Abstract
Type 2 diabetes mellitus (T2DM) has been confirmed as an independent risk factor for colorectal cancer (CRC) in many studies. However, the mechanisms behind T2DM’s role in the progression of CRC remain unclear. This study aims to explore the potential biomarkers and molecular mechanisms involved in T2DM-promoted CRC progression. The limma package was used to identify differentially expressed genes in tumor tissue from CRC patients with or without T2DM. The key biological processes were screened by gene ontology and gene set enrichment analysis. A diagnostic model for co-morbidities was constructed by logistic regression model with least absolute shrinkage and selection operator (Lasso) regularization method. The diagnostic performance was assessed by supplementing external datasets to draw ROC curves on the diagnostic model. The diagnostic model was further screened for key genes by prognostic analysis. The relationship of key genes with immune cells and other cells was evaluated by immune infiltration algorithm and single-cell transcription analysis. Drug prediction was performed by cMAP and the obtained drugs were molecularly docked with the key genes. The differentially expressed genes of T2DM-promoted CRC progression were mainly enriched to O-linked glycosylation-related processes. The diagnostic model constructed based on Lasso logistic regression had good diagnostic performance (AUC > 0.8). COX11 was the key gene for co-morbidities: in tumor tissues, COX11 expression was significantly higher than that in normal colon tissues. However, COX11 gene expression was significantly lower in patients with comorbidities than in patients without T2DM in tumor tissue. External datasets confirmed from both mRNA and protein expression levels that low COX11 expression was significantly associated with poor CRC prognosis. Immune infiltration analysis suggested that its expression related to the proportion of M2 macrophages. Single-cell transcriptome analysis revealed a close association of COX11 expression with endothelial cells and macrophages. The top4 drugs predicted bound well to COX11. Our study revealed that the pathogenesis of T2DM-promoted CRC progression related to O-linked glycosylation. We constructed a diagnostic model for T2DM-CRC co-morbidity. Meanwhile, we identified COX11 as a potential immune-related molecular marker closely associated with T2DM-promoted CRC progression. These mechanisms and molecular markers may provide new ideas for further studies of T2DM-promoted CRC progression and contribute to drug discovery for the treatment of co-morbidities.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC), including colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ), is one of the most common malignancies and a leading cause of cancer deaths worldwide. CRC is susceptible to comorbidities, which can complicate the disease and affect treatment and survival1, as well as create a greater economic and social burden. In a retrospective cohort study of 29,733 CRC patients, nearly 4% of deaths were attributed to diabetes2. This simultaneous presence of two or more chronic diseases in the same individual is known as co-morbidity3. Diseases are often not randomly clustered, and co-morbidities may have common underlying risk factors behind them; or complications or treatments for some diseases lead to the development of another. Exploring disease associations can enhance understanding of co-morbidities, which can further improve disease diagnosis, prognosis, and therapeutic approaches4. Therefore, it is essential to study the molecular mechanisms of co-morbidities.
A number of observational studies have reported an association between type 2 diabetes and CRC risk. In 2006, Paul J Limburg et al. conducted a cohort study5 and found that T2DM was associated with an increased risk of CRC. A multicenter cohort in Korea6 also observed that high fasting glucose and a history of diabetes were linked to increased risk of CRC. In addition to increasing the risk of CRC, diabetes mellitus can affect the prognosis of CRC patients7. In CRC patients with T2DM, unhealthy glucose management is linked to a clinically severe cancer course8. Although most of this evidence comes from epidemiologic studies, which may have limitations due to diagnostic bias and confounding factors, it does suggest an association between T2DM and CRC to some extent. Meanwhile, metformin, a glucose-lowering drug clinically used to treat T2DM, has demonstrated efficacy in lowering the incidence of CRC9,10,11. In 2021, the American Gastroenterological Association (AGA), in a clinical practice update on chemoprevention of colorectal neoplasms12, suggested that: for patients with T2DM, clinicians may think about preventing CRC by using metformin. For patients with co-morbidities of CRC and T2DM, clinicians might think about utilizing metformin to lower mortality. Further basic research has also found that metformin may increase survival in patients with T2DM-CRC co-morbidities by suppressing the epithelial mesenchymalization phenotype13, the immune microenvironment14, and the gut microbiota15. This evidence further supports the association of T2DM-promoting CRC progression.
Some scholars have explored the mechanism of T2DM promotes CRC progression from genetic variation of T2DM susceptibility genes16,17, IGF-1 levels18,19,20,21, and gut microbes22,23. Currently, there are fewer studies exploring molecular mechanisms from the level of the co-morbid transcriptome. Although some bioinformatics studies on T2DM-CRC comorbidities24,25,26,27 have been reported, most of the samples they used were samples with T2DM or CRC alone, rather than samples with co-morbidities. In fact, samples from patients with CRC who have T2DM are more plausible in presenting a co-morbid gene expression profile. We therefore designed, and conducted, the present study to explore the molecular mechanisms by which T2DM promotes CRC progression.
