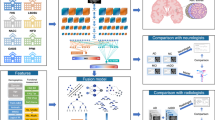
Abstract
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder that significantly impacts cognitive function, posing a major global health challenge. Despite its rising prevalence, particularly in low and middle-income countries, early diagnosis remains inadequate, with projections estimating over 55 million affected individuals by 2022, expected to triple by 2050. Accurate early detection is critical for effective intervention. This study presents Neuroimaging-based Early Detection of Alzheimer’s Disease using Deep Learning (NEDA-DL), a novel computer-aided diagnostic (CAD) framework leveraging a hybrid ResNet-50 and AlexNet architecture optimized with CUDA-based parallel processing. The proposed deep learning model processes MRI and PET neuroimaging data, utilizing depthwise separable convolutions to enhance computational efficiency. Performance evaluation using key metrics including accuracy, sensitivity, specificity, and F1-score demonstrates state-of-the-art classification performance, with the Softmax classifier achieving 99.87% accuracy. Comparative analyses further validate the superiority of NEDA-DL over existing methods. By integrating structural and functional neuroimaging insights, this approach enhances diagnostic precision and supports clinical decision-making in Alzheimer’s disease detection.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is the leading type of dementia, characterized by a gradual decline of brain function primarily observed in older individuals1. Compared to the patient’s prior state of working, there is an indication of a decline in memory and other cognitive skills, which is symptomatic of primary dementia syndrome. Alzheimer’s disease causes about two-thirds of cases of progressive cognitive decline in the elderly population of Saudi Arabia, with an estimated prevalence of over 130,000 individuals2. According to data from 2006, the global prevalence of Alzheimer’s disease (AD) was estimated at 26.6 million individuals3. It is projected that this figure will increase twofold every two decades. Researchers predict that by the year 2046, Alzheimer’s disease will affect about 1.2% of the global population4,5. Early identification of mild cognitive impairment (MCI) plays a crucial role in detecting AD at an early stage, as MCI often precedes full-fledged dementia. Although MCI does not significantly impact daily activities, it carries a high risk of progressing to AD or other types of dementia6. Early and accurate identification of AD, especially tracking MCI progression to AD, allows for timely interventions, lifestyle modifications, and therapeutic strategies that could potentially slow disease progression7.
Diagnosing AD in its early stages remains challenging due to the gradual nature of neurodegeneration and the overlap of symptoms with normal aging. Neuroimaging plays a crucial role in identifying AD-related changes in the brain (see Table 1). MRI and PET scans provide structural and functional insights, helping clinicians detect early biomarkers of AD. Neuronal degeneration and subsequent cell death lead to progressive memory loss, motor impairments, and ultimately, total amnesia. The World Health Organization reported that 50 million people were affected by AD in 2017, with the number expected to rise exponentially. AD progresses through four distinct stages, beginning with pre-dementia (MCI), followed by early-stage, moderate-stage, and advanced-stage dementia. Early diagnosis and staging are crucial, as each phase presents varying degrees of cognitive decline and requires tailored interventions.
Traditional machine learning (ML) approaches, such as support vector machines (SVM), decision trees, and k-nearest neighbors, have been employed in computer-aided diagnosis (CAD) of AD. However, these methods often struggle with high-dimensional neuroimaging data, class imbalance, and limited dataset availability25,26,10. Deep learning (DL), particularly convolutional neural networks (CNNs), has demonstrated superior feature extraction capabilities by learning spatial hierarchies of features from neuroimaging data. Unlike conventional ML models that rely on handcrafted features, DL models automatically capture intricate patterns in MRI and PET scans, improving classification accuracy. However, existing deep learning models primarily focus on binary classification (AD vs. non-AD) and fail to classify AD progression stages accurately.
These limitations make it difficult to create solutions that can be applied broadly and with high accuracy, as they may not effectively represent the diversity of cases and conditions in the broader population11. Many existing studies overlook the need for multi-stage classification of AD, limiting their clinical utility. Additionally, few approaches effectively integrate both structural (MRI) and functional (PET) neuroimaging data to enhance diagnostic accuracy. To address these limitations, this study proposes Neuroimaging-based Early Detection of Alzheimer’s Disease using Deep Learning (NEDA-DL).
The primary aim of our study is to identify and diagnose Alzheimer’s disease in its initial stages. We proposed a methodology to identify whether the patient has Alzheimer’s and further classify the disease into the four stages mentioned above. This research scope is the early diagnosis of AD, with a clear classification of the severity of the disease from early to final. In our proposed methodology, we take magnetic resonance imaging (MRI) from a scanner and then preprocess the image for normalization and smoothing to remove anomalies. After that, we divide the MRI image into black and white pixels. Our Convolutional Neural Network extracts the features from the image. We then train our model to classify dementia into four categories: early stage, moderate stage, and advanced stage. The Alzheimer’s Disease Neuroimaging Initiative (ADNI)12 Data set has been used for this proposed research. Further, our contribution can be summarized as follows:
-
Propose a novel hybrid deep learning model combining ResNet-50 and AlexNet architectures, optimized with CUDA-based parallel processing for enhanced computational efficiency.
-
Unlike conventional binary classification approaches, our model accurately classifies AD into four stages: very mild, mild, moderate, and non-demented categories.
-
Integration of multi-modal neuroimaging data (MRI + PET), leveraging complementary structural and functional insights to improve classification robustness.
-
Employ depth wise separable convolutions, reducing the number of trainable parameters while maintaining high accuracy.
-
Evaluate the performance of the proposed algorithms against existing state-of-the-art algorithms using both simulated and real-world data sets.
The rest of the paper is organized in such a way that section II presents a comprehensive literature review followed by technology background and related work. Section III presents the proposed NEDA-DL framework in detail. Further, in section IV we describe the implementation and results of the proposed model and compare it with existing state-of-the-art methods. Lastly, we conclude our study in section V.
Literature review
In this section we present background knowledge of emerging technologies used in the proposed NEDA-DL approach followed by comprehensive related work in Sect. 2.2.
