Abstract
Gastrointestinal polyps are observed and treated under endoscopy, so there presents significant challenges to advance endoscopy imaging segmentation of polyps. Current methodologies often falter in distinguishing complex polyp structures within diverse (mucosal) tissue environments. In this paper, we propose the Frequency Attention-Embedded Network (FAENet), a novel approach leveraging frequency-based attention mechanisms to enhance polyp segmentation accuracy significantly. FAENet ingeniously segregates and processes image data into high and low-frequency components, enabling precise delineation of polyp boundaries and internal structures by integrating intra-component and cross-component attention mechanisms. This method not only preserves essential edge details but also refines the learned representation attentively, ensuring robust segmentation across varied imaging conditions. Comprehensive evaluations on two public datasets, Kvasir-SEG and CVC-ClinicDB, demonstrate FAENet’s superiority over several state-of-the-art models in terms of Dice coefficient, Intersection over Union (IoU), sensitivity, and specificity. The results affirm that FAENet’s advanced attention mechanisms significantly improve the segmentation quality, outperforming traditional and contemporary techniques. FAENet’s success indicates its potential to revolutionize polyp segmentation in clinical practices, fostering diagnosis and efficient treatment of gastrointestinal polyps.
Similar content being viewed by others
Introduction
Polyp is a common mucosal lesion in gastrointestinal tract (commonly occured in colon, stomach, rectum etc). The majority of gastrointestinal polyps are benign with no symptom while only the adenomatous type has the potential to progress into malignancy. Hence, polyp requires early diagnosis and completely excision1,2,3,4. In clinic, endoscopy allows gastrointestinal tract visualized directly, which promote the observation of polyp. Also, except some special types, polyp can be excised under microscopy. However, that requires clinicians well-trained and this manual process is not only labor-intensive but also prone to variability. An efficient alternative is the development of automated polyp segmentation algorithms that assist clinicians in precisely locating and delineating polyp regions for further analysis5,6,7,8.
Polyps exhibit considerable variability in size and shape at different stages of development, and accurately segmenting them is challenging due to their similarity to surrounding mucosa. To address these challenges, several deep learning models have been implemented, showing promise in polyp segmentation. For instance, Akbari et al.9 employed a fully convolutional network (FCN) combined with Otsu thresholding to delineate prominent polyp regions. Sun et al.10 introduced an FCN-based framework that incorporates dilated convolution to capture expansive semantic features without losing resolution. UNet-based11 architectures, particularly those with encoder-decoder structures like ResUNet++12 and UNet++13, have demonstrated effective performance by merging high-level features from the decoder with low-level features from the encoder through skip connections, thereby integrating semantic and contextual information efficiently. Nevertheless, these methods often focus predominantly on segmenting the polyp’s entire area, occasionally overlooking crucial boundary details. Recent advancements have included area-boundary constraints14 and multi-task frameworks to enhance boundary delineation, significantly improving segmentation accuracy15. Furthermore, Fan et al.16 introduced a reverse attention module to leverage boundary cues for more precise polyp segmentation.
Despite notable progress in automatic polyp segmentation, significant challenges remain. Flat lesions or inadequate bowel preparation can obscure polyp margins, compromising segmentation accuracy. It is crucial to utilize boundary-specific information to improve the delineation between polyps and adjacent tissues. Image processing typically associates inner body structures with low-frequency components, while edges correlate with high-frequency components. Figure 1 illustrates the different frequency components in the red, green, and blue bands of a colonoscopy image. The high-frequency components (LH, HL and HH) contain rich edge-related information, whereas the low-frequency component (LL) captures the inner body features of a polyp. This differentiation underscores that effective models must integrate both low and high-frequency details to achieve a comprehensive understanding of both the inner consistency and the edge details of polyps.
In summary, two primary challenges persist in polyp segmentation using CNN-based models. Firstly, despite their commendable performance, CNNs are restricted by local receptive fields, which limit their ability to capture comprehensive contextual information crucial for accurate inference. Furthermore, while attention-enhanced networks substantially improve by modeling long-range dependencies, they frequently generate blurred polyp boundaries, resulting in considerable misclassification of adjacent pixels. Secondly, conventional learning paradigms in Euclidean space cannot effectively capture the high-frequency details which are vital for representing edge distributions.