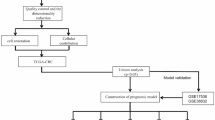
In this study, we focused on key genes linked to T2DM-promoted CRC progression. We analyzed a high-throughput data sequencing dataset (GSE115313) downloaded from the GEO database. An integrated bioinformatics and systems biology approach was used to identify the common differentially expressed genes and their functions in tumor tissues in patients with T2DM-combined CRC and CRC patients without T2DM, and to explore the unique phenotype of T2DM-promoted CRC progression. In addition, sequencing data from patients’ tumor tissues and normal tissues were combined to screen gene clusters and construct diagnostic models by Lasso logistic regression. The performance of the diagnostic models was confirmed using an external dataset (TCGA-COREAD). Key genes in the model were screened by prognostic analysis and evaluated for correlation with pathological staging and immunization. Cells with key gene COX11 expression were localized by analyzing colon cancer single-cell transcriptome data (GSE231559). Finally, drug prediction and molecular docking were performed. The flow of this study is shown in Fig. 1.
Materials and methods
Download of datasets
The main datasets used in this study were as follows: 84 samples were collected in GSE115313, including tumor and normal colon tissues from T2DM-CRC and NoT2DM-CRC patients. 10× single-cell sequencing data from tumor and normal colon tissues from CRC patients were collected in GSE231559. The above datasets were downloaded from the GEO database (www.ncbi.nlm.nih.gov/geo). The dataset for TCGA-COREAD was acquired from UCSC Xena. Other datasets were downloaded from CPTAC (https://gdc.cancer.gov/about-gdc/contributed-genomic-data-cancer-research/clinical-proteomic-tumor-analysis-consortium-cptac), HPA (https://www.proteinatlas.org/) and TISIDB (http://cis.hku.hk/TISIDB/).
Obtaining T2DM-CRC DEGs
The limma package was utilized to perform differential analysis, and identify the differentially expressed genes (DEGs) in tumor tissues of patients with T2DM-CRC and NoT2DM-CRC. The genes with p < 0.05 were taken as differentially expressed genes, in which log2FoldChange > 0 was up-regulated differentially expressed genes and log2FoldChange < 0 was down-regulated differentially expressed genes. The obtained groups of differentially expressed genes were plotted as venn plots to take the intersection. The downstream differentially expressed gene clusters of T2DM promoting CRC development were identified. Volcano plots were drawn to show the results of differential analysis.
Enrichment analysis
DEGs were obtained and performed Gene ontology enrichment analysis using DAVID28 and metaScape. These results were visualized by drawing bubble plots. Using GSEA 4.3.3 software, Gene Set Enrichment Analysis (GSEA) was carried out on the gene expression matrix. These p < 0.05 enrichment results were considered statistically significant.
Least absolute shrinkage and selection operator regression to construct a diagnostic model
Least absolute shrinkage and selection operator (Lasso) regression is a machine learning algorithm that has been widely used to screen signature genes and construct models for various diseases, such as gastrointestinal diseases29. We construct a logistic regression model (lasso-logistic regression) with Lasso regularization. The analysis was performed using the glmnet package, setting the number of random seeds, and after inputting the gene expression matrix, setting up ten-fold cross-validation to train the model.
Plotting ROC curves
Receiver Operating Characteristic (ROC) curves are most typically used in medicine as a technique of assessing diagnostic tests30. The TCGA-COREAD and internal datasets were analyzed using the pROC package, and ROC curves were generated to evaluate the model’s diagnostic performance.
Prognostic analysis and validation
Using GEPIA 231 for survival analysis of each gene in the diagnostic model. In the meantime, an analysis was conducted on the gene expression levels of various disease stages. The correlation between the genes in the diagnostic model and CRC prognosis was assessed. After screening to obtain genes significantly correlated with CRC prognosis, the expression levels (mRNA and protein relative expression levels) and prognostic significance of the key genes were again verified using the GSE223119, GSE25724, GSE20966, GSE184050 datasets, HPA database, and CPTAC database.
Immune correlation analysis
Cibersoft32 is an algorithm for analyzing the proportion of immune cells that has been widely used to assess the level of immune infiltration in oncologic and non-oncologic diseases. Twenty-two immune cell ratios from sequencing data were assessed using Cibersoft. Correlations between key gene expression levels and immune cell ratios were calculated and correlation heatmaps were drawn. Evaluate the correlation between key genes and immunity based on the TISIDB database.