Technology background
Computer-aided diagnostic (CAD)
Computer-aided diagnostic (CAD) systems play a crucial role in modern healthcare by assisting healthcare professionals in accurate and efficient diagnosis of various medical conditions. These systems leverage advanced algorithms and machine learning techniques to analyze medical data, such as images, signals, and patient records, and provide valuable insights to aid in decision-making. One significant aspect of CAD systems is their ability to enhance diagnostic accuracy and reduce human error, leading to improved patient outcomes and optimized treatment plans. CAD systems also enable early detection of diseases, facilitating timely interventions and potentially saving lives. Moreover, CAD systems can enhance workflow efficiency in healthcare settings by automating repetitive tasks and assisting healthcare providers in managing large volumes of data effectively. CAD systems offer invaluable support in tracking and managing individuals’ health conditions. By continuously analyzing various health parameters, such as vital signs, physiological signals, and biomarkers, CAD systems can detect deviations from normal patterns and alert users or healthcare professionals to potential health risks or abnormalities. This proactive approach to health monitoring enables early intervention and preventive measures, thereby promoting better overall health and well-being. Additionally, CAD systems can facilitate remote monitoring, allowing individuals to monitor their health status from the comfort of their homes and reducing the need for frequent hospital visits. Overall, CAD systems play a vital role in revolutionizing health monitoring practices, empowering individuals to take control of their health and enabling healthcare providers to deliver more personalized and proactive care.
Deep learning
Deep learning (DL), a subset of artificial intelligence (AI), has emerged as a transformative technology across various fields. In recent years, its applications have expanded rapidly, fueled by the availability of large datasets and computational power. Deep learning algorithms, inspired by the structure and function of the human brain’s neural networks, excel at learning intricate patterns and features from data. This capability has revolutionized numerous industries, including finance, automotive, and entertainment, by enabling tasks such as image recognition, natural language processing, and autonomous driving. In the healthcare sector, deep learning holds immense promise, particularly in the realm of Alzheimer’s disease research and diagnosis. Alzheimer’s, a complex neurodegenerative disorder, presents significant challenges for accurate diagnosis and treatment monitoring. Deep learning algorithms can analyze vast amounts of medical data, including neuro-imaging scans and genetic information, to identify patterns indicative of the disease’s progression. By leveraging these insights, healthcare professionals can improve early detection rates and tailor personalized treatment plans for patients. Additionally, deep learning techniques contribute to the development of computer-aided diagnostic systems, enhancing the efficiency and accuracy of Alzheimer’s disease diagnosis. Overall, deep learning stands poised to revolutionize healthcare by enabling more precise, timely interventions in the fight against Alzheimer’s disease and other neurological disorders.
ResNet-50
The ResNet-50 model, a variant of Residual Network architecture, is widely employed across various domains for tasks such as image classification, object detection, and feature extraction. With its 50 layers, including residual blocks, the ResNet-50 model addresses the challenge of vanishing gradients in deep neural networks, allowing for the training of extremely deep architectures. This model’s usage spans diverse applications, from identifying objects in images to diagnosing medical conditions from scans. In image classification tasks, ResNet-50 achieves state-of-the-art performance due to its ability to learn intricate features from data. Moreover, its pertaining weights on large-scale image datasets, such as ImageNet, make it highly suitable for transfer learning, where the model can be fine-tuned on smaller, ___domain-specific datasets. In healthcare, ResNet-50’s feature extraction capabilities have been leveraged for disease diagnosis and medical image analysis, aiding in the detection of abnormalities and assisting healthcare professionals in making informed decisions. Overall, the ResNet-50 model’s versatility and effectiveness make it a go-to choice for a wide range of machine learning and computer vision tasks.
Alex net
Alex Net, introduced by Alex Krizhevsky, Ilya Sutskever, and Geoffrey Hinton in 2012, marked a significant milestone in the field of deep learning as the winner of the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) in that year. The architecture of AlexNet comprises eight layers, including five convolutional layers and three fully connected layers as shown in Fig. 113.
It features a novel design with large kernel sizes, local response normalization, and dropout regularization, which were groundbreaking at the time. The convolutional layers in AlexNet are followed by max-pooling layers to down sample the feature maps and increase the network’s receptive field. The fully connected layers serve as a classifier, mapping the high-level features extracted by the convolutional layers to class labels. AlexNet’s architecture demonstrated superior performance in image classification tasks, significantly advancing the state of the art and paving the way for the widespread adoption of deep convolutional neural networks in computer vision. Its impact extended beyond academic research, influencing the design of subsequent architectures and catalyzing the rapid progress of deep learning in various domains.
Related work
Alzheimer’s is a neurodegenerative condition characterized by a gradual death of neuronal cells, leading to memory loss, cognitive functions, behavioral patterns, and language skills. Approximately 5.4 million individuals in the United States are affected by this condition, mainly affecting 5.2 million elderly people. Numerous research studies highlight the significance of quickly and accurately diagnosing Alzheimer’s disease, as it enables timely treatment, appropriate symptom control, implementation of safety precautions, and cost-effective therapies. Research endeavors in Alzheimer’s disease diagnosis have employed diverse computer-aided multi-class diagnosis methodologies with varying levels of achievement. Most research conducted in computer-aided diagnosis for Alzheimer’s disease mostly concentrates on binary classification, specifically determining whether a patient has been diagnosed with Alzheimer’s or not. In this study, Karami et al. (2011) provides a comprehensive analysis of the current methodologies used in computer-aided diagnosis systems (CADS) for Alzheimer’s disease (AD) utilizing neuroimaging techniques. Dementia refers to a decline in cognitive abilities and brain functioning. Alzheimer’s disease (AD) is a form of dementia that often develops in adulthood. It is characterized by a progressive and degenerative nature, leading to a steady loss of memory function. The issue is caused by the fact that AD diagnosis often occurs during old stages, reducing the effectiveness of possible treatment approaches. The growing global population of patients has led to increased focus on the early detection of Alzheimer’s disease (AD). Brain neuroimaging analysis is currently used in the comprehensive diagnostic process for Alzheimer’s disease (AD).
Various neuroimaging methodologies are employed, including axial tomography, structural imaging utilizing MRI, and positron emission tomography (PET). Single-photon emission computed tomography (SPECT) is a medical imaging technique that utilizes the emission of gamma rays from a radiopharmaceutical to create three-dimensional images of the internal structures and functions of the utilization of MRI is presently employed as a technique for investigating brain morphology. Various MRI sequences exist. The two most often employed sequences in medical imaging are T1-weighted (T1-w) and T2-weighted (T2-w) scans, with the third commonly utilized sequence being fluid-attenuated inversion recovery. Diffusion-weighted imaging and diffusion tensor imaging (DTI) are utilized to detect diffusion-related phenomena. Most of the study analysis involves the utilization of diverse image modalities such as structural MRI, fluorodeoxyglucose-positron emission tomography (FDG-PET), single-photon emission computed tomography (SPECT), diffusion tensor imaging (DTI), and arterial spin labelling combined with DTI datasets (ASL + DTI). Various image modalities allow the observer to perceive certain occurrences in specific types while being inaccessible in other modalities. As stated in their study, the authors primarily utilized structural magnetic resonance imaging (sMRI), with a particular emphasis on T1-weighted (T1-W) images. sMRI has demonstrated efficacy in diagnostic applications by effectively delineating the contrasting characteristics of healthy and pathological soft tissues within organs. The dataset utilized in this work was acquired from the PET (positron emission tomography) database. The study revealed that support vector machines (SVM) emerged as the most effective classifier, exhibiting an accuracy rate of 90.91%. One disadvantage of this study is its exclusive focus on binary categorization of Alzheimer’s disease (AD), without considering the potential for multiclassification of Alzheimer’s disease.