To overcome these limitations, we introduce a Frequency Attention Model (FAM) designed to preserve edge details more effectively during feature map encoding and decoding. FAM operates by segregating feature maps into high and low-frequency components, enhancing them within and across components through Intra-Component Attention (ICA) and Cross-Component Attention (CCA), respectively. This process not only refines the features within each frequency band but also enhances inter-component interactions, significantly improving feature distinguishability. The main contributions are as follows:
-
We propose a FAM, which projects feature maps into the frequency ___domain, enhancing edge detail preservation through a dual attention mechanism. This mechanism refines both high and low-frequency components, substantially sharpening polyp edges in segmentation tasks.
-
We design the Frequency Attention-Embedded Network (FAENet) for polyp segmentation, which incorporates FAMs instead of original convolution layers to refine feature maps across various scales efficiently.
-
Through extensive testing on two challenging datasets, FAENet demonstrates superior performance over state-of-the-art models. Our results are further validated by an ablation study that highlights the advantages of FAM over convolution layer and conventional self-attention models.
Related works
Semantic segmentation of medical images
In medical imaging, segmentation is critical for delineating relevant areas within computer-aided diagnosis systems17,18. Conventional segmentation methods, such as random walk and cantilever beam approaches, have been applied successfully in medical image analysis due to their simplicity and low computational cost19,20,21. These methods rely on probabilistic boundary delineation or physical models, making them efficient and effective in scenarios with limited computational resources or simpler segmentation tasks, such as single-organ contour extraction. However, their performance is often constrained in complex scenarios involving irregular structures, subtle boundaries, or varying imaging conditions. In contrast, FAENet incorporates frequency-based attention mechanisms, which significantly enhance segmentation accuracy by refining edge details and capturing contextual information. While this comes at the cost of increased computational requirements, it positions FAENet as a more robust solution for demanding applications like polyp segmentation, where precision and adaptability are paramount.
CNN-based approaches have shown promising results, with UNet achieving widespread success. Innovations such as ResUNet++12, which integrates additional layers into UNet22, and H-DenseUNet23, which combines 2D and 3D networks for feature extraction and context aggregation, exemplify advancements aimed at refining segmentation accuracy. UNet++13 further optimizes this by redesigning skip connections to minimize the semantic gap between the encoder and decoder stages. Deep learning networks have often matched the precision of manual delineation in segmenting medical images, yet accurately defining certain critical regions, such as organ boundaries with low contrast, continues to pose challenges24,25,26,27. To address these issues, several innovative strategies have been introduced. One effective technique integrates attention mechanisms into the network architecture, enhancing the model’s focus on salient features within the medical images28. This integration aids in accentuating important areas while diminishing the impact of non-essential data, thereby boosting the accuracy of segmentation29,30. Another strategy leverages advanced imaging processing techniques to further refine the detection and delineation of subtle features in medical scans31. These approaches collectively contribute to overcoming inherent limitations in traditional imaging segmentation, particularly in areas where contrast is minimal and detail is crucial32,33.
In this study, we propose to segregate feature maps into high and low-frequency bands and apply targeted enhancements with ICA and CCA. ICA focuses on refining the details within each frequency band, while CCA facilitates interactions between the bands, thereby enhancing the overall distinguishability of features. This dual approach ensures that both the nuances within each frequency range and the synergy between them are optimized, leading to more precise feature representation.
Learning in frequency ___domain
Leveraging the intricate frequency and spatial representation strengths of wavelet transforms, these have been integrated into deep neural networks to enhance semantic segmentation, with various approaches documented34,35,36,37,38. Common methodologies typically utilize wavelet transforms for preprocessing or post-processing tasks, or as replacements for certain CNN operations like up- and down-sampling. Preprocessing techniques play a vital role in enhancing the suitability of input data for advanced machine learning models. Studies have demonstrated that preprocessing can improve feature extraction by balancing data distributions and reducing input noise39,40. In the context of frequency-based segmentation, preprocessing steps such as normalization and denoising could align input images more effectively with the requirements of frequency transformations, leading to more accurate attention maps and segmentation results. Despite their effectiveness, many of these methods tend to be object-specific, thus limiting their broader applicability37.