Single-cell transcriptome analysis
10 × single-cell transcriptome data were extracted using the Seurat package, and the data were normalized after removing mitochondrial genes with expression greater than 20% and genes with expression less than 200. After PCA downscaling, the appropriate number of Clusters were filtered by JackStraw and ElbowPlot methods to draw umap sub-cluster plots. Individual Clusters were annotated using singleR package with celldex package. The annotated cell clusters were differentiated into normal and tumor tissues, and the cells expressing key genes were located at the same time.
Drug prediction & molecular docking
DEGs from tumor samples of patients with T2DM-CRC co-morbidities versus CRC-only patients were uploaded to cMAP33, and the reference cell line was HT29 (colon cancer cell line) for drug prediction. Structure files of predicted drugs were obtained from Pubchem. Structure files of key genes were downloaded from AlphaFoldDB database (https://alphafold.ebi.ac.uk/). Molecular docking was performed using CB-DOCK234 to assess the level of drug binding to key genes.
Results
T2DM-CRC differentially expressed genes analysis
A total of 995 DEGs were obtained by differential analysis (Fig. 2A). The expression levels of these differentially expressed genes between the two groups were displayed using the clustering heat map (Fig. 2B).
T2DM promotes CRC progression and is strongly associated with O-linked glycosylation. (A) Volcano plot of differential gene analysis of tumor tissues from CRC patients without T2DM versus CRC patients with T2DM. (B) Heatmap of expression clustering of differentially expressed genes. (C,D) O-linked glycosylation-associated processes were significantly enriched by GO-BP versus KEGG35,36,37 enrichment analysis (DAVID). (E) Go BP enrichment analysis (METESCAPE). (F) O-glycan-related processes were significantly upregulated in T2DM-CRC co-morbidities (GSEA).
T2DM promotes CRC progression associated with O-linked glycosylation
The 995 DEGs were subjected to enrichment analysis by the DAVID database. These DEGs are mainly associated with O-linked glycosylation biological processes, such as protein O-linked glycosylation via serine or via threonine (Fig. 2C). Meanwhile, from the results of KEGG enrichment, we observed that these DEGs are mainly involved in O-glycan biosynthesis, endocytosis and other pathways (Fig. 2D). Considering the limitations of a single database, enrichment analysis was supplemented using the metascape database. Enrichment of these differential genes to O-linked glycosylation biological processes was similarly observed (Fig. 2E). The gene expression data were uploaded to GSEA, which revealed a significant upregulation of O-linked glycosylation-related biological processes (Fig. 2F).
Diagnostic model construction and validation
Tumor samples and healthy samples of CRC patients without T2DM were differentially analyzed, and tumor samples and healthy samples of CRC patients with combined T2DM were differentially analyzed. The differentially expressed genes of combined T2DM combined CRC were taken as intersection, and a total of 276 genes were obtained (Fig. 3A–C). These genes were closely associated with T2DM promoting CRC development.
Constructing diagnostic models based on Lasso logistic regression. (A–C) Tumor tissues and normal tissues of CRC patients were analyzed for differentially expressed genes, and the intersection set was taken to screen the set of differentially expressed genes for T2DM-CRC co-morbidities. (D) Lasso logistic regression was used to screen the key variables and construct the diagnostic models. (E) ROC curves were plotted to assess the diagnostic performance using GSE115313 as an internal dataset. (F) ROC curves were plotted to assess the diagnostic performance using TCGA-COREAD as an external validation dataset.
Diagnostic models were constructed for these genes using Lasso logistic regression. After setting the number of random seeds and ten-fold cross-validation, diagnostic models constructed for 24 genes were obtained (Fig. 3D). The diagnostic performance was validated using internal dataset, and its model had an excellent performance. The external dataset validation supplementing TCGA also had a good performance (AUC > 0.8) (Fig. 3E,F).
Screening COX11 as a key gene in T2DM for CRC progression
Analysis of the survival prognosis and pathological correlations of 24 genes in the diagnostic model by GEPIA 2 revealed that only COX11 was associated with prognosis and pathological staging, and the expression trend was consistent with that in T2DM combined with CRC (Supplementary Figs. 1–2). In contrast to normal tissues, the mRNA expression level of COX11 was significantly increased with or without combined T2DM (Fig. 4A). In contrast, in tumor tissues, a significant decrease in COX11 expression was observed in samples with combined T2DM. In the proteomic dataset, COX11 protein expression levels were found to be significantly higher in tumor samples from CRC patients. This effect was also observed on immunohistochemical staining (Fig. 4B,C). In CRC tumor tissues, there was an increase in the mRNA expression level of COX11 in the external dataset, whereas in T2DM patients, there was a notable drop in the mRNA expression level of COX11 (Fig. 4D).
