Abstract
Platelet-rich fibrin (PRF) and Enamel Matrix Derivatives (EMD) can support the local regenerative events in periodontal defects. There is reason to suggest that PRF and EMD exert part of their activity by targeting the blood-derived cells accumulating in the early wound healing blastema. However, the impact of PRF and EMD on blood cell response remains to be discovered. To this aim, we have exposed human peripheral blood mononucleated cells (PBMCs) to PRF lysates prepared by a swing-out rotor and EMD, followed by bulk RNA sequencing. A total of 111 and 8 genes are up- and down-regulated by PRF under the premise of an at least log2 two-fold change and a minus log10 significance level of two, respectively. Representative is a characteristic IFN response indicated by various human leukocyte antigens (HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQA2, HLA-DRA, HLA-DRB1, HLA-DRB5), gamma Fc receptors (FCGR1A, FCGR1B, FCGR3B), chemokines (CXCL9-11), and calprotectin (S100A8/9 and S100A12), complement (C1QA/B, C2) and interferon-induced guanylate-binding proteins (GBP1, GBP5). With EMD, 67 and 29 genes are up- and down-regulated, respectively. Characteristic of the upregulated genes are tensins (TNS1 and TNS3). Among the genes downregulated by EMD were epsilon Fc receptors (FCER1A; FCER2), Fc receptor-like proteins (FCRL1, FCRL3) and CX3CR1. Genes commonly upregulated by PRF and EMD were most noticeably NXPH4 and MN1, as well as FN1, MMP14, MERTK, and AXL. Our findings suggest that PRF provokes an inflammatory response, while EMD dampens IgE signaling in peripheral mononucleated blood cells.
Similar content being viewed by others
Introduction
The periodontium connects the tooth with the surrounding alveolar bone and is the functional link between two hard tissues required for eating and speaking1. The periodontium, particularly the junctional epithelium with the periodontium ligament lying underneath, is exposed to the oral cavity with its microbial burden2and is prone to damage by masticatory forces3,4. Periodontal homeostasis, therefore, requires an efficient defense system plus a continuous renewal of the periodontal tissues and their stem cell pool5. However, insufficient oral hygiene overwhelms the defense and renewal capacity, initiating the catabolic process of chronic inflammation with its clinical hallmarks of inflammatory osteolysis and soft tissue degeneration. These events culminate in the formation of periodontal pockets that even worsen the catabolic process2. Ultimately, tooth loss stops the dissemination of microorganisms, thus protecting the organism. However, tooth loss is not the solution we seek; the overall goal is to maintain periodontal health or, if there is already a periodontal pocket with furcation defects, to support its regeneration by taking advantage of the intrinsic capacity of the periodontium to renew under homeostasis5,6.
Regeneration of the periodontium, particularly the renewal of the junctional epithelium and the periodontium ligament5,6, is challenging. It requires removing the inflammatory cause and supporting the body’s intrinsic healing capacity – ideally targeting the remaining stem cells and forcing their expansion and differentiation towards the specialized periodontal tissues required to reconnect the tooth with the surrounding alveolar bone5,6. Today’s attempts to support periodontal regeneration are not explicitly designed to target potential stem cells but are trying to mimic tooth development or support wound healing, and the latter is not specific to periodontal cells7,8,9,10,11,12. However, any regeneration follows a conserved sequence of events that starts with bleeding and the presence of blood cells entrapped in the fibrin-rich matrix of the clot. Thus, peripheral blood cells, including the neutrophils, monocytes, and lymphocytes accumulating in the blood clot, define sites of early repair and regeneration13. It would thus be interesting to understand how peripheral blood cell signatures change during periodontal therapies.
Today’s attempts to support periodontal regeneration include but are not limited to the local use of an enamel matrix derivative (EMD), an extract of tooth germs from piglets that is applied in a carrier of propylene glycol alginate (PGA)7,12. This concept was introduced to periodontology in 199714,15, and since then, it has become commercially available. Its introduction was followed by numerous studies trying to support its clinical use, including attempts to understand the cellular and molecular mechanism of how EMD supports periodontal regeneration16. In vitro attempts have focused on how EMD affects the behavior of gingival and periodontal fibroblasts17, adipocytes18, oral epithelial cells19, and cells of the hematopoietic lineage, e.g., their differentiation into bone-resorbing osteoclasts20and macrophage polarization21,22. Omics technology helped to understand how EMD changes the genetic signature of oral fibroblasts17, allowing us to identify strongly regulated such as IL11, NOX4, and PRG4 that could be traced back to the TGF-β activity of EMD17. The TGF-β activity is also involved in hematopoietic cell responses, at least in a mouse setting20,21. The present study’s search for EMD target genes continues but with a new focus: the human peripheral blood mononucleated cells.
The local use of platelet-rich fibrin (PRF) is another attempt to support periodontal regeneration10,11. Independent of the various protocols to optimize the size ratio and the yield of platelets and other cells23, PRF is the coagulated plasma fraction separated from the red clot and squeezed between metal plates. We have used the H-PRF concept based on the fractionation of blood with a horizontal swing-out rotor using 700 g for 8 min23. PRF membranes are then ready to be used in extraction sockets and for bone augmentation procedures24,25or other indications not exclusively related to dentistry – such as diabetic wound healing26and facial aesthetics27. Similar to EMD, basic research focuses on how potential target cells respond to PRF, including various sources of mesenchymal cells such as gingival fibroblasts28,29and bone marrow-derived macrophages30that can form osteoclasts31. RNAseq was also implemented to study how PRF lysates affect the genetic signature of gingival fibroblasts, again revealing that IL11, NOX4, and PRG4 are the most strongly expressed genes28. We now study PRF target genes in human peripheral blood mononucleated cells.
There are similarities in the response of gingival fibroblasts to PRF and EMD, which can be explained by the TGF-β activity both preparations have in common; however, peripheral blood cells presumably respond differently than fibroblasts. There is thus a demand to understand the change of the genetic signature of PBMCs to EMD and PRF – not only to identify possible similarities but, more generally, to understand how potential therapies support healing on a cellular and molecular basis. An RNAseq approach with PBMCs exposed to PRF and EMD seems to be an appropriate first step.
Materials and methods
Ethical considerations
Blood donation for preparing PBMCs was approved by the Medical University of Vienna ethics committee (1539/2017), and informed consent was obtained from all three healthy male donors aged between 25 and 29 years. Blood donation for preparing PRF following our established protocol32,33 was approved (1644/2018), and informed consent was obtained from all donors. All experiments were performed in accordance with “Good Scientific Practice” – Ethics in Science and Research – Guidelines of the Medical University of Vienna.