Many countries, including developed ones, are experiencing a demographic shift towards an ageing population. Within this context, Alzheimer’s disease (AD) has emerged as a prominent form of cognitive decline, with its prevalence exhibiting a notable correlation with advancing age. In contemporary times, computer-aided design (CAD) software has gained significant popularity among architects and has emerged as a prominent area of investigation within medical imaging and diagnostic radiology.
In another research project14, the researchers studied a chronic neurological ailment called Alzheimer’s disease. Alzheimer’s disease is a common type of dementia characterized by a range of cognitive impairments, including memory loss, decreased cognitive functioning, and difficulties with learning, among others. This work aims to use unsupervised learning technology to diagnose Alzheimer’s disease (AD). Unsupervised learning agents aim to acquire knowledge autonomously by detecting patterns in data without knowing the associated output values. The primary objective of this work is to present a novel unsupervised learning approach for the automated prediction of Alzheimer’s disease in patients using MRI imaging data. Unsupervised deep learning involves the absence of labelled data, leading researchers to propose the utilization of Convolutional Neural Networks (CNNs) for feature learning from input data.
Additionally, k-means clustering has been suggested as a technique for unsupervised classification methods, ultimately leading to final classification outcomes. In this study, the researchers initially utilized an unsupervised Convolutional Neural Network (CNN) called PCANet to extract the learned features from the MRI pictures. Subsequently, they used the k-means algorithm to get the final prediction for Alzheimer’s Disease (AD). The k-means clustering algorithm partitions a dataset into k distinct clusters through a two-phase process. In the first phase, it calculates the centroid for each cluster. In the second phase, it assigns each data point to the nearest centroid within the dataset. The experimental findings demonstrate that the proposed methodology attains an average prediction accuracy of 92.5% for Alzheimer’s disease (AD) prediction over 1075 patients. A limitation of this study refers to the binary classification employed, specifically the difference between individuals with Alzheimer’s disease (AD) and mild cognitive impairment (MCI), as well as between those with MCI and normal cognition (NC). The dataset utilized in this study was sourced from the ADNI (Alzheimer Disease Neuroimaging Initiative) database12.
In a recent study15, researchers have presented a methodology based on deep learning for diagnosing Alzheimer’s disease. In this scholarly publication, the researchers used a convolutional neural network (CNN) analysis to illustrate the neurological patterns. The researchers used the MRI method to visually represent the convolutional neural network (CNN) patterns, capturing images of both the white and grey matter within the brain. In the CNN framework, detecting patterns utilize multilayer computation, wherein inputs are processed, and their corresponding outputs are examined. One of the limitations of this study article is the usage of binary classification. Researchers discovered that 97% of patients diagnosed with Alzheimer’s (AD) and 88.7% without the disease had specific features.
Furthermore, the average accuracy rate of the diagnostic method used was 87.76%. This article employs three convolutional neural network (CNN) layers. The dataset used in this paper is the Alzheimer’s Disease Neuroimaging Initiative (ADNI), supplemented with positron emission tomography (PET) data.
Alzheimer’s is a neurodegenerative disease that affects the human brain. The damage that is done is irreversible. In this paper16, researchers propose a method based on a 3D convolutional network consisting of eight layers, five of which are used to extract features from the MRI scan and the other three for the classification of Alzheimer’s disease. Our results have shown that the proposed scheme is simple and effective on the ADNI dataset, which contains 1198 MRI brain scans from 199 AD patients and 141 NC subjects. Using the random data partitioning strategy, the proposed scheme has yielded 98.74% accuracy on the test dataset, with 100% accuracy for AD detection and a false alarm rate of 2.4%. The limitation of the proposed method, binary classification, only detects whether the patient has Alzheimer’s. The dataset used in the research is ADNI. Lian, Chunfeng, et al.17 use the convolutional neural network method to pre-determine informative locations in MRIs. This atrophy localization is useful for the feature’s extraction and classifier construction. The problem statement of this paper is that the researchers try to propose a hierarchical, fully convolutional neural network (H-FCN) that can automatically identify the local patches and regions of interest for MRI, through which multiple features are learned, and construct a hierarchical classification model for Alzheimer’s disease. This H-FCN method proved very beneficial for brain disease diagnosis, with some limitations. First, in their implementation, the size of input patches was fixed for all the ___location proposals. Secondly, they use the aggressive network pruning strategy; thirdly, a non-linear registration step was required to establish voxel-wise anatomical correspondence across different subjects. Fourth, the ___location proposal module is isolated from the subsequent network. The limitation used in this paper is binary classification in comparison with AD vs. NC and AD vs. MCI. The researchers have achieved an accurate rate of 89.31%. The dataset used in this paper is obtained from the ADNI (Alzheimer’s Disease Neuroimaging Initiative).