Low expression of COX11 in tumor tissues of patients with T2DM-CRC co-morbidities was significantly associated with poor prognosis. (A) Expression of COX11 in tumor tissues and normal tissues of patients with CRC. (B,C). Proteomic data and HPA databases confirmed that the relative expression level of COX11 protein is significantly up-regulated in CRC tumor tissues. (D) External datasets confirmed at the transcriptional level that COX11 is highly expressed in tumor tissues and low expressed in T2DM pancreatic islets. (E) COX11 was significantly correlated with pathological stage and N stage, and not significantly correlated with MSI stability and T and M stage. tissues and low expression in T2DM islets. (F–H). Low expression of COX11 (at transcriptional level) in TCGA-COREAD(F)/TCGA-COAD(G)/TCGA-READ(H) is suggestive of poor prognosis. (I) COX11 survival curves based on CPTAC data (protein level). (J) COX11 survival curves obtained based on HPA (transcript level) database analysis.
COX11 is strongly associated with CRC prognosis
Extraction and analysis of the data in TCGA suggested that COX11 mRNA expression levels were not significantly correlated with microsatellite instability (MSI) and T and M staging (Fig. 4E). COX11 mRNA expression levels were significantly correlated with pathological staging and N staging. The highest COX11 expression level was observed in COAD in HM-indel, whereas this correlation was not observed in READ. In the combined COAD-READ dataset (Fig. 4F), COAD (Fig. 4G) dataset, READ (Fig. 4H) dataset, increased COX11 mRNA expression levels were observed as a feature of good prognosis. Protein expression levels and prognosis, extracted and analyzed from the CPTAC dataset (CPTAC-2 Prospective, Cell 2019)38 of protein expression data for prognostic analysis (Fig. 4I). We similarly observed that low COX11 expression levels were associated with poor prognosis, not statistically significant (p = 0.088). The HPA database similarly suggested that COX11 is a good prognostic biomarker for CRC (Fig. 4J).
Analysis of COX11 and immune correlations
In tumor tissues from patients with combined T2DM-CRC, there was a notable rise in the percentage of M2 macrophages in the COX11 high-expression group. But correlation thermograms did not observe a correlation between COX11 expression levels and M2 macrophage proportions (Fig. 5A,B). In tumor tissues from TCGA-COREAD patients, the COX11 high-expression group showed a significant drop in activated dendritic cells and a large rise in the number of M0 macrophages (Fig. 5C). Significantly lower expression of COX11 than the other immune subtypes was observed in COAD in C3 (inflammation), and C6 (TGF-β dominant), which was not observed in READ (Fig. 5D,E). In COX11 expression correlated with CD4 T cells in a certain positive proportion (Fig. 5F).
COX11 is one of immune correlation gene in T2DM-CRC. (A) Proportion of immune cells in tumor tissues of patients with co-morbidities in the COX11 low-expression versus high-expression group. (B) Correlation between COX11 expression level and immune cells in tumor tissues of patients with co-morbidities. (C) Proportion of immune cells in tumor samples of patients with TCGA-COREAD in the COX11 low-expression versus high-expression group. (D,E) In COAD, READ, the Correlation of COX11 expression level with immune subtypes. (F) A heat map of immune cells and tumor diseases constructed based on MISIDB revealed that the percentage of CD4 T cells and the amount of COX11 expression in COAD, READ, were favorably associated.
Single-cell transcriptome analysis
After annotation, tumor tissues were mainly divided into 8 cell clusters with normal colon tissues (Fig. 6A,B). Among them, Fig. 6C demonstrated part of the marker that distinguished the cell clusters. In tumor tissues, a trend of increasing COX11 expression levels was observed in cells such as endothelial cells and macrophages (Fig. 6D,E). There was an increase in the proportion of CD8 + T lymphocytes to epithelial cells in tumor tissues (Fig. 6F).
Single-cell transcriptome validation of COX11 expression in tumor versus normal colon tissues of CRC patients. (A,B) Cellular clustering of tumor tissues versus normal colon tissues. (C) Heatmap of marker genes in cellular clusters. (D,E) Expression of COX11 in tumor tissues versus normal colon tissues of different cells. (F) Percentage of different cells in tumor tissues versus normal colon tissues.
COX11 binds well to predictive drugs
Tumor tissue sequencing matrix differentially expressed genes from co-morbid versus CRC-only patients were uploaded to the cMAP database, with the reference cell line HT29 (colon cancer cell line). HDAC inhibitors were found to have the highest percentage (12%) of drugs with a predicted score of < -90 (Fig. 7A). The top 4 drugs with negative scores obtained were JNJ-16259685, topotecan, mocetinostat, and aristolochic-acid. The molecular docking results (Fig. 7B) suggested that they could dock well in Table 1 (Vina score < -4).
