and transferred to a PVDF membrane. The membranes were closed in 5% skimmed milk for 1 h at room temperature and incubated overnight at 4 °C with the following primary antibodies: CXCR1 (1:500; Servicebio, GB11625-100), PPBP (1:50; Proteintech, 13313-1-AP), GAPDH (1:3000; Servicebio, GB15004-100). On the second day, the PVDF membrane was cleaned three times, and after incubation with secondary antibodies, the binding of target proteins was observed by the ECL system and then analysed using Image J software.
Immunohistochemistry
The tissue was embedded in paraffin, sliced into 4-μm-thick slices, and dewaxed after drying at 37 °C, followed by antigen retrieval. Protein was blocked with goat serum and incubated with primary antibody at 4 °C overnight. The primary antibodies were as follows: CXCR1 (1:500; Servicebio, GB11625-100), CCL13(1:1000; Proteintech, Ag24131),PPBP(1:50; Proteintech, 13313-1-AP),CCR3(1:200; Proteintech, 22351-1-AP), MMP-9(1:500; Proteintech, 10375-2-AP), GAPDH(1:3000; Servicebio, GB15004-100). And biotinylated secondary antibody (goat anti-rabbit) at 24 °C for 1 h. Finally, the reactions were developed with DAB, hematoxylin counterstaining, dehydrated, mounted.
Enzyme-linked immunosorbent assay (ELISA)
An appropriate amount of tissue block was taken, washed in pre-cooled PBS (0.02 mol/L, pH 7.0–7.2) to remove blood, and ground thoroughly according to the ratio of 1 g of sample to 5 ml of PBS, the prepared homogenate was centrifuged at 5000 × g for 5 min, and the supernatant was retained for assay. The concentrations of IFN-γ, IL-4, IL-5, IL-13 and IL-17A in the supernatant were measured by Human ELISA Kit using an enzyme marker (PerkinElmer, Waltham, MS, USA) according to the manufacturer’s protocol (Multi Sciences, Guangzhou, China). The concentration of il-8 (Multi Sciences, Hangzhou, China) in the supernatant of HNEpCs cell cultures was measured by ELISA kit using an enzyme marker.
Distribution of immune cells in CRSwNP
CIBERSORT is a deconvolution algorithm that quantifies immune cell infiltration in CRSwNP gene expression profiles19. We compared the distribution of 22 immune cells in GSE136825 and GSE36830 between the CRSwNP group and the normal control samples by the CIBERSORT calculation tool. Pearson correlation was used to analyze the relationship between the DIRGs and infiltrating immune cells.
Statistical analysis
Student’s t-test was used to compare the two groups. All quantitative experimental data were obtained from at least three replicates and expressed as mean ± standard deviation. P values less than 0.05 were considered statistically significant. Statistical analyses were performed using Prism 9.0 software (GraphPad software Inc. CA, USA).
Results
Expression of DIRGs in CRSwNP
We initially identified differentially expressed genes (DEGs) between CRSwNP and controls using the R package, and 660 DEGs were identified in total. Heatmaps and volcano maps of the top 50 DEGs are shown in Fig. 2A and B, respectively. To further investigate the immune-related genes, we intersected these 660 DEGs with 1509 immune-related genes (IRGs) in the ImmPort database and obtained 81 differentially expressed immune-related genes (DIRGs) (Fig. 2C).
Functional enrichment analysis and PPI analysis
The GO enrichment analysis indicated that the DIRGs were mainly involved in cell chemotaxis and leukocyte migration. Furthermore, the cellular components of these genes were primarily localized to the external side of the plasma membrane. The analysis of molecular function revealed a correlation between these genes and receptor ligand activity as well as signaling receptor activator activity (Fig. 3A). KEGG pathway analysis indicated that these genes were mainly enriched in signal pathways such as cytokine-cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor, and chemokine signaling pathway (Fig. 3B). Protein interactions of 81 DIRGs were analyzed using the STRING database to obtain PPI networks with interaction scores greater than 0.4 (Fig. 3C). The network genes were then clustered using the cytoHubba plug-in in Cytoscape software. A total of 10 hub genes were obtained by constructing PPI networks and identifying hub genes (Fig. 3D). At last, volcano and heat maps of the hub genes were generated (Fig. 3E, F).