For instance, Aerial LaneNet41, a symmetric CNN architecture augmented with wavelet transform, is specifically tailored for lane-marking segmentation in aerial images. Similarly, the Constrained Wavelet Neural Network (CWNN) employs wavelet-constrained pooling layers as a substitute for traditional pooling operations, focusing on synthetic aperture radar image segmentation42. Wavelet Segmentation Network (WaveSNet)43 integrates wavelet transforms to enhance detail during down-sampling and employs inverse transforms to restore details in the up-sampling phase. In contrast, we use wavelet transform to generate low-frequency and high-frequency images as dual-branch input to extract corresponding features.
Method
Overview
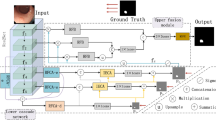
As shown in Fig. 2, FAENet inherits the design of U-shape network with its stable capability for medical image segmentation. The architecture is designed to process an input image through multiple convolution layers (batch normalization and ReLU are omitted in this paper), each followed by a pooling operation to reduce spatial dimensions and capture higher-level features. These features are then intricately processed through our novel FAM, which enhances feature representation by focusing on both low-frequency and high-frequency components separately. After processing through the FAM, features from both branches are merged using concatenation followed by element-wise summation, ensuring that both detailed textural information and broader contextual details are preserved and emphasized. This combination allows the network to maintain sharpness in boundaries and improve the segmentation accuracy, particularly in challenging areas of the image. Subsequently, the concatenated features undergo several convolution layers to refine the feature maps, which are then progressively up-sampled back to the original image dimensions. Each up-sampling stage is paired with a convolution operation to further refine details and maintain spatial consistency. The final output is generated through a Softmax layer that classifies each pixel into the appropriate label, producing the segmented output.
Frequency attention model
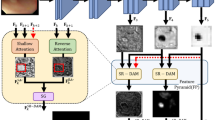
The Frequency Attention Model (FAM) is the core module of FAENet, designed to extract and process both low-frequency and high-frequency details in parallel. As illustrated in Fig. 3, using the Discrete Wavelet Transform (DWT), the input feature map \(F_{input}^{fa} \in {\mathbb {R}}^{H \times W \times C}\), where H, W, and C represent height, width, and number of channels, is decomposed into four components: \(F_{LL}, F_{LH}, F_{HL}, F_{HH}\). These components capture the low-frequency (contextual) and high-frequency (edge) features at half the spatial resolution \(\frac{H}{2} \times \frac{W}{2} \times C\).
Low-frequency features: The low-frequency component \(F_{LL}\), which contains global contextual information, is processed through a \(3 \times 3\) convolution layer, followed by element-wise summation and a \(1 \times 1\) convolution layer, resulting in the initialized low-frequency feature \(F_{lc} \in {\mathbb {R}}^{\frac{H}{2} \times \frac{W}{2} \times C}\).
High-frequency features: The high-frequency components \(F_{LH}, F_{HL}, F_{HH}\), which represent boundary and texture details, are concatenated and compressed into a single feature map using a \(1 \times 1\) convolution layer. The resulting high-frequency feature \(F_{hc} \in {\mathbb {R}}^{\frac{H}{2} \times \frac{W}{2} \times C}\) preserves critical edge details for further enhancement.
Intra-component attention (ICA): ICA is applied to independently enhance \(F_{lc}\) and \(F_{hc}\). The process includes:
-
Generating three feature maps \(F_{1}^{ica}, F_{2}^{ica}, F_{3}^{ica} \in {\mathbb {R}}^{H \times W \times C}\) using \(1 \times 1\) convolutions.
-
Computing position-wise similarity via attention maps:
$$\begin{aligned} AM_{ica} = \text {Softmax}\left( \left( F_{1}^{ica}\right) ^\top \times F_{2}^{ica}\right) , \end{aligned}$$(1)where \(AM_{ica} \in {\mathbb {R}}^{(HW) \times (HW)}\).