Most of the research focused on the binary classification of AD, Bhatkoti, P., & Paul, M18. employed a deep learning framework incorporating a modified ksparse auto encoder (ıKSA) classification method to identify regions of neural degeneration in brain MRI, as well as low levels of amyloid beta 1–42 in cerebrospinal fluid (CSF) and amyloid imaging through positron emission tomography (PET). The dataset consisted of 150 images for each modality. The findings indicate a significant association between the computational demarcation of diseased zones and the corresponding photographs. The utilization of the ıKSA algorithm in boosting classification has been found to enhance the classifier’s overall performance. The KSA approach, in conjunction with a deep learning framework, is employed for the initial time to yield precise outcomes in diagnosing Alzheimer’s disease. Similarly, Khosravi, M., Parsaei, H. and Rezaee, K19. proposed a Cascade Attention Model-CNN (CAM-CNN), a deep learning-based approach for early Alzheimer’s disease classification using MRI scans. The model integrates CNN with an attention mechanism, incorporating Satisfied Rank Loss and Cross-Network Similarity Loss to enhance feature extraction and classification accuracy. Evaluated on the Kaggle AD dataset, the model achieves an accuracy of 97.07%, outperforming traditional CNNs in both accuracy and computational efficiency. The CAM-CNN framework enhances early detection and subtype classification, aiding in patient care and targeted therapies. Despite its effectiveness, future improvements could involve MRI preprocessing techniques like skull alignment and subtraction methods and integrating patient medical history for more comprehensive diagnostic insights. Another study proposed by Kaya, M. and Çetin-Kaya, Y20., a particle swarm optimization (PSO)-based CNN framework for classifying Alzheimer’s disease (AD) severity using MRI scans. The model optimizes key CNN hyperparameters, including the number of convolutional layers and filters, to improve classification accuracy and efficiency. Evaluated on a public AD dataset, the optimized CNN achieved 97.53% accuracy and a 96.63% F1-score, surpassing existing models in both performance and computational efficiency. The proposed model addresses common issues such as overfitting21, high inter-class similarity, and the computational burden of hyperparameter tuning22. However, its effectiveness is influenced by the quality and size of available datasets, and future improvements may include larger datasets, additional hyperparameter tuning, and integration of inception and attention modules to further enhance generalizability and disease detection accuracy. This lightweight framework has the potential to assist clinicians in early diagnosis, patient monitoring, and personalized treatment planning, ultimately improving healthcare efficiency and patient outcomes.
Similarly, recent works have explored various optimization and hybrid deep learning strategies to improve diagnostic performance in medical imaging. For instance, Saber et al. (2025)23 proposed an ensemble model enhanced by meta-heuristic algorithms for breast tumor classification. Similarly, Alnowaiser et al. (2024)24 combined an adaptive CNN with the Grey Wolf Optimizer to improve breast cancer detection. Additionally, Bhatnagar et al. (2024)25 introduced a Vision Transformer model with the Lion Optimizer for accurate skin lesion classification. These studies reflect the growing importance of optimization-driven and hybrid models, which our proposed NEDA-DL approach also aims to build upon by fusing ResNet-50 with AlexNet and leveraging CUDA for real-time processing. A comparative analysis of existing state of the art methods is presented in Table 2 as follows.
Most of the research work done in the computer aided diagnosis of Alzheimer disease is only focused on the Binary classification whether the patient has Alzheimer or not, very few go beyond from binary to multi classification one of the major limitations observed in literature is the non-availability of publically available dataset. The limited size of these datasets may not adequately capture the whole spectrum of Alzheimer’s disease patients, perhaps resulting in overfitting and a biased performance of the deep learning model (see Table 2). One prominent issue observed in numerous research publications pertaining to computer-aided diagnosis (CAD) for Alzheimer’s disease utilizing deep learning techniques is the absence of real-world validation. These studies frequently rely on limited datasets and fail to incorporate comprehensive validation in real-world scenarios. This implies that the findings presented may lack applicability or generalizability across a wide range of patient demographics, clinical environments, or varying data collection methodologies.
Proposed model
The primary objective of this study is to present a computer-aided diagnostic (CAD) system for Alzheimer’s disease which includes a precise classification of the disease’s current stage. For this purpose, we propose NEDA-DL Neuroimaging based Early Detection of Alzheimer’s disease with Deep Learning. Before discussing the NEDA-DL model, first we describe the used dataset as follows.
Dataset
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) is longitudinal multi-site observational research of older people with normal cognition, mild cognitive impairment (MCI), or AD. The National Institutes of Health (NIH) and industry support it jointly through the NIH Foundation. The purpose of this research is to compare the effectiveness of using information obtained from MRI, fludeoxyglucose positron emission tomography (18 F) (FDG PET), urine, serum, and cerebrospinal fluid (CSF) biomarkers, as well as clinical and neuro psychometric assessments, to track disease progression in the three groups mentioned above of the elderly people. The reason to use MRI and PET scan data is because of widely used neuroimaging modalities for Alzheimer’s disease diagnosis due to their ability to capture structural and functional brain changes. Further the inclusion/exclusion criteria of selected datasets is described in Table 3.
MRI provides high-resolution anatomical details, allowing for precise identification of brain atrophy and structural abnormalities associated with disease progression. PET, on the other hand, enables metabolic and functional analysis by detecting amyloid and tau protein accumulations, which are key biomarkers of Alzheimer’s pathology. The combination of MRI and PET enhances diagnostic accuracy by integrating structural and functional insights, improving early-stage detection. In our research, we utilized both MRI and PET data to leverage their complementary strengths, ensuring a comprehensive assessment of neurodegenerative patterns. MRI was primarily used for volumetric and texture-based feature extraction, while PET contributed to functional analysis through metabolic activity mapping. This multimodal approach enhances model robustness, providing a more accurate classification of Alzheimer’s disease stages. The integration of MRI and PET data allows for a deeper understanding of disease progression, making them preferable choices over other imaging techniques. Therefore, 55 collaborating centres in North America collect imaging, clinical, and biologic samples from 200 cognitively normal seniors, 400 individuals with MCI, and 200 individuals with AD across multiple time points.
Table 4 provides details of the dataset used for binary classification, where the task involves distinguishing between individuals with dementia (Demented) and those without (Non Demented). The table indicates that there are 2561 samples labeled as Demented and 2560 samples labeled as Non Demented, ensuring a balanced distribution between the two classes. This dataset is crucial for training and evaluating machine learning models aimed at detecting dementia based on various features, such as neuroimaging data or clinical assessments.
In contrast, Table 5 presents the dataset details for multi-class classification, where samples are classified into four distinct classes: Very Mild Demented, Mild Demented, Moderate Demented, and Non-Demented.
The table specifies the number of samples corresponding to each class label, revealing that there are 1792 samples labeled as Very Mild Demented, 717 samples labeled as Mild Demented, 52 samples labeled as Moderate Demented, and 2560 samples labeled as Non-Demented, summing up to 5121 samples in total. This dataset provides a more nuanced understanding of the severity of dementia, enabling machine learning models to differentiate between different stages of the disease with varying degrees of impairment. The distribution of samples across different Alzheimer’s disease stages in our dataset is highly imbalanced, with the “Moderate Demented” class containing only 52 samples, compared to 2,560 in the “Non-Demented” class. To mitigate this imbalance and prevent biased predictions, we implemented a combination of data augmentation, weighted loss functions, and resampling techniques. Sample MRI images from the dataset are as follows (see Fig. 2):
Workflow of proposed model
Our proposed methodology consists of five phases including dataset collection & preprocessing, training, validating, and testing of the model. In the last phase we analyze our results with different models. Figure 3 presents the block diagram of proposed methodology.