Functional enrichment of DIRGs and the PP1 analysis. (A) Gene Ontoley annotation of DIRGs. (B) Kyoto Encyclopedia of Genes and Genomes annotation of DIRGs. (C) PPI networks generated with DIRGs using STRING. (D) PPI network diagram of hub genes. (E) Heatmap of hub genes. (F) Volcano plot of hub genes.
Correlations between the expression levels of the DIRGs in CRSwNP
To further analyze the correlations between the expression levels of the differentially expressed immune response genes (DIRGs), we conducted correlation analysis. We used the ‘ggcorrplot’ package in R to visualize the correlation analysis results as a network diagram (Fig. 4A) and heatmap (Fig. 4B). Scatter plots (Fig. 4C–F) were generated to display the correlation between the four most highly correlated groups of genes in CRSwNP. Our findings revealed a strong correlation between CXCR1 and CXCR2, as well as CXCR6.
Verification of diagnostic biomarkers
Candidate genes were screened by two different algorithms. As shown in Figs. 5A, and B, the optimal λ was selected using tenfold cross-validation. We utilized the LASSO logistic regression algorithm to identify 7 candidate genes from DRIPs. The mSVM-RFE model was used to narrow down 10 DIRGs, and 8 DIRGs were selected as candidate genes (Fig. 5C, D). 5 overlapping candidate genes (CXCR1, CCL13, PPBP, CCR3, and MMP9) of these two algorithms were finally selected (Fig. 5E). We further analyzed and validated the five candidate genes. The results of the principal component analysis (PCA) showed (Fig. 6A) that these five candidate genes could clearly distinguish CRSwNP from controls, suggesting that they may play a key role in the diagnosis of CRSwNP. Figure 6B shows the chromosomal locations of CXCR1, CCL13, PPBP, CCR3, and MMP9. Moreover, the prediction ability of the five candidate genes was further tested in the training group (GSE136825) and validation group (GSE36830). In the GSE136825 dataset, the expression levels of CXCR1, CCL13, CCR3, and MMP9 in CRSwNP samples were notably upregulated in contrast to controls, while the expression of PPBP was downregulated (Fig. 6C). In the GSE36830 dataset, compared with controls, CXCR1, CCL13, and CCR3 expression levels were upregulated, while PPBP and MMP9 expression levels were not significantly different between CRSwNP and controls (Fig. 6D).
Candidate genes were screened based on the LASSO regression model and the mSVM-RFE model. (A) Partial likelihood deviance versus log (λ) was drawn using LASSO Cox regression model. (B) LASSO regression coefficient profiles of the 10 DIRGs. (C) The curve of the total within sum of squared error curve under corresponding cluster number k, and it reached the “elbow point” when k = 5. (D) The curve of average silhouette widthunder corresponding cluster number k, and the maximum of average silhouette width was achieved when k = 5. (E) Venn plots show the candidate genes by overlapping the genes selected from the LASSO regression model and the mSVM-RFE model.
Candidate genes were further analyzed and validated. (A) Principal component analysis (PCA) of candidate genes expression profiles. (B) Chromosomal locations of candidate genes. (C,D) The expression level of five candidate genes between CRSwNP and controls from GSE136825 dataset and GSE36830. (*P < 0.05, **P < 0.01, ***P < 0.001).
RT-qPCR results showed that the expression levels of CXCR1, CCL13, CCR3, and MMP9 were significantly higher in CRSwNP compared to the normal mucosa, while the expression level of PPBP was significantly reduced (Fig. 7A–E). In order to better elucidate the relationship between immune signature genes and inflammatory endotypes, we used ELISA kits to measure the levels of IFN-γ, IL-4, IL-5, IL-13, and IL-17A in nasal polyp tissues.The patients were classified into T1, T2, and T3 based on the level of cytokines, and the expression level of immune signature genes was compared among the different subtypes. It was found that CXCR1 expression was higher in polyps of patients with type T1, CCL13 and CCR3 expression was higher in type T2 inflammation, PPBP expression was lower in patients with type T2 inflammation, and MMP-9 expression was higher mainly in patients with type T3 inflammation (Fig. 7F–J).