Preparation and stimulation of PBMCs
As recently described34, heparinized blood was diluted 1:2 in Hanks balanced salt solution (HBBS, Lonza, Basel, Switzerland) and subjected to fractionation for 15 min at 800 g at room temperature by Ficoll‒Paque (G.E. Healthcare Bio-Sciences AB, Sweden). PBMCs were washed in HBSS and resuspended in a CellGro serum-free medium (CellGenix, Freiburg, Germany; 25 × 106 cells/ml) with no antibiotics added. PBMCs from the three donors were incubated for 20 h without or with 300 µg/mL of enamel derivative matrix (EMD; Straumann AG, Basel, Switzerland) or three independent pools of 30% PRF lysate. PBMCs exposure was performed in an incubator at 37 °C, 95% humidity, 5% CO2.
Preparation of PRF lysates
PRF membranes were prepared from blood using glass tubes (Bio-PRF, Venice, FL, USA) with centrifugation at 700 g for 8 min (swing-out rotor; Z306 Hermle, Universal Centrifuge, Wehingen, Germany). Based on our setting with a swing-out rotor, the terminology H-PRF would be appropriate23,35. The yellow PRF clot was separated from the remaining red thrombus and compressed between metal plates. The resulting PRF membranes were transferred to a serum-free medium (1 cm PRF membrane/1 mL) and subjected to two cycles of freeze–thawing followed by sonication on ice (Sonopuls 2000.2, Bandelin electronic, Berlin, Germany). After centrifugation at 15,000× g for 10 min, the PRF lysates were used to stimulate the PBMCs. These PRF lysates were reported to be active in a series of different bioassays28. To rule out large donor variation, PRF lysates from at least three donors were pooled to obtain one PRF lysate to stimulate one PBMC donor. The other PBMCs were exposed to other independent pools of PRF lysates. Thus, the setting is strictly allogenic with three individual PBMCs exposed to three different pools of PRF lysates.
Total RNA isolation
Following stimulation of PBMCs, total RNA was isolated (Trizol® Reagent, Invitrogen, Carlsbad, CA) and quantified using a spectrophotometer (NanoDrop-1000, Peglab, Erlangen, Germany). RNA quality was examined by an Agilent 2100 Bioanalyzer (Agilent, Böblingen, Germany) and used at an integrity score between 5.7 and 10. A total of 9 samples were generated from the three different donors.
RNA sequencing and data analysis
Similar to what we have reported recently19,36, sequencing libraries were prepared at the Core Facility Genomics, Medical University of Vienna, using the NEBNext Poly(A) mRNA Magnetic Isolation Module and the NEBNext UltraTM II Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) with unique dual indices (UDI). Libraries were QCchecked (Bioanalyzer 2100, Agilent Technologies, Santa Clara, CA) using a High Sensitivity DNA Kit for correct insert size and quantified using Qubit dsDNA H.S. Assay (Invitrogen, Waltham, MA). Pooled libraries were sequenced on one flowcell of a NextSeq2000 instrument (Illumina, San Diego, CA) in 1 × 75 bp single-end sequencing mode. On average, 23 million reads per sample were generated. Reads in fastq format were generated using the Illumina bcl2fastq command line tool (v2.19.1.403), incl. trimming of the sequencing adapters. Reads in fastq format were aligned to the human reference genome version GRCh38 (1) with Gencode 29 annotations (2) using STAR aligner37 version 2.6.1a in 2-pass mode. Raw reads per gene were counted by STAR. Batch correction was performed using ComBat from sva version 3.35.2. Differential gene expression was analyzed using DESeq2 38version 1.22.2. TPM were generated by RSEM39.
Gene set enrichment analysis, volcano plot generation, and gene clustering
For volcano plot generation, we used VolcaNoseR, a web-based tool40. The up- and down-regulated genes were used for further analysis under the premise of an at least log2 two-fold change (4-fold) and a minus log10 significance level of two (p= 0.01)40. InteractiVenn, another web-based tool, was implemented to analyze gene sets through Venn diagrams41. The STRING algorithm MCL was used to cluster the genes displayed in the network42. g: Profiler was used as a functional enrichment analysis tool that integrates many databases, including Gene Ontology and KEGG43. Figures are based on g: Profiler but not KEGG pathway images, thus there is no requirement to obtain copyright permission of KEGG44. Transcription factor enrichment analysis (TFEA) was based on the web-based platform45. The original gene counts are presented in Table S1. RNAseq data supporting this study’s findings have been deposited in the GEO with the accession number GSE266995. https://www.ncbi.nlm.nih.gov/geo/.
Results
PCA and heat map of gene expression changes by PRF and EMD
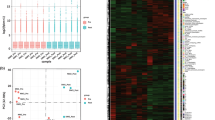
To investigate the degree to which PRF and EMD alter the transcriptional signature of human peripheral blood cells, we conducted bulk RNA sequencing of PBMCs from three donors treated ex vivo with PRF lysates and EMD for 6 h. A principal component analysis (PCA) of the first two principal components demonstrates high reproducibility within the replicates, whereas the different treatments could be separated within the two dimensions (Fig. 1A). We performed a heatmap analysis to visualize further the gene expression pattern of PRF and EMD-treated PBMCs. The transcriptional heterogeneity between untreated and PBMCs exposed to PRF and EMD was confirmed in a heatmap based on the average expression of cluster-defining genes (Fig. 1B).
Principal component and heat map analysis for differentially expressed genes in PBMCs treated with PRF lysates and EMD. (A) The plot shows the projection of the samples onto the two-dimensional space spanned by the covariance matrix’s first and second principal components. The expression levels used as input are normalized log CPM values. The analysis shows the homogeneity of individual donors in each treatment group and the heterogenicity caused by the treatment of the cells with PRF and EMD. (B) Heatmap of cluster-defining marker genes of normalized gene expression showing distinct gene patterns. Each row represents a gene, and three columns represent the independent donor of PBMCs. Red indicates high levels of expression, while blue indicates low levels. Expression levels are indicated by darker and lighter shades of red and blue. Genes with an adjusted p-value < 0.05 and an average log2 fold change ≥ 1 or ≤ − 1 were considered in this analysis.
Volcano analysis of gene expression changes by PRF and EMD
We next performed a Volcano analysis showing 111 and 8 genes that are up-and down-regulated by PRF under the premise of at least Log2 two-fold change and a minus Log10 significance level of two, respectively (Fig. 2, Table S2). In the case of EMD, Volcano analysis revealed 67 and 29 genes that are up-and down-regulated with a Log2 two-fold change and a minus Log10 significance, respectively (Fig. 2, Table S2). Based on a Manhattan distance > 10, 38 genes were upregulated by PRF including the clusters HLA-DQA1, HLA-DQA2, HLA-DPA1, HLA-DRA, and FCGR1, FCGR1B, FCGR3B as well as CXCL9, CXCL10, CXCL11 and S1008, S1009, S10012 and also C1QA, C1QB, C2. Among the downregulated genes were TNFRSF4, E2F1 NCR3, KCNH3, IL24, FCRL5 and NETO1. Through EMD, 35 genes were upregulated, including APOE and APOC1, as well as TNS1 and TNS3. EMD downregulated seven genes with a Manhattan distance > 10, including FCRL1 and FCRL3 but also MMP10. Interestingly, among the strongly regulated genes being increased by both PRF lysates and EMD are NXPH4 and MN1.