-
Aggregating enhanced features:
$$\begin{aligned} F_{output}^{ica} = F_{input}^{ica} + AM_{ica} \times F_{3}^{ica}, \end{aligned}$$(2)resulting in \(F_{output}^{ica} \in {\mathbb {R}}^{H \times W \times C}\) with enriched feature representation.
Cross-component attention (CCA): CCA refines the interactions between low-frequency (\(F_{lc}^{ica}\)) and high-frequency (\(F_{hc}^{ica}\)) features:
-
Attention maps are computed as:
$$\begin{aligned} AM_{cca} = \text {Softmax}\left( \left( F_{lc}^{ica}\right) ^\top \times F_{hc}^{ica}\right) , \end{aligned}$$(3)where \(AM_{cca} \in {\mathbb {R}}^{(HW) \times (HW)}\).
-
Refined feature maps \(F_{loutput}^{cca}\) and \(F_{houtput}^{cca}\) are produced, maintaining the dimensions \(H \times W \times C\).
Reconstruction: The inverse DWT (iDWT) integrates the refined low-frequency and high-frequency features back into the spatial ___domain. This process ensures minimal distortion while preserving both contextual and edge-specific details.
Summary: FAM is a modular frequency-based enhancement block that processes and enriches feature maps without altering their dimensions. By integrating both contextual and boundary information, FAM ensures robust feature representation, making it highly effective for segmentation tasks.
Detailed pipelines of the Frequency Attention Model (FAM), Intra-Component Attention (ICA), and Cross-Component Attention (CCA). The Discrete Wavelet Transform (DWT) decomposes the input into frequency components (LL, LH, HL, HH). ICA enriches features within each frequency band, while CCA enhances interactions between low-frequency and high-frequency components. The refined features are integrated into the spatial ___domain using the Inverse DWT (iDWT).
Experiments and results
Datasets
We evaluated the segmentation capabilities of FAENet on polyps using the publicly available Kvasir-SEG and CVC-ClinicDB datasets, which comprise optical colonoscopy images. As listed in Table 1, the dataset is partitioned at a ratio of 3:1:1 for training, validation and test. The Kvasir-SEG dataset, sourced from Vestre Viken Health Trust in Norway, includes 1,000 images with resolutions ranging from 487 \(\times\) 332 to 1920 \(\times\) 1072 pixels. These images were annotated by a medical doctor and validated by an experienced gastroenterologist. The CVC-ClinicDB, comprising 612 frames with a resolution of 288 \(\times\) 368 pixels, was derived from video sequences captured during standard optical colonoscopy procedures from 13 patients.
Evaluation metrics
In this study, we evaluated the performance of our predictions on the test set using four standard evaluation metrics:
where TP, TN, FP, and FN denote the counts of true positives, true negatives, false positives, and false negatives, respectively. Moreover, mDice and mIoU are calculated over all test set.
Implement details
FAENet and benchmark models were implemented on a Linux platform using PyTorch, accelerated by an NVIDIA A40 GPU. We employed data augmentation techniques such as random flipping and cropping to improve model robustness. Batch size is set to 64 for training with the sub-patch is with a size of \(256\times 256\). The training regimen set the initial learning rate at 0.02 across a maximum of 500 epochs, using the SGD optimizer with a polynomial decay learning strategy and a momentum of 0.9. Moreover, the Softmax cross-entropy is selected as the loss function. The model version with the lowest validation loss was selected for subsequent evaluations. FAENet was benchmarked against leading models such as SegNet44, UNet22, UNet++13, PraNet16, ResUNet++12, SSCNet35 and XNet45, ensuring consistent settings for all comparisons.
Comparisons with state-of-the-art methods
The results presented in Table 2 demonstrate the performance of FAENet against a variety of state-of-the-art segmentation models, including CNN-based (e.g., UNet, PraNet) and Transformer-based (e.g., Swin-Unet, MedT) methods. FAENet achieves the highest Dice coefficient (0.917 on Kvasir-SEG and 0.933 on CVC-ClinicDB) and mIoU (0.863 on Kvasir-SEG and 0.883 on CVC-ClinicDB), outperforming all baseline methods. Notably, Transformer-based methods like Swin-Unet and MedT, despite their advanced architectures, show marginally lower performance (e.g., MedT achieves 0.847 Dice on Kvasir-SEG) compared to FAENet. This highlights the effectiveness of FAENet’s frequency attention mechanisms in preserving edge details and handling diverse polyp morphologies.