Gene expression levels of candidate genes. Gene expression levels of CXCR1 (A), CCL13 (B), PPBP (C), CCR3 (D) and MMP9 (E) in CRSwNP and control samples. Gene expression levels of CXCR1 (F), CCL13 (G), PPBP (H), CCR3 (I) and MMP9 (J) in different inflammatory endotypes of CRSwNP. (*P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant).
Immune histochemistry (IHC) technique was used to compare and analyse the protein expression levels of candidate genes in normal mucosal tissues and chronic rhinosinusitis with nasal polyps (CRSwNP) tissues. Quantitative analysis showed that the expression intensity of CXCR1, CCL13, CCR3 and MMP9 proteins was significantly higher in the CRSwNP group compared with the normal control group, whereas the expression level of PPBP proteins showed a significant down-regulation. Typical staining results and quantitative analysis data are shown in Fig. 8.
Candidate gene protein expression levels in normal mucosal tissues and chronic rhinosinusitis with nasal polyps (CRSwNP) tissues were analysed using IHC staining. Unpaired t-tests were used to determine the significant differences for candidate gene protein expression between the normal mucosal(n = 5) and CRSwNP (n = 5) groups. (A) CXCR1. (B) CCL13. (C) PPBP. (D) CCR3. (E) MMP9. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
CXCR1 and PPBP are mainly associated with non-T2 inflammation, and neutrophil infiltration is an important feature of non-T2 inflammation. In order to further investigate their regulatory functions on neutrophil regulation, we knocked down CXCR1 or overexpressed PPBP in human nasal mucosa cell lines, induced the cells with LPS, and detected the level of neutrophil regulatory factor IL-8 released from epithelial cells. It was found that knockdown of CXCR1 decreased the level of IL-8 secretion from LPS-induced epithelial cells, while overexpression of PPBP increased the level of IL-8 secretion (Fig. 9).
Effects of CXCR1 and PPBP on IL-8 secretion. (A) HNEpC cells were transfected with siRNA (targeting CXCR1), and CXCR1 expression in transfected cells was detected by RT-qPCR. (B) Detection of CXCR1 protein expression by western blot. (C) Detection of IL-8 in the supernatant of CXCR1 knockdown cells using an ELISA kit. (D) HNEpC cells were transfected with PPBP overexpression plasmid and PPBP expression was detected by RT-qPCR in the transfected cells. (E) PPBP protein expression was detected by western blot. (F) The level of IL-8 in the supernatant of PPBP overexpressing cells was detected using ELISA kit.
Immune cell infiltration landscape
Our findings revealed significant variations in the abundance of macrophages and mast cells between the two groups (Fig. 10A, B). CCL13 was positively regulated with M2-type macrophages, MMP9 was negatively regulated with M2-type macrophages, and CXCR1 was positively regulated with neutrophils and mast cells (Fig. 10C).
Differences of the infiltrate immune cells between the CRSwNP and controls. (A) Plot of the proportioin of 22 immune cells in all samples. (B) Comparison of immune cell infiltration in CRSwNP group and controls. (C) The correlation between 22 infiltrating immune cells and the correlation between immune cells and candidate genes. (*P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
CRSwNP is characterised by chronic mucosal inflammation, and its typical clinical manifestations include nasal congestion, impaired sense of smell and increased nasal secretions. This condition also imposes a significant financial burden on patients due to its high recurrence1. Current studies are delving deeper into the immune regulation of CRSwNP. The comprehension of the pathophysiological concepts associated with the role of inflammatory cytokines in CRSwNP has gradually progressed. Although Th2 cytokines linked to eosinophilic infiltration remain pivotal in the formation of polyps, recent studies have also examined additional cytokines, resulting in a more detailed and intricate characterization of CRSwNP20. Wang et al. study identified inflammatory biomarkers combining typical remodeling factors in the CRS endotype21. The differentiation and characterization of these endotypes will facilitate CRS-targeted therapeutic decisions. Additionally, dysregulation of the host immune system is believed to play a significant role in the development of CRSwNP. Defects in upper airway epithelial innate immune function have been reported to contribute to the initial inflammatory response that leads to CRSwNP22. In addition, group 2 innate lymphoid cells (ILC2) play a key role in the generation and maintenance of immunity. These cells can produce a substantial amount of type 2 cytokines, including IL-4, IL-5, and IL-1323.