Volcano plot analysis of 119 and 96 differentially expressed genes in PBMCs treated with PRF and EMD, respectively. Volcano plot analysis identified upregulated (red) and downregulated (blue) genes in PBMCs treated with PRF and EMD. The annotated dots are data points with the largest (Manhattan) distance from the origin and are above the thresholds indicated by the dashed line. The threshold was set at least two-fold change and a significance level of two.
Venn analysis of genes regulated by PRF and EMD
To further reveal differences and similarities in the response of PBMCs to PRF and EMD exposure, we performed a Venn analysis. This analysis showed 92 and 7, as well as 48 and 28 genes being independently increased and decreased by PRF and EMD, respectively (Fig. 3). When comparing the genes being commonly upregulated by PRF and EMD, we identified 19 genes, namely NXPH4, MN1, FPR3, CCDC182, FN1, LAMA3, SMARCD3, MMP14, HTRA1, MERTK, GPNMB, COL23A1, MS4A4A, A4GALT, APOC1, CPNE7, PDPN, RNASE1, AXL; and 1 gene being downregulated NCR3 (Fig. 3).
STRING analysis of gene clusters formed by PRF and EMD
Next, we performed a cluster analysis of the differentially expressed genes by PRF lysates and EMD. STRING analysis revealed PRF upregulated genes are clustering: Cluster I FCGR1A, FCGR1B, FCGR3B, GBP1, GBP5, CD274, CD300LF, IDO1, IFIT3, KCNJ2, LAP3, LILRB4, MUC1, PDCD1LG2, RSAD2, SIGLEC1, STAT1 and TLR8 – or Cluster II with CXCL9, CXCL10, CXCL11, CCL2, ADAMDEC1, BATF2, CALHM6, ETV7, SECTM1 and VAMP5 – or Cluster III with HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQA2, HLA-DRA, HLA-DRB1, HLA-DRB5, CD74, COL2A1, FGL2 – or Cluster IV with C1QA, C1QB, C2, SERPING1 and SLCO2B1 or Cluster V with DAPK1, DOCK1, TLN2 and VCL and Cluster VI with S100A8, S100A9, S100A12, APOBEC3A,. No cluster for PRF downregulated genes was identified (Table S3).
STRING analysis of the 67 EMD upregulated genes showed clustering: Cluster I with C3, PROC, RETN, SERPINE1 - Cluster II with ABCA1, APOE, and NCEH1 - and Cluster III with ADGRG3, MCEMP1, and SLC11A1, etc. also mentioned should be TNS1 and TNS3 (Table S3). Three clusters for EMD downregulated genes were identified: Cluster I: FCER1A, FCER2, CLEC10A, and IL1RL1 – with Cluster II: BATF3, CX3CR1, and TNFRSF9 as well as Cluster III: FCRL1, FCRL3 and NIBAN3 (Table S3). Clustering of genes commonly regulated by PRF and EMD are Cluster I: APOC1, FN1, HTRA1, LAMA3, MMP14 and PDPN, - with Cluster II: AXL, GPNMB, and MERTK, - and Cluster III: FPR3 and MS4A4A (Table S3).
G: profiler analysis of gene expression changes by PRF and EMD
We performed a functional enrichment analysis based on the 119 and 96 differentially expressed genes in PBMCs exposed to PRF lysates and EMD, respectively. Among the PRF upregulated genes, the most significantly enriched G.O. molecular function categories were those related to MHC class II receptor activity. Consistently, analysis of G.O. biological processes and G.O. cellular compartment revealed enriched categories mainly related to immunity and MHC class II protein complex. Enrichment was also observed for cell migration and chemotaxis related to the increased chemokine expression (Fig. 4). No enrichment was observed for the eight downregulated genes.
gProfiler analysis of differentially expressed genes in PBMCs treated with PRF. Functional enrichment analysis of upregulated genes by PRF. The enrichment analysis results are presented in the form of a Manhattan plot, where the x-axis shows the functional terms grouped by the color code of the source database used, while the y-axis shows the enrichment-adjusted p-values in a negative decimal logarithm scale.
For genes upregulated with EMD, G.O. molecular function revealed proteoglycan binding, and G.O. biological processes categories were enriched for inflammatory response and KEGG of cholesterol metabolism. Maybe more impressive is the enrichment for genes downregulated with EMD; G.O. molecular function showed IgE receptor activity binding leucocyte activation, and collagen catabolic process (Fig. 5). Thus, while PRF raises MHC class II receptor activity, EMD lowers IgE receptor activity.
gProfiler analysis of differentially expressed genes in PBMCs treated with EMD. Functional enrichment analysis of (upper) up- and (lower) downregulated genes by EMD. The enrichment analysis results are presented in the form of a Manhattan plot, where the x-axis shows the functional terms grouped by the color code of the source database used, while the y-axis shows the enrichment-adjusted p-values in a negative decimal logarithm scale.
Reactome signaling pathway enrichment analysis
We next focused on the Reactome signaling pathway enrichment analysis of the most strongly regulated genes by PRF and EMD (Fig. 6). Most of the top 10 enriched PRF-regulated genes in PBMCs are strongly enriched for IFNG signaling, as well as activation of T-cells, including PD-1 signaling, ZAP-70 translocation, and costimulation by CD28. No enrichment was found for down-regulated genes. On the other hand, EMD upregulated genes are enriched, even though less strongly, for PTK6 and NTRK2 signaling – and plasma lipoprotein clearance, collagen degradation, and IL10 signaling. Downregulated genes by EMD are enriched for prostanoid ligand receptors and binding.
Differentially expressed genes in PBMCs treated with PRF lysates and EMD. Dot plots of top 10 enriched REACTOME database terms of PRF and EMD regulated genes with enriched terms on y-axes, −log10 of Benjamini-Hochberg method adjusted p-values on x-axes, dots colors encoding Enrichr combined scores, dot sizes indicating gene ratios.
Transcription factor enrichment analysis (TFEA) of gene expression changes by PRF and EMD
Finally, we used TFEA to identify which transcription factors are causally responsible for the observed strong expression changes when PBMCs are exposed to PRF and EMD (Fig. 7). Exposing cells to PRF causes enrichment of AHR, SPI1, CIITA, RFX1, RFXAP, RFX5, RFXANK, IRF1, IRF9, STAT1, and KFL9. For instance, the regulatory factor X (RFX) gene family and CIITA control MHC class II gene expression are related to IFN signaling46. IRF-1 and IRF9 regulate genes have an interferon-stimulated response element (ISRE) in their promoters, and STAT1 belongs to canonical IFN signaling47,48. A different enrichment was observed with EMD revealing, SMAD1, SMAD2, NR4A1, AP1, NCOA1, EGR1, ETV5, TFAP2B, PAX3, and also KLF9. For instance, SMAD1 and SMAD2 belong to the canonical BMPR and TGFBR signaling pathways, respectively49. Both PRF and EMD caused an enrichment for NFYA, BCL3, PAX5, and SREBF2; for instance, SREBF2 and NFY signaling control lipid metabolism50.