Molecular docking plots of drugs with COX11 based on cMAP prediction. (A) Categorical percentage of cMAP-predicted drugs (prediction scores < -90). (B) Docking binding sites are shown on the left, the names of drugs binding to COX11 are shown in the middle region, and the heatmap of the binding scores of each drug binding to COX11 is shown on the right (this diagram was drawn by author: Luo Simin).
Discussion
CRC is currently the second leading cause of cancer deaths worldwide. With the accumulation of clinical data, a large number of studies have reported the association between T2DM and CRC risk. T2DM has received increasing attention as an independent risk factor for CRC. One cohort study39 found that patients with T2DM had a significantly higher risk of cancer death than non-diabetic patients. Another cohort study40 that included 59,202 patients with CRC found that patients with colorectal cancer combined with diabetes also had a higher risk of recurrence than patients without diabetes. Metformin has been reported to significantly improve overall survival41 and disease-free survival42 in patients with T2DM-CRC co-morbidities. Currently, some studies have pointed out that pathological processes inherent to T2DM such as chronic hyperglycemia, insulin resistance, oxidative stress, and inflammation also play an active role in colorectal carcinogenesis and progression43. These may be the molecular mechanisms by which T2DM promotes CRC progression. Some studies24,25,26,27 have also explored the related molecular mechanisms from a transcriptomic perspective. Some of these studies integrated large samples with single-cell transcriptomes and eliminated the batch effect, which has some credibility. However, the sequencing data used were not from co-morbid patient tissues, but samples from single T2DM or CRC patients. It is therefore difficult to accurately characterize the gene expression profile of tumor tissues in the T2DM-CRC co-morbid state. In order to explore the molecular mechanisms of T2DM-promoted CRC progression more comprehensively, we designed the present study.
First, we performed differential expression analysis and enrichment analysis on the normalized sequencing data. The DEGs obtained were mainly enriched for related processes such as O-linked glycosylation (a type of glycosylation). Glycosylation is a way in which proteins and lipids are modified with complex carbohydrates called glycans44. Glycans regulate tumor proliferation, invasion, metastasis, and angiogenesis, among other aspects of tumor growth. Because of this, aberrant glycosylation is a prime contender for cancer diagnostics45. MC38 colon cancer cells cultured under high glucose showed more abundant N-glycans46. These evidences suggest a close association of glycosylation with CRC. Other studies provide further possible directions for the link between O-linked glycosylation and T2DM-promoted CRC progression. For example, O-GlcNAcylation, a post-translational covalent modification of β-N-acetylglucosamine (O-GlcNAc) molecules on protein serine or threonine residues via O-linkage, has been closely associated with both diabetes and cancer47. Upregulation of fatty acid synthase expression increases O-GlcNAc protein glycosylation and promotes colorectal cancer growth48. O-GlcNAcylation negatively regulated by microRNA-101 may promote colorectal cancer metastasis by enhancing the stability and function of EZH2 protein49. Moreover, O-GlcNAcylation destabilizes the active tetramer PKM2, which promotes the Warburg effect and bridges metabolic reprogramming in tumor diseases50. The chemotherapeutic medication 5-fluorouracil (5-FU) impacts O-GlcNAcylation in non-cancerous and malignant colon cells in vitro by reducing O-GlcNAc transferase production. Moreover, Thiamet-G, an O-GlcNAcase inhibitor, and 5-FU together have a synergistic inhibitory effect on tumor development51. The above evidence suggests that upstream regulators can promote CRC progression/metastasis by enhancing O-linked glycosylation. Interestingly, we found that T2DM-CRC patients exhibited stronger levels of O-linked glycosylation. It is then likely that T2DM promotes CRC progression by enhancing O-linked glycosylation. In fact, O-linked glycosylation is indeed strongly associated with T2DM52,53. Because the subunits necessary for protein glycosylation are supplied by glucose. Therefore, it is likely that hyperglycemia induce O-linked glycosylation of certain proteins (e.g., O-GlcNAcylation) in T2DM patients, which in turn promotes the development of CRC and triggers a poor prognosis.