A closer look at sensitivity and specificity metrics reveals FAENet’s ability to balance true positive identification with reduced false positives. On CVC-ClinicDB, FAENet achieves a sensitivity of 0.993 and specificity of 0.960, surpassing Swin-Unet (0.989 sensitivity and 0.883 specificity) and MedT (0.988 sensitivity and 0.889 specificity). These improvements underscore FAENet’s robustness in identifying challenging polyp boundaries, even in edge cases involving small or flat lesions.
As shown in Fig. 4, visual inspections on the Kvasir-SEG dataset illustrate FAENet’s ability to handle complex polyp structures with high precision. Compared to Swin-Unet and MedT, FAENet demonstrates sharper boundary delineation and fewer false positives in flat or small lesions. For example, in cases where lesions are nearly indistinguishable from surrounding mucosa, FAENet’s frequency-based attention mechanisms ensure accurate segmentation, as evident in its ability to preserve edge details without compromising sensitivity.
On the CVC-ClinicDB dataset, FAENet consistently outperforms baseline methods, including Transformer-based models, by correctly segmenting irregular and faint polyp boundaries (see in Fig. 5). Visual comparisons show that FAENet produces results closest to the ground truth, with minimal boundary artifacts and higher consistency across varied lesion sizes. Particularly in edge cases involving extremely small lesions, FAENet’s sensitivity ensures that no significant regions are overlooked, providing robust segmentation even under challenging imaging conditions.
Effects of FAM
The experimental results detailed in Table 2 provide a robust evaluation of the proposed FAENet against its two variants: FAENet-C, where the FAM is replaced with standard convolution layers, and FAENet-S, where FAM is substituted by self-attention layers. These comparisons are crucial to understanding the specific impact and advantage of incorporating the FAM into the network architecture.
FAENet demonstrates superior performance across all metrics on both the Kvasir-SEG and CVC-ClinicDB datasets, illustrating the effectiveness of the frequency attention mechanism. Notably, FAENet achieves a Dice score of 0.917 and an mIoU of 0.863 on Kvasir-SEG, significantly outperforming FAENet-C (Dice: 0.817, mIoU: 0.745) and FAENet-S (Dice: 0.832, mIoU: 0.781). This trend persists across the datasets, where FAENet consistently leads with higher sensitivity and specificity. For instance, on CVC-ClinicDB, FAENet records a specificity of 0.960, considerably higher than that of FAENet-C (0.841) and FAENet-S (0.874). These results underline the efficacy of FAENet in maintaining high precision in segmenting polyps while reducing false positives more effectively than its counterparts.
Effects of ICA and CCA
As shown in Table 2, removing ICA causes a noticeable decline in segmentation performance, with the Dice coefficient dropping from 0.917 to 0.875 on Kvasir-SEG and from 0.933 to 0.887 on CVC-ClinicDB. Similarly, the mIoU decreases by approximately 5%, highlighting the reduced capability to model detailed context within each frequency band. These results underscore that ICA effectively refines frequency-specific features, improving segmentation precision, particularly in scenarios requiring nuanced boundary delineation. Excluding CCA from FAENet results in moderate but significant performance degradation, with the Dice coefficient decreasing to 0.882 on Kvasir-SEG and 0.890 on CVC-ClinicDB. The mIoU also suffers, reflecting reduced synergy between contextual and edge-specific features. These findings highlight the importance of CCA in modeling cross-frequency correlations, which is crucial for accurately segmenting complex structures with diverse spatial and frequency characteristics.