Phase-1: Data Collection & Preprocessing
In this step, data from MRI and PET scans is collected from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. The dataset is then divided into three subsets: a training set, a validation set, and a test set. This division is done in an 80:10:10 ratio, ensuring that each subset adequately represents the distribution of data. To address any class imbalances within the training data, data augmentation techniques are applied, which involves artificially increasing the size of the training dataset by generating new samples through operations like rotation, flipping, or scaling. Before feeding the data into the model, preprocessing steps are performed to ensure consistency and improve model performance. For each image in the balanced training dataset, preprocessing begins by applying techniques for normalization and noise reduction specifically tailored for MRI images. This helps in enhancing the quality of the images and making them more suitable for analysis. Additionally, all images are resized to a fixed size of 64 × 64 × 3, where 64 × 64 represents the height and width of the image, and 3 represents the number of color channels (RGB). Further the detailed description of each step for preprocessing is as follows:
Step-1: Skull Stripping.
Skull stripping is the process of removing non-brain tissues (such as the skull, scalp etc.) from MRI or PET images. This step eliminates unnecessary artifacts that may interfere with feature extraction and classification accuracy. Skull stripping can be formulated as a binary segmentation problem, where each voxel x in an image I(x) is classified as either brain tissue or non-brain tissue described in Eq. 1 as follows:
.
where, \(\:S\left(x\right)\) denotes the skull stripping image, x is a voxel intensity value, 1 represents the brain tissues, while 0 as non-brain tissues. We applied FSL’s Brain Extraction Tool (BET) and AFNI’s 3dSkullStrip to remove non-brain structures from MRI scans. For PET scans, we use thresholding-based skull stripping since PET intensity values for non-brain regions tend to be lower than brain regions.
Step-II: Normalization
The second step is normalization that ensures the images in the dataset have a consistent range of intensity values, removing scanner-dependent variations and making deep learning models more generalizable. Min/max normalization is commonly used for intensity standardization, and can be determined using Eq. 3.
where \(\:I\left(x\right)\) presents the intensity value, \(\:{I}_{max}\:and\:{I}_{min}\) are maximum and minimum intensity values in any image. We apply this normalization mechanism to standardize intensity distributions across subjects. Since PET images contain metabolic intensity variations, therefore, we use min-max normalization to scale values to [0,1].
Step-III: Data Augmentation
Data augmentation artificially increases the dataset size and helps the model generalize better, preventing overfitting. Given the class imbalance in multi-class classification (e.g., only 52 samples in Moderate Demented class), augmentation is necessary. Generally, augmentation transformations include the rotation by θ degrees, scaling by factor s, translation by tx,ty that can be determined by following Eqs. 3,4, and 5.
.
Further, to balance the classes, we apply random rotations, flipping, intensity shifts, and elastic deformations for MRI scans whereas Gaussian noise addition and contrast variations are used to mimic PET scanning.
Step-IV: Segmentation
Segmentation involves dividing the brain image into meaningful anatomical regions like gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF). It helps extract disease-specific biomarkers such as hippocampal atrophy in Alzheimer’s patients. One common approach is K-means clustering, which iteratively updates cluster centroids, and can be determined using Eq. 6.
.
Step 2: Model Training: In this step, we train the proposed model using ResNet-50 with AlexNet architecture9. The selection of ResNet-50 for feature extraction and AlexNet for classification was guided by a balance of computational efficiency, feature extraction strength, and interpretability. ResNet-50, with its residual learning framework, effectively captures hierarchical image features while mitigating the vanishing gradient problem, making it particularly suitable for extracting fine-grained anatomical details from MRI scans. AlexNet, despite being a relatively shallow architecture, offers fast convergence, reduced computational cost, and efficient classification capabilities. To fuse ResNet-50 and AlexNet, we extract feature maps from the final convolutional block of ResNet-50 and concatenate them with intermediate-level features from AlexNet’s third convolutional layer. This hybrid fusion strategy allows the network to combine deep contextual features with low-level abstractions from AlexNet, enhancing classification robustness. To reduce model complexity, we apply depthwise separable convolutions in selected layers first performing channel-wise spatial convolution followed by a 1 × 1 pointwise convolution. Thus reducing the number of parameters and computation time compared to standard convolutions. The entire model is implemented in PyTorch, and CUDA acceleration is used to parallelize matrix operations and tensor computations across multiple GPU threads. CUDA’s grid-block-thread hierarchy enables concurrent processing of convolution operations and batch normalization steps, significantly reducing training time and supporting real-time inference scalability.
Alternative architectures such as VGGNet32 and DenseNet33 were considered, but their significantly higher parameter count, and computational demands made them less suitable for real-time clinical applications. Vision Transformers (ViTs), while promising for medical imaging, require extensive labeled data and computational resources, making them less practical in our scenario. By leveraging ResNet-50’s superior feature extraction capability and AlexNet’s efficient classification framework, our model achieves a strong balance between accuracy, computational feasibility, and clinical applicability.
Therefore, we utilize the ResNet-50 architecture for feature extraction. The deep residual connections in ResNet-50 allow for effective learning of intricate features from the input images. We remove the fully connected layers from ResNet-50, retaining only the convolutional layers. These convolutional layers serve as feature extractors, capturing hierarchical features from the input images. After extracting features using ResNet-50, we feed these features into the AlexNet architecture for further processing and classification. The AlexNet architecture, with its fully connected layers, effectively maps the extracted features to class labels. By leveraging the AlexNet architecture for classification, we benefit from its proven performance in image classification tasks. Once the ResNet-50 and AlexNet architectures are integrated, we fine-tune the combined model using transfer learning that allows us to initialize the model with pre-trained weights from both ResNet-50 and AlexNet. We then continue training the model on our specific ADNI dataset, adjusting the weights to better suit our task.