Subsequently, we constructed a diagnostic model by Lasso logistic regression, and the diagnostic performance was validated by an external dataset. In order to further explore potential targets closely related to prognosis, we performed prognostic analyses of individual genes in the model, and finally identified COX11 as a key molecule. We discovered that COX11 expression levels, both in mRNA and protein, were slightly elevated in tumor tissues. This elevation may be protective in nature. Therefore, when COX11 expression levels were elevated in tumor tissues, we observed a better prognostic outcome in CRC patients. When the prognostic significance was verified using proteomic data, a trend towards a better prognosis with high expression was observed, but no statistical significance was observed (p = 0.088). The reason for this may be that the duration of observation was only 40 months, which is much lower than the duration of observation in databases such as TCGA (150 months). Other studies54 have also found that high levels of COX11 in CRC tumor tissues have a better prognosis. In our validation using an external dataset, we found a significant decrease in COX11 levels in T2DM patients. Related literature also reported that levels of oxidative phosphorylation genes (e.g., COX11) were significantly decreased in T2DM patients with elevated blood glucose55. This implies that low levels of COX11 brought about by T2DM in patients with concomitant CRC may be a key factor triggering the poor prognosis of CRC. COX11 is a copper-transporting protein essential for respiratory growth and is associated with the assembly of the Cu(B) site of cytochrome c oxidase56. Cellular copper levels are closely related to mitochondrial oxidative phosphorylation, and COX11 has been reported to interact with several proteins of the same family involved in copper ion transport and oxidative phosphorylation. For example, COX17, which transports copper from the cytoplasm to MT-CO1/COX1 via COX1157,58. Deletion of COX 19 of the same family may block the COX11 response cycle to a phase that is particularly sensitive to redox, leading to over-oxidation and inactivation of COX1159. These evidences provide a possibility to explain the molecular mechanism by which COX11 is involved in the progression of co-morbidities. We also explored the relationship of COX11 with pathological staging and other subtypes. Although COX11 was associated with pathological staging, its correlation with N staging was only observed. This suggests that COX11 is significantly associated with high-grade pathological staging and the main reason behind this may be lymph node involvement.
After clarifying that COX11 is potentially a key molecule in T2DM-promoted CRC progression, we further explored its association with immunity. In tumor tissues of T2DM-CRC patients, the proportion of M2 macrophages was significantly higher in the COX11 high-expression group. But this effect was not observed in the TCGA-COREAD cohort. The reason for the non-observation may be the lack of T2DM information in the TCGA-COREAD cohort, which does not allow stratification of the T2DM-CRC cohort. We performed a single-cell transcriptome analysis, although the samples were not from co-morbidities but from a single CRC patient. However, it was able to demonstrate to some extent the localization of COX11 and the differences in its expression levels among different cells. We noticed that an increase in the ratio of endothelial cells to CD8 + T cells in tumor tissues, which may suggest that immunological infiltration of CD8 + T cells took place in tumor tissues. Since COX11 is localized to mitochondria, we found that COX11 was somewhat expressed on various cell clusters. In line with the expected trend, in the tumor tissues, we observed an increase in COX11 expression levels in cells such as endothelial cells and macrophages (although no statistical difference was observed). Combined with the results of immune correlation analysis, we suggest that macrophages may also be key cells for the role of COX11 in T2DM-promoted CRC progression.
Finally, we performed drug prediction and found that the drugs with high prediction scores were mainly HDAC inhibitors, which mainly act by inhibiting histone deacetylation. While no study has yet demonstrated that HDAC inhibitors can delay the progression of CRC by influencing T2DM, numerous studies have confirmed their efficacy in delaying the progression of either T2DM or CRC. Initially, HDAC inhibitors were incorporated into preclinical and clinical evaluations for gastrointestinal tumors as promising novel therapeutics as early as 200560. Since then, they have been extensively reported to exert antitumor effects, particularly in combination with 5-fluorouracil (5-Fu)61,62. Recent research has further elucidated that HDAC inhibitors can impede colon cancer progression or metastasis by modulating various cellular processes, including mitosis63, adhesion64, apoptosis, epithelial-mesenchymal transition65, and drug resistance66 in mesenchymal tumor cells. In a separate line of inquiry, a 2014 study identified an association between single nucleotide variants in the HDAC gene and the incidence of T2DM within the Chinese population67. Subsequent findings revealed that HDAC inhibitors could enhance β-cell differentiation, proliferation, and function, as well as ameliorate insulin resistance68,69. Additionally, HDAC inhibitors have been shown to delay the progression of T2DM by mitigating endoplasmic reticulum stress70. Furthermore, HDAC inhibitors have been shown to impede the progression of diabetic complications. For instance, in the context of diabetic cardiomyopathy, HDAC inhibitors have been observed to modulate cardiac PPARs, fatty acid metabolism, and pro-inflammatory cytokines71. These findings suggest the potential involvement of the HDAC in the promotion of CRC progression by T2DM. Notably, insulin resistance is a fundamental mechanism underlying metabolic alterations and is believed to be associated with CRC prognosis72. Given that HDAC inhibitors have the capacity to ameliorate insulin resistance, we propose that these inhibitors may mitigate the progression of T2DM-CRC co-morbidity by enhancing insulin sensitivity. Future research should focus on evaluating the efficacy and safety of HDAC inhibitors in patients with T2DM-CRC co-morbidities, as well as elucidating the specific mechanisms by which the HDAC contributes to CRC progression in the context of T2DM. Molecular docking suggests that all four drugs predicted by our screening can bind well to the key gene COX11, which to some extent demonstrates the potential of COX11 as a therapeutic target.