Heat map analysis of transcription factor binding motif enrichment in PBMCs treated with PRF lysates and EMD. This heatmap represents the enrichment of transcription factor binding motifs for the tsDMRs identified in each tissue. Each row indicates a transcription factor, and three columns represent the independent donor of PBMCs. Red shows high levels of transcription factor enrichment, while blue designates low levels. Expression levels are indicated by darker and lighter shades of red and blue.
Discussion
The present research aims to extend existing knowledge of how cells are potentially involved in periodontal regeneration and wound healing response to PRF and EMD. While it was recently cells of the mesenchymal lineage17,18,28,29, epithelial cells19,51, and mouse bone marrow cells20,21,22,30,31 that were the focus of PRF and EMD research, we now draw out attentiveness to the human blood cells. PBMCs were exposed to PRF and EMD for this aim, followed by an RNAseq analysis and the downstream interpretation of the altered genetic signature. Based on this bulk RNAseq analysis, we have identified a panel of genes being differentially expressed by PBMCs upon exposure to PRF and EMD – with most genes being independently expressed by each treatment. Our first main finding was that PRF lysates provoke an IFN response culminating in the increased expression of characteristic gene clusters such as HLAs, FCGRs, CXCL9-11, the S100s proteins, and complement factors. Our second main finding was that EMD induces the downregulation of FCERs as well as FCRLs and the upregulation of APOC1, APOE and TNS1, TNS3. This research is essential as it shows that human PBMCs are targets for PRF and EMD in vitro. Even though it can be expected, the confirmation of PRF and EMD to regulate any PBMC genes in vivo is pending.
How can our data help us to understand the clinical effects caused by PRF? There is agreement that early and transient inflammation is part of the physiological process of wound healing and bone regeneration. For instance, chemokines are central regulators of wound healing, supporting angiogenesis and the recruitment of inflammatory cells, with their secretome targeting the other repair cells. For example, during the re-epithelialization phase, basal keratinocytes express CXCL10 and CXCL11, suggesting an essential role of these chemokines in the healing phase52. Fibrin functionalized with fibronectin, and CXCL11 supported healing full-thickness mouse wounds53. Moreover, CXCL10 and CXCL11 are lower in chronic diabetic wounds than in acute wounds54. Thus, in response to PRF, accumulating PBMCs become a vital source of these chemokines that, in turn, support re-epithelization and basement membrane regeneration of wound healing. The S100 genes were increased in the regenerating epidermis55, and mice lacking functional S100A8/A9 have impaired phagocyte movement and wound healing56. Impressive was also that PRF lysates increased MHC Class II molecules HLA-DPA1, HLA-DQA1, HLA-DQA2, and HLA-DRA to better interact with immune cells, like the T helper cell. Also of interest is the observation that FCGR1A was increased in PBMCs, similar to the human gingiva transcriptome during wound healing57, suggesting an impact on humoral and innate immunity by releasing cytokines or phagocytosis58. Also, C1q of the complement system controls angiogenesis with therapeutic implications in wound healing59. Taken together, evidence suggests that PRF’s wound healing effects may be attributed to changes in the genetic signature of PBMCs, potentially reflecting blood cells accumulating in a healing blastema.
Transcription factor enrichment analysis suggests that PRF causes activation of IFN signaling, with, for instance, IRF1 and IRF9 regulating genes with an interferon-stimulated response element (ISRE) in their promoters, and STAT1 belongs to canonical IFN signaling. The question is, how can PRF lysates cause the massive IFN response? Proteomic analysis failed to identify detectable amounts of IFNs in PRF lysates, even though PRF contains leucocytes capable of producing interferons28,60. Nevertheless, CXCL9 and CXCL11 are usually secreted by PBMCs61. Leukocytes that predominantly produce CXCL10 are T cells and monocytes62. Thus, our data support the overall findings that PBMCs are a source of CXCL9, CXCL10, and CXCL11, but how PRF lysates drive the increased expression remains to be determined. Similarly, the Ca2+binding S100 protein family S100A8, S100A9, and S100A12 that modulate the inflammatory response are released from neutrophils and monocytes upon IFN signaling63. Neutrophils accumulate cytosolic S100A8 and S100A9, whereas in monocytes, the proteins need to be produced64. For instance, IFNγ increased S100A8 in mouse macrophages65. The same applies to the HLA class II histocompatibility antigens chain or CD74 – and the three genes coding for Fc-gamma receptors: FCGR1A, FCGR1B, and FCGR1C66. FCGR1A is constitutively expressed by monocytes and macrophages but induced in other myeloid cell types, like neutrophils and eosinophils, following stimulation with IFNγ67,68. Thus, our data provide strong evidence for the ability of PRF lysates to provoke an IFN response in PBMCs, but the underlying molecular mechanisms remain to be answered. One mechanism might relate to platelet lipids, mainly lysophosphatidic acid69that cause a proinflammatory-like response in macrophages70, converts monocytes into macrophages71, and targets oral fibroblasts72, requires further attention.
How can our data help us to understand the clinical effects caused by EMD? Among the genes being downregulated by EMD and PRF lysates is CX3CR1, a chemokine receptor present on early myeloid lineage cells, including osteoclast precursors. Mice deficient in CX3CR1 had an increase in bone thickness and reduced osteoclast precursor cells and mature osteoclast number; thus, lowering of CX3CR1 in PBMCs by EMD might support bone regeneration73. However, these mice experience delayed wound closure because of reduced macrophage immigration and reduced fibroblast activation74. Moreover, Steap4, required for iron uptake, is indispensable for osteoclast development and function75. Also, FABP4 can contribute to inflammatory-mediated diseases. Thus, downregulation reduces inflammation76. A lack of MMP10, however, is linked to an impaired resolution of inflammation77and fracture healing in mice78. We further have identified FCER1A and FCER2 to be downregulated by EMD, both receptors allowing high-affinity IgE binding, a process linked to mast cell activation in the context of allergic disorders, but also basophils eosinophils and monocytes express the FCER1. FCER2, with the synonym CD23, is the low-affinity receptor for IgE and is expressed by mature B-cells and monocytes in PBMCs79. Nevertheless, the impact of EMD and its potential to decrease the sensitivity of PBMCs to IgEs remains unclear. FCRL1 and FCRL3 belong to a family of Fc receptor-like proteins with immunoregulatory potential via their tyrosine-based activating/ inhibitory motifs (ITAM/ ITIM) preferentially expressed by B-cells. FCRL1 is a biomarker and therapeutic target in B cell lymphoproliferative disorders80. FCRL3 promotes TLR9-induced B-cell activation and suppresses plasma cell differentiation81, and elevated expression levels of FCRL3 on T-cell subsets were associated with rheumatoid arthritis82. Again, whether the clinical effects of EMD are mediated via the regulation of FCRL1 and FCRL3 in blood cells remains an open question.