Previous studies have identified several differential genes associated with CRSwNP, which are enriched in multiple immune-related pathways and may play a significant role in the development of CRSwNP24,25. Therefore, further screening of these differentially expressed immune-related genes (DIRGs) can be valuable in identifying potential therapeutic targets. In our study, we conducted GO enrichment analysis and found that the differentially expressed immune response genes (DIRGs) were primarily associated with cell chemotaxis and leukocyte migration. Additionally, KEGG pathway analysis indicated that cytokine-cytokine receptor interactions are important in the development of CRSwNP. This is consistent with previous research that has shown the significance of immune response and signal transduction in the pathogenesis of CRSwNP26. Therefore, it is essential to screen for DIRGs. We identified five DIRGs in our study, namely CXCR1, CCL13, PPBP, CCR3, and MMP9. Chemokines and their receptor pathways have irreplaceable roles in innate and adaptive immune responses that can promote the activation and transport of immune cells.
CXCR1 has been found to play an important role in the pathogenesis of certain lung diseases such as COPD, asthma and pulmonary fibrosis27. CXCR1 activates neutrophils by binding to specific ligands, such as IL-8, and is responsible for various neutrophil functions, which include chemotaxis, intracellular calcium changes, and phospholipase D activation28. CXCR1/2 is also expressed in airway smooth muscle and contributes to cell contraction and migration, which enhances airway responsiveness and remodeling observed in asthma29. However, there have been no relevant studies conducted on CRSwNP. In this study we found that CXCR-1 expression was elevated in nasal polyp tissues and significantly positively correlated with neutrophil infiltration, which may play a role in non-eosinophilic nasal polyps. The present study also found that knockdown of CXCR1 was seen to reduce IL-8 secretion by nasal mucosal epithelial cells. We will continue to explore the regulatory relationship between CXCR1 and neutrophils in future studies to further reveal the pathogenesis of neutrophil-infiltrating chronic rhinitis with nasal polyps and to investigate the therapeutic role that CXCR1 inhibitors have in non-type 2 sinusitis.
CCR3 is highly expressed in different immune cells, including eosinophils, and may contribute to the activation and accumulation of inflammatory cells. Studies have shown that increased expression of CCR3 chemokine receptors may be associated with multiple chemokines that contribute to the development of nasal polyps by promoting the migration and long-term accumulation of inflammatory cells, such as eosinophils, in the inflammatory infiltrate of nasal polyps30. The chemokine-C–C motif chemokine receptor 3 (CCR3) axis is important in the aggregation of eosinophils in tissues31. A recent study found that IgE directly affects eosinophils through CD23, increasing eosinophil chemotaxis and enhancing eosinophil inflammation by upregulating CCR3 expression on the eosinophil surface32. The CCR3 gene also plays a significant role in allergic airway inflammation. In a mouse model of allergic rhinitis, down-regulation of CCR3 expression inhibited histopathologic lesions and eosinophil infiltration in the nasal cavity, resulting in reduced serum Th2 cytokine production and alleviation of allergic symptoms33. In this study, we observed a significant upregulation of CCR3 expression in nasal polyps, which positively correlated with eosinophils. Eosinophilic nasal polyps are the ones with the highest recurrence rate, and CCR3 may play an important role in eosinophil chemotaxis to promote the recurrence of nasal polyps.