Utilizing a GPU (Graphics Processing Unit)10 with CUDA (Compute Unified Device Architecture) programming model34 enhances the training efficiency of the system. Leveraging the parallel processing capabilities of the GPU, we employ the CUDA programming model, which allows us to exploit the parallel processing capabilities of NVIDIA GPUs. The parallel nature of deep learning computations, particularly in convolutional neural networks (CNNs), makes GPUs an ideal choice for accelerating training tasks. In CUDA programming, we decompose the neural network operations into parallelizable tasks, such as matrix multiplications and convolutions, which are then executed concurrently on multiple GPU cores. This parallelism is achieved by launching threads, organized into blocks and grids, to perform computations on different subsets of the input data simultaneously. Furthermore, CUDA provides optimized libraries, such as cuDNN35, which are specifically designed for deep learning tasks and offer efficient implementations of common operations like convolutions and pooling. By leveraging cuDNN, we significantly accelerate the training process of the proposed model, reducing the overall training time and enabling faster iterations for model development and optimization.
Step 3: Validating model
Validation of a trained model is a critical step in assessing its performance and ensuring its reliability for real-world deployment. During validation, the trained NEDA-DL model is evaluated on a separate dataset, known as the validation, set, which consists of samples that were not used during the training process. The validation set allows us to assess how well the model generalizes unseen data and provides insights into its ability to make accurate predictions in real-world scenarios. Various metrics, such as accuracy, precision, recall, and F1-score, are computed to quantify the model’s performance across different classes or categories. The validation results are carefully analyzed, and any discrepancies or areas for improvement are identified and addressed through further fine-tuning or optimization.
Step 4: Testing model
Testing the trained NEDA-DL model involves evaluating its performance on a separate test dataset to assess its ability to generalize to unseen data. This process aims to validate the model’s effectiveness in accurately detecting emotions from neurological signals. During testing, the model takes input neurological signals and predicts the corresponding emotional states. Further, the predictions are compared against the ground truth labels to calculate various performance metrics, including accuracy, precision, recall, and F1-score. Additionally, metrics specific to emotion detection tasks, such as confusion matrices and emotion-specific metrics, are used to assess the model’s performance across different emotional states. Further details are given in the results section.
Step 5: Results analysis
In the final step, the results of the model’s performance are analyzed and interpreted. This involves examining metrics such as validation accuracy and test accuracy, which provide insights into how well the model performs in detecting Alzheimer’s disease. By comparing these metrics and considering any observed trends or patterns, conclusions can be drawn regarding the effectiveness of the CNN model for Alzheimer’s disease detection. Additionally, any limitations or areas for improvement may be identified, informing future research directions in this ___domain. Further details are presented in the following section.

Algorithm 1: Alzheimer’s Detection Using NEDA-DL (Proposed Method)
Performance evaluation metrics
Accuracy
In the realm of deep learning, accuracy serves as a crucial metric to assess the performance of a model in classification tasks. Mathematically, accuracy represents the proportion of correctly classified instances over the total number of instances in the dataset. It is typically expressed as a percentage. Let TP denote the number of true positive predictions (correctly classified instances), TN denotes the number of true negative predictions (correctly rejected instances), FP denotes the number of false positive predictions (incorrectly classified instances), and FN denote the number of false negative predictions (incorrectly rejected instances). The accuracy Acc is calculated using the following formula 1:
Sensitivity
Sensitivity (Recall) serves as a crucial metric for evaluating the completeness of a classification model’s predictions, especially in scenarios where it is essential to capture all positive instances. Mathematically, recall represents the proportion of true positive predictions over the total number of actual positive instances in the dataset. Let TP denote the number of true positive predictions (correctly classified instances), and FN denote the number of false negative predictions (incorrectly rejected instances). Sensitivity SEN is calculated using the following Eq. 3:
Precision
In deep learning, precision is a vital metric used to evaluate the quality of a classification model, particularly in scenarios where the focus is on minimizing false positive predictions. Mathematically, precision represents the proportion of true positive predictions over the total number of positive predictions made by the model. Let TP denote the number of true positive predictions (correctly classified instances), and FP denote the number of false positive predictions (incorrectly classified instances). Precision P is computed using the following Eq. 3:
F1-score
The F1-score serves as a comprehensive metric that balances both precision and recall, providing a single numerical value to assess the overall performance of a classification model. Mathematically, the F1-score is the harmonic mean of precision (P) and recall (R). Let P denote precision and R denote recall. The F1-score (F1) is computed using the following formula 4:
Specificity
In deep learning, specificity is a metric used to evaluate the ability of the model to correctly identify negative instances. It complements sensitivity (recall), which measures the model’s ability to correctly identify positive instances. Specificity is particularly important in applications where correctly identifying negative instances is crucial, such as medical testing where correctly identifying healthy individuals is as important as identifying individuals with a disease. To measure specificity in a deep learning model, first need to calculate the true negative rate, which is the proportion of actual negative instances that are correctly identified as negative by the model. Mathematically, specificity (SPC) is defined as in Eq. 5:
Implementation and results
The proposed NEDA-DL model was developed using Python programming language, specifically version 3.11 on Dell XPS 15, RAM 32 GB, HD 1 TB, processor core i9 12th generation, and NVIDIA GPU 3050 Ti 4 GB. We conducted different experiments using ADNI dataset on variety of classifiers including Softmax, VGG-16, reLU, Sigmoid, SVM, and RF. First, we apply the TL on ResNet-50 with AlexNet architecture and remove the fully connected layers from ResNet-50, retaining only the convolutional layers. Once the ResNet-50 and AlexNet architectures are integrated, we fine-tune the combined model using transfer learning that allows us to initialize the model with pre-trained weights from both ResNet-50 and AlexNet. Leveraging the parallel processing capabilities of the GPU, we employ the CUDA programming model, which allows us to exploit the parallel processing capabilities of NVIDIA GPUs. Below Fig. 4, and 5 present the training and testing accuracy, and loss level respectively observed during the experiments.
The confusion matrix presented in Fig. 6 represents the model’s classification performance on the testing dataset, not the training or validation sets. The dataset was split into 80% for training and validation, and 20% for testing, ensuring a representative evaluation of the final trained model. The total 75 samples in the confusion matrix correspond to the test set, which was drawn from the overall dataset after stratified splitting. This ensured that each class was proportionally represented in both training and testing phases, preventing bias in the classification results. The confusion matrix demonstrates the model’s classification accuracy for Normal Control (NC) and Alzheimer’s Disease (AD), highlighting its strong performance with minimal misclassifications.