Among genes being upregulated by EMD, similar to PRF, is FN1, a critical glycoprotein of the extracellular matrix connecting with integrins and other extracellular matrix proteins such as collagen, fibrin, and heparan sulfate proteoglycans83. Thus, FN1 plays a crucial role in wound healing, and EMD supports its expression by PBMCs. Another example is APOC1, a component of lipoproteins84. HTRA1 being secreted by osteoclasts and negatively regulates osteoblast differentiation85, suggesting a bone-protective activity. Also upregulated is oxidized low-density lipoprotein receptor 1 (OLR1, LOX1), which is required for internalization and degradation of oxidized low-density lipoprotein. OLR1-deleted mice showed decreased bone mass because of elevated bone resorption86. APOE plays crucial roles not only in lipid metabolism but also in preserving bone mass87. Also protective are formyl peptide receptors (FPRs); for example, FPR2/3 knockout mice exhibited delayed recovery from acute experimental colitis and biopsy injuries88. Interestingly, EMD, like PRF, also increases MMP14, an essential protease in development and growth as, for instance, mice genetically deficient in MMP14 show severe defects in skeletal development and angiogenesis89,90. Also, a lack of MMP2 delays bone remodeling during fracture repair91, suggesting that a transient increase of MMP14 and MMP2, as observed with EMD, is beneficial for regeneration. Among the other upregulated genes are APOC1, APOE, and TNS1 and TNS3, both cellular adhesion molecules that bind to extracellular fibronectin fibrils92. In general, and to better understand to which extent the changes in gene expression are caused by TGF-β1 in EMD, blocking TGF-β receptor 1 kinase with SB431542 should be performed. Other genes are also commonly upregulated by PRF and EMD, such as the clustering of the TAM family of receptor tyrosine kinases MERTK and AXL. Interestingly, osteoblast-targeted deletion of MERTK increased bone mass in mice93, but global deletion of MERTK and AXL caused more severe antibody-induced arthritis94. Overall, PRF and EMD are critical regulators of the TAM family of receptor tyrosine kinases in PBMCs.
The study has limitations. Since the PBMCs were a bulk-RNA preparation, we cannot conclude which particular cell type - lymphocytes (T cells, B cells, and NK cells), monocytes, and dendritic cells - is responsible for the gene expression changes, thus being the target cell type for PRF and EMD. Future research needs to focus on single-cell RNA seq – which allows us to decipher the primary target cell responsible for the change in the genetic signature of PBMCs. Based on this information, we can identify further similarities and differences between PRF and EMD on how they mediate their activity on a cellular level, possibly understand the line between the local activity of EMD and PRF and how it translates into regeneration and clinical success. Perhaps also human organoid model of periodontal wound healing might serve as bioassay to reveal the complex cell integration when exposed to PRF or EMD95. Another limitation is that the data observed with human PBMCs contrast what we have observed with murine bone marrow cells and murine marrow stromal cells, where PRF lysates effectively reduce a forced expression of cytokines and chemokines30,96,97. From a cellular and molecular perspective, future studies should identify which cell type in PBMCs is mainly responsible for the expression changes caused by PRF and EMD. From a clinical perspective, research needs to show that the beneficial effects caused by PRF and EMD involve the activation of the patient’s blood cells – and if yes, to which extent, for instance, the PRF-induced sharp changes of MHC class II receptor activity, FCGRs, chemokines, S100A proteins, and complement factors play a role in this context. The overall effects of EMD were less pronounced. However, a reduced IgE receptor activity, binding leucocyte activation, and a catabolic collagen process might contribute to the clinical outcomes when treated with EMD. Another open question is which molecular mechanisms are responsive for the 19 genes commonly regulated by PRF and END and if this pattern may point towards a moderate similarity in their clinical effects. Thus, the present study is a fundament for future research, clearly showing that it is worthwhile to consider the patient’s blood cells to be targets for PRF and EMD therapy.
Data availability
RNAseq data that support the findings of this study have been deposited in the GEO with the accession number GSE266995. https://www.ncbi.nlm.nih.gov/geo/. Reinhard Gruber should be contacted to request the data from this study.
References
Nanci, A. & Bosshardt, D. D. Structure of periodontal tissues in health and disease. Periodontol 2000. 40, 11–28. https://doi.org/10.1111/j.1600-0757.2005.00141.x (2006).
Bosshardt, D. D. The periodontal pocket: pathogenesis, histopathology and consequences. Periodontol 2000. 76, 43–50. https://doi.org/10.1111/prd.12153 (2018).
Dutzan, N. et al. On-going mechanical damage from Mastication Drives Homeostatic Th17 cell responses at the oral barrier. Immunity 46, 133–147. https://doi.org/10.1016/j.immuni.2016.12.010 (2017).
Yuan, X. et al. Linking the mechanics of chewing to Biology of the junctional epithelium. J. Dent. Res. 102, 1252–1260. https://doi.org/10.1177/00220345231185288 (2023).
Yuan, X. et al. The junctional epithelium is maintained by a Stem Cell Population. J. Dent. Res. 100, 209–216. https://doi.org/10.1177/0022034520960125 (2021).
Krivanek, J., Buchtova, M., Fried, K. & Adameyko, I. Plasticity of Dental Cell types in Development, Regeneration, and evolution. J. Dent. Res. 102, 589–598. https://doi.org/10.1177/00220345231154800 (2023).
Miron, R. J. et al. Twenty years of enamel matrix derivative: the past, the present and the future. J. Clin. Periodontol. 43, 668–683. https://doi.org/10.1111/jcpe.12546 (2016).
Tavelli, L., Chen, C. J., Barootchi, S. & Kim, D. M. Efficacy of biologics for the treatment of periodontal infrabony defects: an American Academy of Periodontology best evidence systematic review and network meta-analysis. J. Periodontol. 93, 1803–1826. https://doi.org/10.1002/JPER.22-0120 (2022).
Nevins, M. et al. Platelet-derived growth factor promotes periodontal regeneration in localized osseous defects: 36-month extension results from a randomized, controlled, double-masked clinical trial. J. Periodontol. 84, 456–464. https://doi.org/10.1902/jop.2012.120141 (2013).
Miron, R. J. et al. Use of platelet-rich fibrin for the treatment of gingival recessions: a systematic review and meta-analysis. Clin. Oral Investig. 24, 2543–2557. https://doi.org/10.1007/s00784-020-03400-7 (2020).
Miron, R. J. et al. Use of platelet-rich fibrin for the treatment of periodontal intrabony defects: a systematic review and meta-analysis. Clin. Oral Investig. 25, 2461–2478. https://doi.org/10.1007/s00784-021-03825-8 (2021).