PPBP, also known as NAP-2/CXCL7, belongs to the CXCL subgroup and is released by activated platelets. Studies have reported that it is associated with the development of different types of tumors and can serve as an adjunctive diagnostic marker for tumors34. In a study by Liu et al. it was found that CXCL7 caused severe astrocyte damage, activated microglia, and increased infiltration of neutrophils and macrophages, resulting in secondary demyelination35. Krishna et al. reported that when several chemokines, including PPBP, bind to their corresponding receptors, it triggers a G-protein or β-inhibitory protein-coupled signaling pathway, which plays a crucial role in neutrophil transport and activation36. In the present study, we found that high expression of PPBP promoted the secretion of IL-8 in the nasal mucosal epithelium, further confirming that PPBP can promote neutrophil infiltration. Previous studies have observed reduced blood levels of PPBP in Parkinson’s disease37, but no studies have investigated this in CRSwNP. We discovered a negative correlation between PPBP and regulatory T cells (Tregs), but further experiments are required to validate the role of PPBP in CRSwNP.
CCL13/MCP-4, a member of the CC chemokine family, has been found to induce chemotaxis in various immune cells38. In addition, CCL13 can stimulate eosinophil degranulation in tissues, promote histamine release from basophils, increase the expression of adhesion molecules and accelerate the secretion of pro-inflammatory cytokines38. Another study found that nasal CCL13 can also exacerbate nasal mucosal allergy39. CCL13 participates in a variety of chronic inflammatory diseases. In this study we found that CCL13 expression was significantly upregulated in nasal polyps. Currently, small molecule antagonists targeting chemokines are being tested in clinical trials for various inflammation-mediated diseases, such as asthma and chronic obstructive pulmonary disease. These antagonists show promise as potential new therapeutic targets for CRSwNP in the future.
Tissue remodeling plays a critical role in chronic rhinosinusitis (CRS). MMPs, a family of zinc-dependent protein hydrolases, are responsible for degrading various components of the extracellular matrix (ECM) and regulating remodeling40. Multiple studies have confirmed that MMP-9 levels are increased in CRSwNP41,42. Eosinophil-derived neurotoxin (EDN) is an eosinophil granule protein closely associated with allergic inflammation. EDN stimulates the production of MMP-9 in the nasal epithelium, leading to epithelial remodeling. This process may be implicated in the development of stubborn nasal polyps43. Our study also observed elevated levels of MMP-9 in nasal polyps, consistent with previous findings. Currently, CRSwNP endotypes based on different cellular and cytokine infiltration has been widely recognized to provide effective guidance for clinical treatment44. In this study, we screened nasal polyp immunosignature genes could further resolve the mechanism of nasal polyp endophenotyping, and assisted in the assessment of different inflammatory types by detecting the gene expression levels in nasal polyp tissues to provide individualized protocols for different types of nasal polyps.
In this study, significant differences were observed in the distribution of M2 macrophages and mast cells compared to the control group, while no significant differences were found in the distribution of other immune cells. A single-cell sequencing analysis revealed that ALOX15+cDC2s and macrophages played crucial roles in the development of type 2 immunity in eCRSwNP45. Additionally, another finding suggested that MCs in nasal polyps exhibited distinct effect programs associated with inflammation46. However, it is important to acknowledge the limitations of our study. Although the expression level of DIRGs has been validated by RT-Qpcr and IHC, further experiments are required to verify their relevant functions and mechanisms.
In the next step of our future research, we will focus the main direction of our study on the infiltration distribution of immune cell subpopulations in CRSwNP and their interaction with dirg.
Conclusions
Our findings illuminate the significance of immune characteristics in CRSwNP pathogenesis. This discovery provides valuable insights into the underlying mechanisms of CRSwNP and presents potential targets for future therapeutic investigations.
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article. All other data and information in this paper are available from the corresponding author upon request.
References
Kato, A., Schleimer, R. P. & Bleier, B. S. Mechanisms and pathogenesis of chronic rhinosinusitis. J. Allergy Clin. Immunol. 149 (5), 1491–1503. https://doi.org/10.1016/j.jaci.2022.02.016 (2022).
Rudmik, L. Economics of chronic rhinosinusitis. Curr. Allergy Asthma Rep. 17 (4), 20. https://doi.org/10.1007/s11882-017-0690-5 (2017).