Table 6 presents the performance metrics of various classifiers applied to a dataset, particularly focusing on the accuracy, sensitivity (SEN), F1-score, and specificity (SPC) achieved by each classifier. The dataset appears to be named “ADNI,” and the classifiers include ResNet-50, VGG-16 with different activation functions (ReLU, Softmax, and Sigmoid), Support Vector Machine (SVM), and Random Forest (RF). The highest accuracy of 99.57% is attained by the Softmax classifier with ResNet-50, closely followed by VGG-16 with 98.64%. However, in terms of sensitivity, the VGG-16 classifier with Softmax activation function achieves the highest score of 99.24%. Overall, these results provide insights into the performance of different classifiers on the dataset, aiding in the selection of appropriate models for classification tasks. The experimental results show that the proposed NEDA-DL model along with SoftMax outperformed all other classifiers with respect to all measured metrics.
Further, we compare performance metrics with existing state-of-the-art methods12,13,14,17,28, and31 that were evaluated using the similar ADNI dataset. Most of the existing methods used only for binary classification but not multi-classification for Alzheimer. Only18 method used for multi-classification for Alzheimer’s disease detection. Therefore, with the same objective and datasets, we observed that our proposed NEDA-DL approach achieved the highest accuracy 99.87 whereas18 attained maximum 83. 69 as shown in Fig. 7.
According to Fig. 8, the highest sensitivity 99.44% obtained by our approach NEDA-DL indicates its remarkable ability to accurately detect positive instances, making it a promising approach for various classification tasks. This analysis underscores the effectiveness of our proposed method in achieving high sensitivity, a crucial aspect in tasks where correctly identifying positive instances is paramount, such as medical diagnosis or anomaly detection. Similarly, we evaluate the specificity, a key metric in assessing the performance of classification methods, and compare our proposed method, NEDA-DL, with existing approaches referenced in31,12,13,14,17, and18. Specificity measures the ability of a classifier to correctly identify negative instances out of the total number of actual negative instances in the dataset. Across the existing methods, specificity scores ranged from 83.55 to 92.97%. Our proposed method, NEDA-DL, achieved a notably higher specificity score of 99.3% as shown in Fig. 9. These results indicate that NEDA-DL outperforms existing methods in accurately detecting negative instances. With a specificity score well above those of the existing methods, NEDA-DL demonstrates its effectiveness in minimizing false positive predictions and enhancing the reliability of negative classifications. This suggests that NEDA-DL may be particularly well-suited for applications where correctly identifying negative instances is crucial, such as in medical screening or anomaly detection systems.
For future perspective research directions, researchers can work in different directions as our deep learning pipeline involves computationally intensive architectures (ResNet-50 and AlexNet) that require high-end GPUs for optimal training and inference. Deploying this model in hospitals, particularly those with limited computational infrastructure, may be challenging. Additionally, seamless integration with existing hospital information systems is crucial for real-world usability. Researchers can consider working for a model that must be compatible with Picture Archiving and Communication Systems (PACS) and Electronic Health Records (EHRs) to streamline diagnosis and reporting. Developing an intuitive graphical user interface (GUI) for radiologists would further enhance clinical adoption.
Conclusions
This study introduces a system designed for medical image classification and the recognition of Alzheimer’s disease. Referred to as Neuroimaging based Early Detection of Alzheimer’s disease using Deep Learning (NEDA-DL), this paper presents a computer-aided diagnostic (CAD) approach aimed at precisely categorizing the stages of Alzheimer’s disease, thus enhancing diagnostic accuracy. By employing advanced analysis and preprocessing techniques, including annotation and labeling, NEDA-DL surpasses existing methods in processing neuro-imaging datasets. Notably, the architecture of NEDA-DL utilizes depth wise separable convolutions, reducing parameter numbers compared to traditional models and resulting in more lightweight neural networks. We train the proposed model using ResNet-50 with AlexNet architecture Leveraging the parallel processing capabilities of the GPU, we employ the CUDA programming model, which allows us to exploit the parallel processing capabilities of NVIDIA GPUs. We decompose the neural network operations into parallelizable tasks, such as matrix multiplications and convolutions, which are then executed concurrently on multiple GPU cores. This parallelism is achieved by launching threads, organized into blocks and grids, to perform computations on different subsets of the input data simultaneously. Various performance metrics including accuracy, sensitivity (SEN), F1-score, and specificity (SPC) are employed to evaluate the NEDA-DL model with different classifiers such as ReLU, Softmax, and Sigmoid, SVM, and RF, and attained highest accuracy of 99.57% by Softmax classifier with ResNet-50. Future work involves implementing additional pre-trained models for classification and investigating patient progression between Alzheimer’s disease stages. While the proposed NEDA-DL model demonstrates promising results on the ADNI dataset, we acknowledge the need for clinical validation through real-world testing and physician involvement. To support clinical decision-making in practice, future work will involve collaboration with radiologists and neurologists to test the system in real hospital settings and assess usability within clinical workflows. Additionally, interpretability of deep learning models remains a key challenge. To address this, techniques such as Gradient-weighted Class Activation Mapping (Grad-CAM) and SHAP (SHapley Additive exPlanations) will be explored to highlight the brain regions that contribute most to the model’s predictions. This will help clinicians understand and trust the model’s decisions, thereby facilitating clinical adoption.
Data availability
The datasets used and/or analysed during the current study available from the link https://adni.loni.usc.edu/data-samples/adni-data/#AccessData.
References
Alzheimer’s Disease,World Alzheimer report 2022, Last accessed: 8th September, (2023). https:alzint.org/u/World-Alzheimer-Report-2022.pdf
Alorfi, N. M. Public awareness of alzheimer’s disease: A Cross-Sectional study from Saudi Arabia. Int. J. Gen. Med. 15, 7535–7546 (2022).
Mehmood, A. et al. A transfer learning approach for early diagnosis of alzheimer’s disease on MRI images. Neuroscience 460, 43–52 (2021).
Understanding beliefs and treatment of Dementia in Pakistan, Last accessed: 1st January, (2020).
Brookmeyer, R., Johnson, E., Ziegler-Graham, K. & Arrighi, H. M. Forecasting the global burden of Alzheimer’s disease, Alzheimer’s & dementia, vol. 3, pp. 186–191, (2007).
Gauthier, S. et al. Mild Cogn. Impairment Lancet, 367, 1262–1270, (2006).
Roberson, E. D. & Mucke, L. 100 years and counting: prospects for defeating Alzheimer’s disease, Science, vol. 314, pp. 781–784, (2006).
Hassan, E., Saber, A. & Elbedwehy, S. Knowledge distillation model for Acute Lymphoblastic Leukemia Detection: Exploring the impact of nesterov-accelerated adaptive moment estimation optimizer. Biomedical Signal Processing and Control, 94, p.106246. (2024).