Esposito, M., Grusovin, M. G., Papanikolaou, N., Coulthard, P. & Worthington, H. V. Enamel matrix derivative (Emdogain(R)) for periodontal tissue regeneration in intrabony defects. Cochrane Database Syst. Rev. 2009, CD003875. https://doi.org/10.1002/14651858.CD003875.pub3 (2009).
Singer, A. J. & Clark, R. A. Cutaneous wound healing. N Engl. J. Med. 341, 738–746. https://doi.org/10.1056/NEJM199909023411006 (1999).
Hammarstrom, L. The role of enamel matrix proteins in the development of cementum and periodontal tissues. Ciba Found. Symp. 205, 246–255. https://doi.org/10.1002/9780470515303.ch17 (1997). discussion 255–260.
Hammarstrom, L. Enamel matrix, cementum development and regeneration. J. Clin. Periodontol. 24, 658–668. https://doi.org/10.1111/j.1600-051x.1997.tb00247.x (1997).
Roccuzzo, A. et al. Enamel matrix derivative as adjunctive to non-surgical periodontal therapy: a systematic review and meta-analysis of randomized controlled trials. Clin. Oral Investig. 26, 4263–4280. https://doi.org/10.1007/s00784-022-04474-1 (2022).
Stahli, A., Bosshardt, D., Sculean, A. & Gruber, R. Emdogain-regulated gene expression in palatal fibroblasts requires TGF-betaRI kinase signaling. PLoS One. 9, e105672. https://doi.org/10.1371/journal.pone.0105672 (2014).
Gruber, R. et al. Enamel matrix derivative inhibits adipocyte differentiation of 3T3-L1 cells via activation of TGF-betaRI kinase activity. PLoS One. 8, e71046. https://doi.org/10.1371/journal.pone.0071046 (2013).
Panahipour, L., Botta, S., Abbasabadi, A. O., Afradi, Z. & Gruber, R. Enamel Matrix Derivative Suppresses Chemokine Expression in Oral Epithelial Cells. Int. J. Mol. Sci. https://doi.org/10.3390/ijms241813991 (2023).
Gruber, R. et al. TGF-betaRI kinase activity mediates Emdogain-stimulated in vitro osteoclastogenesis. Clin. Oral Investig. 18, 1639–1646. https://doi.org/10.1007/s00784-013-1129-6 (2014).
Panahipour, L., Sordi, M. B., Kargarpour, Z. & Gruber, R. TGF-beta Signalling Mediates the Anti-Inflammatory Activity of Enamel Matrix Derivative In Vitro. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23179778 (2022).
Sordi, M. B., Cabral da Cruz, A. C., Panahipour, L. & Gruber, R. Enamel Matrix Derivative Decreases Pyroptosis-Related Genes in Macrophages. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23095078 (2022).
Miron, R. J., Fujioka-Kobayashi, M., Sculean, A. & Zhang, Y. Optimization of platelet-rich fibrin. Periodontol 2000. https://doi.org/10.1111/prd.12521 (2023).
Castro, A. B., Van Dessel, J., Temmerman, A., Jacobs, R. & Quirynen, M. Effect of different platelet-rich fibrin matrices for ridge preservation in multiple tooth extractions: a split-mouth randomized controlled clinical trial. J. Clin. Periodontol. 48, 984–995. https://doi.org/10.1111/jcpe.13463 (2021).
Temmerman, A. et al. The use of leucocyte and platelet-rich fibrin in socket management and ridge preservation: a split-mouth, randomized, controlled clinical trial. J. Clin. Periodontol. 43, 990–999. https://doi.org/10.1111/jcpe.12612 (2016).
Pinto, N. R. et al. Leucocyte- and platelet-rich fibrin (L-PRF) as a regenerative medicine strategy for the treatment of refractory leg ulcers: a prospective cohort study. Platelets 29, 468–475. https://doi.org/10.1080/09537104.2017.1327654 (2018).
Karimi, K. & Rockwell, H. The benefits of Platelet-Rich Fibrin. Facial Plast. Surg. Clin. North. Am. 27, 331–340. https://doi.org/10.1016/j.fsc.2019.03.005 (2019).
Di Summa, F. et al. TGFbeta activity released from platelet-rich fibrin adsorbs to titanium surface and collagen membranes. Sci. Rep. 10, 10203. https://doi.org/10.1038/s41598-020-67167-3 (2020).
Kargarpour, Z., Nasirzade, J., Panahipour, L., Miron, R. J. & Gruber, R. Relative Centrifugal Force (RCF; G-Force) Affects the Distribution of TGF-beta in PRF Membranes Produced Using Horizontal Centrifugation. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21207629 (2020).
Nasirzade, J., Kargarpour, Z., Hasannia, S., Strauss, F. J. & Gruber, R. Platelet-rich fibrin elicits an anti-inflammatory response in macrophages in vitro. J. Periodontol. 91, 244–252. https://doi.org/10.1002/JPER.19-0216 (2020).
Kargarpour, Z. et al. Platelet-rich fibrin suppresses in vitro osteoclastogenesis. J. Periodontol. 91, 413–421. https://doi.org/10.1002/JPER.19-0109 (2020).
Panahipour, L., Croci, R., Guarnieri, S. & Gruber, R. PRF Lysates Enhance the Proliferation and Migration of Oral Squamous Carcinoma Cell Lines. Dentistry J. https://doi.org/10.3390/dj11100242 (2023).
Sordi, M. B., Panahipour, L., Kargarpour, Z. & Gruber, R. Platelet-Rich Fibrin Reduces IL-1β Release from Macrophages Undergoing Pyroptosis. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23158306 (2022).
Hacker, S. et al. Paracrine factors from irradiated peripheral blood mononuclear cells improve skin regeneration and angiogenesis in a Porcine burn Model. Sci. Rep. 6, 25168. https://doi.org/10.1038/srep25168 (2016).
Quirynen, M. et al. Instructions for the use of L-PRF in different clinical indications. Periodontol 2000. https://doi.org/10.1111/prd.12564 (2024).
Panahipour, L. et al. Bone Allograft Acid Lysates Change the Genetic Signature of Gingival Fibroblasts. Int. J. Mol. Sci. https://doi.org/10.3390/ijms242216181 (2023).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323. https://doi.org/10.1186/1471-2105-12-323 (2011).
Goedhart, J. & Luijsterburg, M. S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 10, 20560. https://doi.org/10.1038/s41598-020-76603-3 (2020).
Heberle, H., Meirelles, G. V., da Silva, F. R., Telles, G. P. & Minghim, R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 16, 169. https://doi.org/10.1186/s12859-015-0611-3 (2015).
Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. https://doi.org/10.1093/nar/gkac1000 (2023).
Kolberg, L. et al. G:profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 51, W207–W212. https://doi.org/10.1093/nar/gkad347 (2023).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Rubin, J. D. et al. Transcription factor enrichment analysis (TFEA) quantifies the activity of multiple transcription factors from a single experiment. Commun. Biol. 4, 661. https://doi.org/10.1038/s42003-021-02153-7 (2021).