Efficiency analysis
To evaluate the computational efficiency of FAENet, we measured the inference time per image on the Kvasir-SEG and CVC-ClinicDB datasets and compared it with state-of-the-art segmentation methods. As shown in Table 3, FAENet achieves an average inference time of 11.0 ± 0.4 ms for 256\(\times\)256 images from the Kvasir-SEG dataset and 12.9 ± 0.5 ms for 384\(\times\)288 images from the CVC-ClinicDB dataset. These times are competitive with other methods, such as UNet and PraNet, while being faster than transformer-based models like Swin-Unet (15.4 ± 0.7 ms on Kvasir-SEG) and MedT (16.2 ± 0.6 ms on Kvasir-SEG). This balance of efficiency and accuracy highlights FAENet’s suitability for real-time clinical applications, where computational performance is as critical as segmentation accuracy.
Discussion and conclusion
The extensive comparison and evaluations demonstrate that the FAENet sets a new benchmark in polyp segmentation across challenging datasets. By innovatively integrating frequency-based attention mechanisms, FAENet addresses common limitations in polyp segmentation such as capturing fine-grained details and handling complex textures and shapes. The distinct use of frequency attention to refine both low and high-frequency components allows the network to perform robustly under diverse imaging conditions, making it particularly effective in scenarios where precision and reliability are paramount.
Real-time segmentation is essential for clinical applications, where computational delays can hinder decision-making. Recent advancements in parallel computation, such as heterogeneous system-on-chip architectures and FPGA-based accelerations, have shown promise in reducing computational complexity for tasks like blood flow simulation48,49,50. Incorporating similar parallelization techniques into FAENet could optimize its computational efficiency, enabling faster inference without sacrificing accuracy. Future work will explore these avenues to further enhance FAENet’s clinical utility.
FAENet, while effective, has several limitations that warrant attention. First, its computational complexity and memory requirements may hinder deployment in real-time or resource-constrained environments. Moreover, the method’s sensitivity to hyperparameters and its reliance on high-quality input images suggest the need for further optimization and preprocessing strategies to ensure consistent performance across diverse datasets and imaging modalities. Future work will focus on reducing computational demands through model optimization and exploring automated hyperparameter tuning methods. Additionally, we aim to validate FAENet on broader medical imaging tasks and diverse datasets to enhance its generalizability and clinical applicability.
Data availability
The datasets and source codes used and/or analysed during the current study available from the first author on reasonable request.
References
Bernal, J., Sánchez, J. & Vilarino, F. Towards automatic polyp detection with a polyp appearance model. Pattern Recogn. 45, 3166–3182 (2012).
Navarro, M., Nicolas, A., Ferrandez, A. & Lanas, A. Colorectal cancer population screening programs worldwide in 2016: An update. World J. Gastroenterol. 23, 3632 (2017).
Liu, G. et al. Cafe-net: Cross-attention and feature exploration network for polyp segmentation. Expert Syst. Appl. 238, 121754 (2024).
Liu, Y., Yang, Y., Jiang, Y. & Xie, Z. Multi-view orientational attention network combining point-based affinity for polyp segmentation. Expert Syst. Appl. 249, 123663 (2024).
Zhang, R., Zheng, Y., Poon, C. C., Shen, D. & Lau, J. Y. Polyp detection during colonoscopy using a regression-based convolutional neural network with a tracker. Pattern Recogn. 83, 209–219 (2018).
Lee, G.-E., Cho, J. & Choi, S.-I. Shallow and reverse attention network for colon polyp segmentation. Sci. Rep. 13, 15243 (2023).
Sikkandar, M. Y. et al. Utilizing adaptive deformable convolution and position embedding for colon polyp segmentation with a visual transformer. Sci. Rep. 14, 7318 (2024).
Lewis, J., Cha, Y.-J. & Kim, J. Dual encoder-decoder-based deep polyp segmentation network for colonoscopy images. Sci. Rep. 13, 1183 (2023).
Akbari, M. et al. Polyp segmentation in colonoscopy images using fully convolutional network. In 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 69–72 (IEEE, 2018).
Sun, X., Zhang, P., Wang, D., Cao, Y. & Liu, B. Colorectal polyp segmentation by u-net with dilation convolution. In 2019 18th IEEE International Conference on Machine Learning and Applications (ICMLA), 851–858 (IEEE, 2019).
Sang, D. V. et al. Ag-curesnest: A novel method for colon polyp segmentation. Preprint at http://arxiv.org/abs/2105.00402 (2021).