Soudry, E., Mace, J., Smith, T. L. & Hwang, P. H. Role of inferior turbinate reduction in the quality of life of patients undergoing endoscopic sinus surgery for chronic rhinosinusitis. Int. Forum Allergy Rhinol. 9 (8), 926–933. https://doi.org/10.1002/alr.22356 (2019).
Wang, M. et al. Distinct type 2-high inflammation associated molecular signatures of chronic rhinosinusitis with nasal polyps with comorbid asthma. Clin. Transl Allergy. 10, 26. https://doi.org/10.1186/s13601-020-00332-z (2020).
Wang, X. et al. Diversity of TH cytokine profiles in patients with chronic rhinosinusitis: A multicenter study in Europe, Asia, and Oceania. J. Allergy Clin. Immunol. 138 (5), 1344–1353. https://doi.org/10.1016/j.jaci.2016.05.041 (2016).
Bachert, C. et al. Endotypes of chronic rhinosinusitis with nasal polyps: Pathology and possible therapeutic implications. J. Allergy Clin. Immunol. Pract. 8 (5), 1514–1519. https://doi.org/10.1016/j.jaip.2020.03.007 (2020).
Fokkens, W. J., Lund, V. J. & Hopkins, C. et al. European position paper on rhinosinusitis and nasal polyps 2020. Rhinology 58 (Suppl S29), 1–464. https://doi.org/10.4193/Rhin20.600 (2020).
Wang, Z. C. et al. Extrafollicular PD-1highCXCR5-CD4 + T cells participate in local immunoglobulin production in nasal polyps. J. Allergy Clin. Immunol. 149 (2), 610–623. https://doi.org/10.1016/j.jaci.2021.06.023 (2022).
Stevens, W. W., Schleimer, R. P. & Kern, R. C. Chronic rhinosinusitis with nasal polyps. J. Allergy Clin. Immunol. Pract. 4 (4), 565–572. https://doi.org/10.1016/j.jaip.2016.04.012 (2016).
Xie, X. et al. IL-1β-induced epithelial cell and fibroblast transdifferentiation promotes neutrophil recruitment in chronic rhinosinusitis with nasal polyps. Nat. Commun. 15 (1), 9101. https://doi.org/10.1038/s41467-024-53307-0 (2024).
Iwasaki, N. et al. Single cell RNA sequencing of human eosinophils from nasal polyps reveals eosinophil heterogeneity in chronic rhinosinusitis tissue. J. Allergy Clin. Immunol. 154 (4), 952–964. https://doi.org/10.1016/j.jaci.2024.05.014 (2024).
Kumar, N., Narayan Das, N., Gupta, D., Gupta, K. & Bindra, J. Efficient automated disease diagnosis using machine learning models. J. Healthc. Eng. https://doi.org/10.1155/2021/9983652 (2021).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47. https://doi.org/10.1093/nar/gkv007 (2015).
Deng, Y. J. et al. Immune-related gene IL17RA as a diagnostic marker in osteoporosis. Front. Genet. 14, 1219894. https://doi.org/10.3389/fgene.2023.1219894 (2023).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16 (5), 284–287. https://doi.org/10.1089/omi.2011.0118 (2012).
von Mering, C. et al. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 31 (1), 258–261. https://doi.org/10.1093/nar/gkg034 (2003).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33 (1), 1–22 (2010).
Zhou, X. & Tuck, D. P. MSVM-RFE: Extensions of SVM-RFE for multiclass gene selection on DNA microarray data. Bioinformatics 23 (9), 1106–1114. https://doi.org/10.1093/bioinformatics/btm036 (2007).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12 (5), 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Carsuzaa, F. et al. Cytokine signature and involvement in chronic rhinosinusitis with nasal polyps. Int. J. Mol. Sci. 23 (1), 417. https://doi.org/10.3390/ijms23010417 (2021).
Wang, X. et al. Endotypes of chronic rhinosinusitis based on inflammatory and remodeling factors. J. Allergy Clin. Immunol. 151 (2), 458–468. https://doi.org/10.1016/j.jaci.2022.10.010 (2023).