Ting, J. P. & Trowsdale, J. Genetic control of MHC class II expression. Cell 109 Suppl, 21–33. https://doi.org/10.1016/s0092-8674(02)00696-7 (2002).
Feng, H., Zhang, Y. B., Gui, J. F., Lemon, S. M. & Yamane, D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 17, e1009220. https://doi.org/10.1371/journal.ppat.1009220 (2021).
Ramana, C. V., Gil, M. P., Schreiber, R. D. & Stark, G. R. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 23, 96–101. https://doi.org/10.1016/s1471-4906(01)02118-4 (2002).
Blank, U. & Karlsson, S. The role of Smad signaling in hematopoiesis and translational hematology. Leukemia 25, 1379–1388. https://doi.org/10.1038/leu.2011.95 (2011).
Shimano, H. & Sato, R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710–730. https://doi.org/10.1038/nrendo.2017.91 (2017).
Panahipour, L., Croci, R., Guarnieri, S. & Gruber, R. PRF Lysates Enhance the Proliferation and Migration of Oral Squamous Carcinoma Cell Lines. Dent. J. (Basel) https://doi.org/10.3390/dj11100242 (2023).
Yates, C. C. et al. Delayed reepithelialization and basement membrane regeneration after wounding in mice lacking CXCR3. Wound Repair. Regen. 17, 34–41. https://doi.org/10.1111/j.1524-475X.2008.00439.x (2009).
Tortelli, F., Pisano, M., Briquez, P. S., Martino, M. M. & Hubbell, J. A. Fibronectin binding modulates CXCL11 activity and facilitates wound healing. PLoS One. 8, e79610. https://doi.org/10.1371/journal.pone.0079610 (2013).
Bekeschus, S. et al. Distinct cytokine and chemokine patterns in chronic diabetic ulcers and acute wounds. Exp. Dermatol. 26, 145–147. https://doi.org/10.1111/exd.13215 (2017).
Nuutila, K. et al. Human skin transcriptome during superficial cutaneous wound healing. Wound Repair. Regen. 20, 830–839. https://doi.org/10.1111/j.1524-475X.2012.00831.x (2012).
Vogl, T. et al. MRP8 and MRP14 control microtubule reorganization during transendothelial migration of phagocytes. Blood 104, 4260–4268. https://doi.org/10.1182/blood-2004-02-0446 (2004).
Wang, Y. & Tatakis, D. N. Human gingiva transcriptome during wound healing. J. Clin. Periodontol. 44, 394–402. https://doi.org/10.1111/jcpe.12669 (2017).
Ben Mkaddem, S., Benhamou, M. & Monteiro, R. C. Understanding fc receptor involvement in Inflammatory diseases: from mechanisms to New Therapeutic Tools. Front. Immunol. 10, 811. https://doi.org/10.3389/fimmu.2019.00811 (2019).
Bossi, F. et al. C1q as a unique player in angiogenesis with therapeutic implication in wound healing. Proc. Natl. Acad. Sci. U S A. 111, 4209–4214. https://doi.org/10.1073/pnas.1311968111 (2014).
Hermida-Nogueira, L. et al. Deciphering the secretome of leukocyte-platelet rich fibrin: towards a better understanding of its wound healing properties. Sci. Rep. 10, 14571. https://doi.org/10.1038/s41598-020-71419-7 (2020).
Farber, J. M. A macrophage mRNA selectively induced by gamma-interferon encodes a member of the platelet factor 4 family of cytokines. Proc. Natl. Acad. Sci. U S A. 87, 5238–5242. https://doi.org/10.1073/pnas.87.14.5238 (1990).
Proost, P. et al. Microbial toll-like receptor ligands differentially regulate CXCL10/IP-10 expression in fibroblasts and mononuclear leukocytes in synergy with IFN-gamma and provide a mechanism for enhanced synovial chemokine levels in septic arthritis. Eur. J. Immunol. 33, 3146–3153. https://doi.org/10.1002/eji.200324136 (2003).
Wang, S. et al. S100A8/A9 in inflammation. Front. Immunol. 9, 1298. https://doi.org/10.3389/fimmu.2018.01298 (2018).
Schiopu, A. & Cotoi, O. S. S100A8 and S100A9: DAMPs at the crossroads between innate immunity, traditional risk factors, and cardiovascular disease. Mediators Inflamm 2013, 828354. https://doi.org/10.1155/2013/828354 (2013).
Xu, K. & Geczy, C. L. IFN-gamma and TNF regulate macrophage expression of the chemotactic S100 protein S100A8. J. Immunol. 164, 4916–4923. https://doi.org/10.4049/jimmunol.164.9.4916 (2000).
Ernst, L. K., van de Winkel, J. G., Chiu, I. M. & Anderson, C. L. Three genes for the human high affinity fc receptor for IgG (fc gamma RI) encode four distinct transcription products. J. Biol. Chem. 267, 15692–15700 (1992).
Li, Y. et al. Increased expression of FcgammaRI/CD64 on circulating monocytes parallels ongoing inflammation and nephritis in lupus. Arthritis Res. Ther. 11, R6. https://doi.org/10.1186/ar2590 (2009).
Uciechowski, P. et al. IFN-gamma induces the high-affinity fc receptor I for IgG (CD64) on human glomerular mesangial cells. Eur. J. Immunol. 28, 2928–2935. (1998).
Bosetti, M. et al. The Number of Platelets in Patient’s Blood Influences the Mechanical and Morphological Properties of PRP-Clot and Lysophosphatidic Acid Quantity in PRP. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21010139 (2019).
Fransson, J. et al. Activation of macrophages by Lysophosphatidic Acid through the Lysophosphatidic Acid receptor 1 as a Novel mechanism in multiple sclerosis pathogenesis. Mol. Neurobiol. 58, 470–482. https://doi.org/10.1007/s12035-020-02130-x (2021).
Ray, R. & Rai, V. Lysophosphatidic acid converts monocytes into macrophages in both mice and humans. Blood 129, 1177–1183. https://doi.org/10.1182/blood-2016-10-743757 (2017).
Gruber, R., Kandler, B., Jindra, C., Watzak, G. & Watzek, G. Dental pulp fibroblasts contain target cells for lysophosphatidic acid. J. Dent. Res. 83, 491–495. https://doi.org/10.1177/154405910408300611 (2004).
Wojdasiewicz, P., Turczyn, P., Dobies-Krzesniak, B., Frasunska, J. & Tarnacka, B. Role of CX3CL1/CX3CR1 Signaling Axis Activity in Osteoporosis. Mediators Inflamm. 2019, 7570452. https://doi.org/10.1155/2019/7570452 (2019).