In this study, we identified the differentially expressed genes, key genes, and predicted the related molecular mechanisms of T2DM-promoted CRC progression, which helps to further explore the pathogenesis of T2DM-promoted CRC progression. However, our study has some limitations. First, although we used the co-morbidity dataset for our analyses the sample size of this study was only 84 cases, which is on the low side and may affect the credibility of the results. More large samples are needed for validation in the future. Second, although we used an external RNA-seq dataset with a proteomic dataset for validation, we lacked sufficient clinical co-morbidity samples to confirm our findings. Finally, in the future, we will further refine our ex vivo and in vivo experiments to deeply investigate the role COX11 plays in T2DM-promoted CRC progression.
Conclusions
In summary, our study revealed that the pathogenesis of T2DM-promoted CRC progression may be related to O-linked glycosylation, and constructed a diagnostic model of T2DM-CRC co-morbidity with certain diagnostic performance. Meanwhile, we identified COX11 as a potential immune-related molecular marker closely related to T2DM-promoted CRC progression, and found that it might be closely related to macrophages in tumor tissues by immune infiltration and single-cell transcriptome analysis. Overall, these mechanisms and molecular markers may provide new ideas for further mechanistic studies and contribute to drug discovery for its treatment.
Data availability
All datasets analyzed in this study are housed in public databases. The RNA-seq data were deposited into the Gene Expression Omnibus (GEO) database under accession number GSE115313, GSE231559, GSE25724, GSE20966, GSE184050 and are available at the following URL: https://www.ncbi.nlm.nih.gov/geo. The TCGA RNA-seq data were deposited into the UCSC Xena database under accession number TCGA.COADREAD.sampleMap/HiSeqV2 and are available at the following URL: https://xena.ucsc.edu. The protein datasets analysed during the current study were deposited into the cBioPortal for Cancer Genomics under accession number Colon Cancer (CPTAC-2 Prospective, Cell 2019) and are available at the following URL: https://www.cbioportal.org/study/summary?id=coad_cptac_2019.
References
Qiu, H. et al. Comorbidity patterns in patients newly diagnosed with colorectal cancer: Network-based study. JMIR Public Health Surveill. 9, e41999 (2023).
Gross, C. et al. Multimorbidity and survival in older persons with colorectal cancer. J. Am. Geriatr. Soc. 54(12), 1898–1904 (2006).
Barnett, K. et al. Epidemiology of multimorbidity and implications for health care, research, and medical education: a cross-sectional study. Lancet 380(9836), 37–43 (2012).
Iida, M., Iwata, M. & Yamanishi, Y. Network-based characterization of disease-disease relationships in terms of drugs and therapeutic targets. Bioinformatics 36(Suppl_1), i516–i524 (2020).
Limburg, P. J. et al. Clinically confirmed type 2 diabetes mellitus and colorectal cancer risk: a population-based, retrospective cohort study. Am. J. Gastroenterol. 101(8), 1872–1879 (2006).
Park, H. et al. Fasting glucose and risk of colorectal cancer in the Korean Multi-center Cancer Cohort. PLoS ONE 12(11), e0188465 (2017).
Bella, F. et al. Impact of diabetes on overall and cancer-specific mortality in colorectal cancer patients. J. Cancer Res. Clin. Oncol. 139(8), 1303–1310 (2013).
Siddiqui, A. A. et al. Elevated HbA1c is an independent predictor of aggressive clinical behavior in patients with colorectal cancer: A case-control study. Dig. Dis. Sci. 53(9), 2486–2494 (2008).
Lee, M. S. et al. Type 2 diabetes increases and metformin reduces total, colorectal, liver and pancreatic cancer incidences in Taiwanese: a representative population prospective cohort study of 800,000 individuals. BMC Cancer 11, 20 (2011).
Sehdev, A. et al. Metformin for primary colorectal cancer prevention in patients with diabetes: A case-control study in a US population. Cancer 121(7), 1071–1078 (2015).
He, X. K. et al. Metformin is associated with slightly reduced risk of colorectal cancer and moderate survival benefits in diabetes mellitus: A meta-analysis. Medicine (Baltimore) 95(7), e2749 (2016).