Singh, I., Goyal, G. & Chandel, A. AlexNet architecture based convolutional neural network for toxic comments classification. J. King Saud University-Computer Inform. Sci. 34 (9), 7547–7558 (2022).
Brodtkorb, A. R., Hagen, T. R. & Sætra, M. L. Graphics processing unit (GPU) programming strategies and trends in GPU computing. J. Parallel Distrib. Comput. 73 (1), 4–13 (2013).
Willemink, M. J. et al. Preparing Med. Imaging Data Mach. Learn. Radiol. 295.1 : 4–15. (2020).
Jack, R. Jr et al. The alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging: Official J. Int. Soc. Magn. Reson. Med. 27 (4), 685–691 (2008).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Adv. Neural. Inf. Process. Syst. 25, 1–9 (2012).
Bi, X. et al. Computer aided Alzheimer’s disease diagnosis by an unsupervised deep learning technology. Neurocomputing (2019).
Ji, H. et al. Early Diagnosis of Alzheimer’s Disease Using Deep Learning. Proceedings of the 2nd International Conference on Control and Computer Vision. (2019).
Bäckström, K. et al. An efficient 3D deep convolutional network for Alzheimer’s disease diagnosis using MR images. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE, (2018).
Lian, C. et al. Hierarchical fully convolutional network for joint atrophy localization and Alzheimer’s disease diagnosis using structural MRI. IEEE Trans. Pattern Anal. Mach. Intell. 42(4), 880–893 (2018).
Bhatkoti, P. and Manoranjan Paul. Early diagnosis of Alzheimer’s disease: A multi-class deep learning framework with modified k-sparse autoencoder classification. 2016 international conference on image and vision computing New Zealand (IVCNZ). IEEE, 2016.S. Stefan, M. Neun, and A. Edwardes, Foundations of ___location based services. Lecture Notes on LBS 1, vol. 1, no. 272, pp. 1–28, (2006).
Khosravi, M., Parsaei, H. & Rezaee, K. Novel Classification Scheme for Early Alzheimer’s Disease (AD) Severity Diagnosis Using Deep Features of the Hybrid Cascade Attention Architecture: Early Detection of AD on MRI Scans (Tsinghua Science and Technology, 2024).
Kaya, M. & Çetin-Kaya, Y. A Novel Deep Learning Architecture Optimization for Multiclass Classification of Alzheimer’s Disease Level (IEEE Access, 2024).
Hassan, E. Enhancing coffee bean classification: a comparative analysis of pre-trained deep learning models. Neural Comput. Appl. 36 (16), 9023–9052 (2024).
Elbedwehy, S., Hassan, E., Saber, A. & Elmonier, R. Integrating neural networks with advanced optimization techniques for accurate kidney disease diagnosis. Scientific Reports, 14(1), p.21740. (2024).
Saber, A., Elbedwehy, S., Awad, W. A. & Hassan, E. An optimized ensemble model based on meta-heuristic algorithms for effectively detecting and classifying breast tumors. Neural Comput. Appl. 37 (6), 4881–4894 (2025).
Alnowaiser, K., Saber, A., Hassan, E. & Awad, W. A. An optimized model based on adaptive convolutional neural network and grey Wolf algorithm for breast cancer diagnosis. PloS ONE. 19 (8), e0304868 (2024).
Bhatnagar, R., Hassan, E., Saber, A., Shams, M. Y. & Elbedwehy, S. Enhanced Vision Transformer with Lion Optimizer for Accurate Classification of Monkeypox Skin Images. In 2024 4th International Conference on Technological Advancements in Computational Sciences (ICTACS) (pp. 1897–1902). IEEE. (2024), November.
Karami, V., Nittari, G. & Amenta, F. Neuroimaging computer-aided diagnosis systems for alzheimer’s disease. Int. J. Imaging Syst. Technol. 29 (1), 83–94 (2019).
Wehenkel, M. et al. Computer Aided Diagnosis System Based on Random Forests for the Prognosis of Alzheimer’s Disease. 1st HBP Student Conference-Transdisciplinary Research Linking Neuroscience, Brain Medicine and Computer Science. Frontiers Media SA, (2018).
Fang, C. et al. A Novel Gaussian Discriminant Analysis-based Computer Aided Diagnosis System for Screening Different Stages of Alzheimer’s Disease. 2017 IEEE 17th International Conference on Bioinformatics and Bioengineering (BIBE). IEEE, (2017).
Bron, E. E. et al. Multiparametric computer-aided differential diagnosis of alzheimer’s disease and frontotemporal dementia using structural and advanced MRI. Eur. Radiol. 27 (8), 3372–3382 (2017).
Bhatkoti, P. and Manoranjan Paul. Early diagnosis of Alzheimer’s disease: A multi-class deep learning framework with modified k-sparse autoencoder classification. 2016 International Conference on Image and Vision Computing New Zealand (IVCNZ). IEEE, (2016).
Payan, A. & Montana, G. Predicting Alzheimer’s disease: a neuroimaging study with 3D convolutional neural networks. arXiv preprint arXiv:1502.02506 (2015).
Irrigisetty, H., Madhavi, K. R., Avanija, J., Thokala, R. R. & Varma, K. R. July. Enhancing Alzheimer’s Disease Diagnosis Through Deep Learning-Leveraging VGG-16 Architecture and Adaptive Boosting. In 2024 5th International Conference on Image Processing and Capsule Networks (ICIPCN) (pp. 457–464). IEEE. (2024).
Awang, M. K. et al. Classification of alzheimer disease using DenseNet-201 based on deep transfer learning technique. Plos One. 19 (9), e0304995 (2024).
Dehal, R. S., Munjal, C., Ansari, A. A. & Kushwaha, A. S. Gpu computing revolution: Cuda. In 2018 International Conference on Advances in Computing, Communication Control and Networking (ICACCCN) (pp. 197–201). IEEE. (2018), October.
Jorda, M., Valero-Lara, P. & Pena, A. J. Performance evaluation of Cudnn Convolution algorithms on Nvidia Volta Gpus. IEEE Access. 7, 70461–70473 (2019).
Acknowledgements
The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University, KSA for funding this work through Small Research Project under grant number RGP.1/290/46.
Author information
Authors and Affiliations
Contributions
The authors state that they have read the manuscript and equally contributed. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alasiry, A., Shinan, K., Alsadhan, A.A. et al. A novel neuroimaging based early detection framework for alzheimer disease using deep learning. Sci Rep 15, 23011 (2025). https://doi.org/10.1038/s41598-025-05529-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-05529-5