Jha, D. et al. Resunet++: An advanced architecture for medical image segmentation. In 2019 IEEE International Symposium on Multimedia (ISM), 225–2255 (IEEE, 2019).
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N. & Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 39, 1856–1867 (2019).
Fang, Y., Chen, C., Yuan, Y. & Tong, K.-y. Selective feature aggregation network with area-boundary constraints for polyp segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part I 22, 302–310 (Springer, 2019).
Murugesan, B. et al. Psi-net: Shape and boundary aware joint multi-task deep network for medical image segmentation. In 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 7223–7226 (IEEE, 2019).
Fan, D.-P. et al. Pranet: Parallel reverse attention network for polyp segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 263–273 (Springer, 2020).
Jia, H., Yap, P.-T. & Shen, D. Iterative multi-atlas-based multi-image segmentation with tree-based registration. Neuroimage 59, 422–430 (2012).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017).
Dakua, S. & Sahambi, J. Automatic contour extraction of multi-labeled left ventricle from cmr images using cb and random walk approach. Cardiovasc. Eng. 10, 30–43 (2010).
Dakua, S. P. & Sahambi, J. S. Lv contour extraction from cardiac mr images using random walks approach. In 2009 IEEE International Advance Computing Conference, 228–233 (IEEE, 2009).
Dakua, S. P. & Sahambi, J. Detection of left ventricular myocardial contours from ischemic cardiac mr images. IETE J. Res. 57, 372–384 (2011).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18, 234–241 (Springer, 2015).
Li, X. et al. H-denseunet: hybrid densely connected unet for liver and tumor segmentation from ct volumes. IEEE Trans. Med. Imaging 37, 2663–2674 (2018).
Xu, M., Guo, H., Zhang, J., Yan, K. & Lu, L. A new probabilistic v-net model with hierarchical spatial feature transform for efficient abdominal multi-organ segmentation. Preprint at http://arxiv.org/abs/2208.01382 (2022).
Jin, D. et al. Deeptarget: Gross tumor and clinical target volume segmentation in esophageal cancer radiotherapy. Med. Image Anal. 68, 101909 (2021).
Seo, H., Huang, C., Bassenne, M., Xiao, R. & Xing, L. Modified u-net (mu-net) with incorporation of object-dependent high level features for improved liver and liver-tumor segmentation in ct images. IEEE Trans. Med. Imaging 39, 1316–1325 (2019).
Petit, O. et al. U-net transformer: Self and cross attention for medical image segmentation. In Machine Learning in Medical Imaging: 12th International Workshop, MLMI 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, September 27, 2021, Proceedings 12, 267–276 (Springer, 2021).
Ma, J. et al. Loss odyssey in medical image segmentation. Med. Image Anal. 71, 102035 (2021).
Cheng, J. et al. Resganet: Residual group attention network for medical image classification and segmentation. Med. Image Anal. 76, 102313 (2022).
Jin, Q., Meng, Z., Sun, C., Cui, H. & Su, R. Ra-unet: A hybrid deep attention-aware network to extract liver and tumor in ct scans. Front. Bioeng. Biotechnol. 8, 605132 (2020).
Wang, Z., Zou, N., Shen, D. & Ji, S. Non-local u-nets for biomedical image segmentation. Proc. AAAI Conf. Artif. Intell. 34, 6315–6322 (2020).
Shi, F. et al. Deep learning empowered volume delineation of whole-body organs-at-risk for accelerated radiotherapy. Nat. Commun. 13, 6566 (2022).
Ye, X. et al. Comprehensive and clinically accurate head and neck cancer organs-at-risk delineation on a multi-institutional study. Nat. Commun. 13, 6137 (2022).
Yin, X. & Xu, X. A method for improving accuracy of deeplabv3+ semantic segmentation model based on wavelet transform. In International Conference in Communications, Signal Processing, and Systems, 315–320 (Springer, 2021).
Li, X. et al. Sscnet: A spectrum-space collaborative network for semantic segmentation of remote sensing images. Remote Sens. 15, 5610 (2023).