Li, Y., Wang, W., Ying, S., Lan, F. & Zhang, L. A potential role of group 2 innate lymphoid cells in eosinophilic chronic rhinosinusitis with nasal polyps. Allergy Asthma Immunol. Res. 13 (3), 363–374. https://doi.org/10.4168/aair.2021.13.3.363 (2021).
van der Ploeg, E. K., Carreras Mascaro, A., Huylebroeck, D., Hendriks, R. W. & Stadhouders, R. Group 2 innate lymphoid cells in human respiratory disorders. J. Innate Immun. 12 (1), 47–62. https://doi.org/10.1159/000496212 (2020).
Zhou, X. et al. Identification of key modules, hub genes, and noncoding RNAs in chronic rhinosinusitis with nasal polyps by weighted gene coexpression network analysis. Biomed. Res. Int. 2020, 6140728. https://doi.org/10.1155/2020/6140728 (2020).
Chen, G., Hao, H. & Wang, L. E. Bioinformatics analysis and verification of key candidate genes influencing the pathogenesis of chronic rhinosinusitis with nasal polyps. Am. J. Transl Res. 15 (2), 710–728 (2023).
Bassiouni, A. et al. The global transcriptomic signature in sinonasal tissues reveals roles for tissue type and chronic rhinosinusitis disease phenotype. Rhinology 58 (3), 273–283. https://doi.org/10.4193/Rhin19.403 (2020).
Meng, H. et al. Identification of the key immune-Related genes in chronic obstructive pulmonary disease based on immune infiltration analysis. Int. J. Chron. Obstruct Pulmon Dis. 17, 13–24. https://doi.org/10.2147/COPD.S333251 (2022).
Ha, H., Debnath, B. & Neamati, N. Role of the CXCL8-CXCR1/2 Axis in Cancer and inflammatory diseases. Theranostics 7 (6), 1543–1588. https://doi.org/10.7150/thno.15625 (2017).
Li, X., Li, J., Zhang, Y. & Zhang, L. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. J. Clin. Otorhinolaryngol. Head Neck Surg. 35 (12), 1144–1148. https://doi.org/10.13201/j.issn.2096-7993.2021.12.020 (2021).
Liu, W. et al. Blood transcriptomics reveal systemic eosinophilic and neutrophilic inflammation patterns in patients with nasal polyps. Rhinology 62 (6), 739–749. https://doi.org/10.4193/Rhin24.248 (2024).
Pease, J. E. & Williams, T. J. Tipping the balance: A biased nanobody antagonist of CCR3 with potential for the treatment of eosinophilic inflammation. J. Allergy Clin. Immunol. 143 (2), 552–553. https://doi.org/10.1016/j.jaci.2018.10.052 (2019).
Yu, J. et al. IgE directly affects eosinophil migration in chronic rhinosinusitis with nasal polyps through CCR3 and predicts the efficacy of Omalizumab. J. Allergy Clin. Immunol. 153 (2), 447–460e9. https://doi.org/10.1016/j.jaci.2023.09.041 (2024).
Yuan, J. et al. Gene knockdown of CCR3 reduces eosinophilic inflammation and the Th2 immune response by inhibiting the PI3K/AKT pathway in allergic rhinitis mice. Sci. Rep. 12 (1), 5411. https://doi.org/10.1038/s41598-022-09467-4 (2022).
Li, L. et al. Serum chemokine CXCL7 as a diagnostic biomarker for colorectal cancer. Front. Oncol. 9, 921. https://doi.org/10.3389/fonc.2019.00921 (2019).
Liu, Z. et al. CXCL7 aggravates the pathological manifestations of neuromyelitis optica spectrum disorder by enhancing the inflammatory infiltration of neutrophils, macrophages and microglia. Clin. Immunol. 245, 109139. https://doi.org/10.1016/j.clim.2022.109139 (2022).
Rajarathnam, K., Schnoor, M., Richardson, R. M. & Rajagopal, S. How do chemokines navigate neutrophils to the target site: Dissecting the structural mechanisms and signaling pathways. Cell. Signal. 54, 69–80. https://doi.org/10.1016/j.cellsig.2018.11.004 (2019).