Ishida, Y., Gao, J. L. & Murphy, P. M. Chemokine receptor CX3CR1 mediates skin wound healing by promoting macrophage and fibroblast accumulation and function. J. Immunol. 180, 569–579. https://doi.org/10.4049/jimmunol.180.1.569 (2008).
Zhou, J., Ye, S., Fujiwara, T., Manolagas, S. C. & Zhao, H. Steap4 plays a critical role in osteoclastogenesis in vitro by regulating cellular iron/reactive oxygen species (ROS) levels and cAMP response element-binding protein (CREB) activation. J. Biol. Chem. 288, 30064–30074. https://doi.org/10.1074/jbc.M113.478750 (2013).
Yao, F. et al. FABP4 inhibitor attenuates inflammation and endoplasmic reticulum stress of islet in leptin receptor knockout rats. Eur. Rev. Med. Pharmacol. Sci. 24, 12808–12820. https://doi.org/10.26355/eurrev_202012_24182 (2020).
Koller, F. L. et al. Lack of MMP10 exacerbates experimental colitis and promotes development of inflammation-associated colonic dysplasia. Lab. Invest. 92, 1749–1759. https://doi.org/10.1038/labinvest.2012.141 (2012).
Valdes-Fernandez, J. et al. Molecular and Cellular mechanisms of delayed fracture Healing in Mmp10 (Stromelysin 2) knockout mice. J. Bone Min. Res. 36, 2203–2213. https://doi.org/10.1002/jbmr.4403 (2021).
Nakamura, K., Okubo, Y., Minami, M., Furue, M. & Ishibashi, Y. Phenotypic analysis of CD23 + peripheral blood mononuclear cells in atopic dermatitis. Br. J. Dermatol. 125, 543–547. https://doi.org/10.1111/j.1365-2133.1991.tb14791.x (1991).
Mamidi, M. K. et al. FCRL1 immunoregulation in B cell development and malignancy. Front. Immunol. 14, 1251127. https://doi.org/10.3389/fimmu.2023.1251127 (2023).
Li, F. J., Schreeder, D. M., Li, R., Wu, J. & Davis, R. S. FCRL3 promotes TLR9-induced B-cell activation and suppresses plasma cell differentiation. Eur. J. Immunol. 43, 2980–2992. https://doi.org/10.1002/eji.201243068 (2013).
Bajpai, U. D. et al. A functional variant in FCRL3 is associated with higher fc receptor-like 3 expression on T cell subsets and rheumatoid arthritis disease activity. Arthritis Rheum. 64, 2451–2459. https://doi.org/10.1002/art.34457 (2012).
Potts, J. R. & Campbell, I. D. Fibronectin structure and assembly. Curr. Opin. Cell. Biol. 6, 648–655. https://doi.org/10.1016/0955-0674(94)90090-6 (1994).
Fuior, E. V. & Gafencu, A. V. Apolipoprotein C1: Its Pleiotropic Effects in Lipid Metabolism and Beyond. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20235939 (2019).
Wu, X. et al. HtrA1 is upregulated during RANKL-induced osteoclastogenesis, and negatively regulates osteoblast differentiation and BMP2-induced Smad1/5/8, ERK and p38 phosphorylation. FEBS Lett. 588, 143–150. https://doi.org/10.1016/j.febslet.2013.11.022 (2014).
Nakayachi, M. et al. Lectin-like oxidized low-density lipoprotein receptor-1 abrogation causes resistance to inflammatory bone destruction in mice, despite promoting osteoclastogenesis in the steady state. Bone 75, 170–182. https://doi.org/10.1016/j.bone.2015.02.025 (2015).
Noguchi, T. et al. Apolipoprotein E plays crucial roles in maintaining bone mass by promoting osteoblast differentiation via ERK1/2 pathway and by suppressing osteoclast differentiation via c-Fos, NFATc1, and NF-kappaB pathway. Biochem. Biophys. Res. Commun. 503, 644–650. https://doi.org/10.1016/j.bbrc.2018.06.055 (2018).
Birkl, D. et al. Formyl peptide receptor 2 regulates monocyte recruitment to promote intestinal mucosal wound repair. FASEB J. 33, 13632–13643. https://doi.org/10.1096/fj.201901163R (2019).
Zhou, Z. et al. Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metalloproteinase I. Proc. Natl. Acad. Sci. U S A. 97, 4052–4057. https://doi.org/10.1073/pnas.060037197 (2000).
Holmbeck, K. et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell 99, 81–92. https://doi.org/10.1016/s0092-8674(00)80064-1 (1999).
Lieu, S. et al. Impaired remodeling phase of fracture repair in the absence of matrix metalloproteinase-2. Dis. Model. Mech. 4, 203–211. https://doi.org/10.1242/dmm.006304 (2011).
Wang, Z. et al. TNS1: Emerging Insights into Its Domain Function, Biological Roles, and Tumors. Biology (Basel) https://doi.org/10.3390/biology11111571 (2022).
Engelmann, J. et al. Regulation of bone homeostasis by MERTK and TYRO3. Nat. Commun. 13, 7689. https://doi.org/10.1038/s41467-022-33938-x (2022).
Gao, L. et al. Receptor tyrosine kinases Tyro3, Axl, and Mertk differentially contribute to antibody-induced arthritis. Cell. Commun. Signal. 21, 195. https://doi.org/10.1186/s12964-023-01133-0 (2023).
Santos, A. J. M. et al. A human autoimmune organoid model reveals IL-7 function in coeliac disease. Nature 632, 401–410. https://doi.org/10.1038/s41586-024-07716-2 (2024).
Kargarpour, Z., Nasirzade, J., Panahipour, L., Miron, R. J. & Gruber, R. Platelet-Rich Fibrin Decreases the Inflammatory Response of Mesenchymal Cells. Int. J. Mol. Sci. https://doi.org/10.3390/ijms222111333 (2021).
Kargarpour, Z., Panahipour, L., Mildner, M., Miron, R. J. & Gruber, R. Lipids of Platelet-Rich Fibrin Reduce the Inflammatory Response in Mesenchymal Cells and Macrophages. Cells https://doi.org/10.3390/cells12040634 (2023).
Acknowledgements
We thank the team of the Core Facility Genomics at the Medical University of Vienna for their support.
Funding
ITI Research Grants 1729–2023 and 1709–2022.
Author information
Authors and Affiliations
Contributions
Conceptualization, L.P., Z.K. and R.G.; formal analysis, L.P., M.M., H.K. and R.G.; methodology, L.P., M.M., H.K. and R.G.; project administration, L.P.; visualization, L.P., M.M., H.K. and R.G.; writing—original draft, L.P., M.M. and R.G.; writing—review and editing, L.P., Z.K., M.M., H.K. and R.G.; All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Panahipour, L., Kargarpour, Z., Mildner, M. et al. RNAseq of peripheral blood mononucleated cells exposed to platelet-rich fibrin and enamel matrix derivatives. Sci Rep 15, 3661 (2025). https://doi.org/10.1038/s41598-025-86791-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-86791-5