Upadhyay, K., Agrawal, M. & Vashist, P. Wavelet based fine-to-coarse retinal blood vessel extraction using u-net model. In 2020 International Conference on Signal Processing and Communications (SPCOM), 1–5 (IEEE, 2020).
Zhao, C. et al. Multi-scale wavelet network algorithm for pediatric echocardiographic segmentation via hierarchical feature guided fusion. Appl. Soft Comput. 107, 107386 (2021).
Li, X. et al. A spectral-spatial context-boosted network for semantic segmentation of remote sensing images. Remote Sens. 16, 1214 (2024).
Chandrasekar, V. et al. Investigating the use of machine learning models to understand the drugs permeability across placenta. IEEE Access 11, 52726–52739 (2023).
Ansari, M. Y., Chandrasekar, V., Singh, A. V. & Dakua, S. P. Re-routing drugs to blood brain barrier: A comprehensive analysis of machine learning approaches with fingerprint amalgamation and data balancing. IEEE Access 11, 9890–9906 (2022).
Azimi, S. M., Fischer, P., Körner, M. & Reinartz, P. Aerial lanenet: Lane-marking semantic segmentation in aerial imagery using wavelet-enhanced cost-sensitive symmetric fully convolutional neural networks. IEEE Trans. Geosci. Remote Sens. 57, 2920–2938 (2018).
Duan, Y., Liu, F., Jiao, L., Zhao, P. & Zhang, L. Sar image segmentation based on convolutional-wavelet neural network and markov random field. Pattern Recogn. 64, 255–267 (2017).
Li, Q. & Shen, L. Wavesnet: Wavelet integrated deep networks for image segmentation. In Chinese Conference on Pattern Recognition and Computer Vision (PRCV), 325–337 (Springer, 2022).
Badrinarayanan, V., Kendall, A. & Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 2481–2495 (2017).
Zhou, Y., Huang, J., Wang, C., Song, L. & Yang, G. Xnet: Wavelet-based low and high frequency fusion networks for fully-and semi-supervised semantic segmentation of biomedical images. In Proc. of the IEEE/CVF International Conference on Computer Vision, 21085–21096 (2023).
Cao, H. et al. Swin-unet: Unet-like pure transformer for medical image segmentation. In European Conference on Computer Vision, 205–218 (Springer, 2022).
Valanarasu, J. M. J., Oza, P., Hacihaliloglu, I. & Patel, V. M. Medical transformer: Gated axial-attention for medical image segmentation. In Medical image computing and computer assisted intervention–MICCAI 2021: 24th international conference, Strasbourg, France, September 27–October 1, 2021, proceedings, part I 24, 36–46 (Springer, 2021).
Esfahani, S. S. et al. Lattice-Boltzmann interactive blood flow simulation pipeline. Int. J. Comput. Assist. Radiol. Surg. 15, 629–639 (2020).
Zhai, X. et al. Heterogeneous system-on-chip-based Lattice-Boltzmann visual simulation system. IEEE Syst. J. 14, 1592–1601 (2019).
Zhai, X. et al. Zynq soc based acceleration of the lattice Boltzmann method. Concurr. Comput. Pract. Exp. 31, e5184 (2019).
Funding
This research was funded in part by the Science and Technology Research and Development Project of Henan under Grant No. 242102211053, and National Natural Science Foundation of China under Grant No. 82074580 and 82004498.
Author information
Authors and Affiliations
Contributions
Rui Tang: conceptualization, formal analysis, writing-original draft, writing-review and editing. Hejing Zhao: writing-review and editing, data analysis. Yao Tong: methodology, validation, writing-original draft. Yuqiang Wang: writing-review and editing. Shuhao Zhang: validation, writing-original draft. Yao Zhao: writing-original draft. Ruihui Mu: reviewing, supervision. Min Zhang: data analysis. Yilin Liu: methodology, validation, writing-original draft. Jianbo Gao: conceptualization, formal analysis, writing-original draft, writing-review and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tang, R., Zhao, H., Tong, Y. et al. A frequency attention-embedded network for polyp segmentation. Sci Rep 15, 4961 (2025). https://doi.org/10.1038/s41598-025-88475-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-88475-6