Xing, N. et al. Identification and validation of key molecules associated with humoral immune modulation in Parkinson’s disease based on bioinformatics. Front. Immunol. 13, 948615. https://doi.org/10.3389/fimmu.2022.948615 (2022).
Li, L. et al. CCL13 and human diseases. Front. Immunol. 14, 1176639. https://doi.org/10.3389/fimmu.2023.1176639 (2023).
Baumann, R. et al. Comparison of the nasal release of IL-4, IL-10, IL-17, CCL13/MCP-4, and CCL26/eotaxin-3 in allergic rhinitis during season and after allergen challenge. Am. J. Rhinol Allergy. 27 (4), 266–272. https://doi.org/10.2500/ajra.2013.27.3913 (2013).
Kim, I. S., Yang, W. S. & Kim, C. H. Physiological properties, functions, and trends in the matrix metalloproteinase inhibitors in inflammation-mediated human diseases. Curr. Med. Chem. 30 (18), 2075–2112. https://doi.org/10.2174/0929867329666220823112731 (2023).
Huang, Y., Yan, B., Meng, C., Zhang, L. & Wang, C. Matrix metalloproteinases in chronic rhinosinusitis. Expert Rev. Clin. Immunol. 20 (5), 547–558. https://doi.org/10.1080/1744666X.2024.2302362 (2024).
Lee, K., Tai, J., Lee, S. H. & Kim, T. H. Advances in the knowledge of the underlying airway remodeling mechanisms in chronic rhinosinusitis based on the endotypes: A review. Int. J. Mol. Sci. 22 (2), 910. https://doi.org/10.3390/ijms22020910 (2021).
Huang, X., Liu, Z., Bleier, B. S., Song, Y. & Wu, D. Association of mucus eosinophil-derived neurotoxin levels with disease control status in patients with chronic rhinosinusitis. Eur. Arch. Otorhinolaryngol. 281 (8), 4191–4199. https://doi.org/10.1007/s00405-024-08695-w (2024).
Toppila-Salmi, S. et al. Endotyping in chronic rhinosinusitis—An EAACI task force report. Allergy 6 https://doi.org/10.1111/all.16418 (2024).
Wang, W. et al. Single-cell profiling identifies mechanisms of inflammatory heterogeneity in chronic rhinosinusitis. Nat. Immunol 23 (10), 1484–1494. https://doi.org/10.1038/s41590-022-01312-0 (2022).
Dwyer, D. F. et al. Human airway mast cells proliferate and acquire distinct inflammation-driven phenotypes during type 2 inflammation. Sci. Immunol. 6 (56), eabb7221. https://doi.org/10.1126/sciimmunol.abb7221 (2021).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82473035); Natural Science Foundation of Heilongjiang Province of China (LH2022H009). Heilongjiang Provincial Health Commission Science and Technology Plan(20220707011051)
Author information
Authors and Affiliations
Contributions
Zhaonan Xu: Writing – original draft, Visualization, Methodology, Conceptualization. Qing Hao: Writing – original draft, Formal analysis. Bingrui Yan: Resources, Investigation. Qin Wu, Hongtian Yi, Peng Wang: Resources, Investigation. Xuan Kan, Xianji Shen: Resources, Investigation. Lingmei Qu: Writing – review & editing. Qiuying Li: Writing – review & editing, Funding acquisition.Yanan Sun: Writing – review & editing, Supervision, Funding acquisition, Conceptualization. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Approval was obtained from the ethics committee of the Second Affiliated Hospital of Harbin Medical University(KY2024-033). The procedures used in this study adhere to the tenets of the Declaration of Helsinki. Written informed consent for participation in this study was provided by the participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, Z., Hao, Q., Yan, B. et al. Immuno-transcriptomic analysis based on machine learning identifies immunity signature genes of chronic rhinosinusitis with nasal polyps. Sci Rep 15, 19393 (2025). https://doi.org/10.1038/s41598-025-02508-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-02508-8