Liang, P. S., Shaukat, A. & Crockett, S. D. AGA clinical practice update on chemoprevention for colorectal neoplasia: Expert review. Clin. Gastroenterol. Hepatol. 19(7), 1327–1336 (2021).
Wang, Y., Wu, Z. & Hu, L. Epithelial-mesenchymal transition phenotype, metformin, and survival for colorectal cancer patients with diabetes mellitus II. Gastroenterol. Res. Pract. 2017, 2520581 (2017).
Saito, A. et al. Metformin changes the immune microenvironment of colorectal cancer in patients with type 2 diabetes mellitus. Cancer Sci. 111(11), 4012–4020 (2020).
Huang, Q. Y. et al. Role of gut microbiome in regulating the effectiveness of metformin in reducing colorectal cancer in type 2 diabetes. World J. Clin. Cases 8(24), 6213–6228 (2020).
Sainz, J. et al. Effect of type 2 diabetes predisposing genetic variants on colorectal cancer risk. J. Clin. Endocrinol. Metab. 97(5), E845–E851 (2012).
Ishimaru, S. et al. Increased risk for CRC in diabetic patients with the nonrisk allele of SNPs at 8q24. Ann. Surg. Oncol. 19(9), 2853–2858 (2012).
Cao, H. et al. Changes in serum IGF-1 level and tumor VEGF expression in mice with colorectal cancer under hyperglycemic conditions. Mol. Med. Rep. 7(4), 1361–1365 (2013).
Chen, Z. & Hong, Q. Correlation of serum IGF-1, AGEs and their receptors with the risk of colorectal cancer in patients with type 2 diabetes mellitus. Front. Oncol. 13, 1125745 (2023).
Chen, B. et al. Non-coding RNAs in IGF-1R signaling regulation: The underlying pathophysiological link between diabetes and cancer. Cells 8(12), 1638 (2019).
Ding, J. et al. Higher expression of proteins in IGF/IR axes in colorectal cancer is associated with type 2 diabetes mellitus. Pathol. Oncol. Res. 22(4), 773–779 (2016).
Yang, Y. et al. Metagenomic and targeted metabolomic analyses reveal distinct phenotypes of the gut microbiota in patients with colorectal cancer and type 2 diabetes mellitus. Chin. Med. J. (Engl.) 136(23), 2847–2856 (2023).
Noureldein, M. et al. Intestinal microbiota regulates diabetes and cancer progression by IL-1β and NOX4 dependent signaling cascades. Cell Mol. Life Sci. 79(9), 502 (2022).
Peng, W. F. et al. The key genes underlying pathophysiology association between the type 2-diabetic and colorectal cancer. J. Cell Physiol. 233(11), 8551–8557 (2018).
Elek, Z. et al. Correlation between expression profiles of key signaling genes in colorectal cancer samples from type 2 diabetic and non-diabetic patients. Life (Basel) 10(9), 216 (2020).
Liu, X. et al. Identification of the shared gene signature and biological mechanism between type 2 diabetes and colorectal cancer. Front Genet. 14, 1202849 (2023).
Luo, Y. et al. Exploring the molecular mechanism of comorbidity of Type 2 diabetes mellitus and colorectal cancer: Insights from bulk omics and single-cell sequencing validation. Biomolecules 14(6), 693 (2024).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50(W1), W216–W221 (2022).
Ali, H. et al. Application and impact of Lasso regression in gastroenterology: A systematic review. Indian J. Gastroenterol. 42(6), 780–790 (2023).
Carter, J. V., Pan, J., Rai, S. N. & Galandiuk, S. ROC-ing along: Evaluation and interpretation of receiver operating characteristic curves. Surgery 159(6), 1638–1645 (2016).
Tang, Z. et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47(W1), W556–W560 (2019).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12(5), 453–457 (2015).
Subramanian, A. et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171(6), 1437-1452.e17 (2017).
Liu, Y. et al. CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 50(W1), W159–W164 (2022).
Kanehisa, M. et al. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53(D1), D672–D677 (2025).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28(11), 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30 (2000).
Vasaikar, S. et al. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell 177(4), 1035-1049.e19 (2019).
Dehal, A. N. et al. Impact of diabetes mellitus and insulin use on survival after colorectal cancer diagnosis: the Cancer Prevention Study-II Nutrition Cohort. J. Clin. Oncol. 30(1), 53–59 (2012).
Hsu, H. Y. et al. Diabetic severity and oncological outcomes of colorectal cancer following curative resection: A population-based cohort study in Taiwan. Cancer 129(24), 3928–3937 (2023).
Garrett, C. R. et al. Survival advantage observed with the use of metformin in patients with type II diabetes and colorectal cancer. Br. J. Cancer 106(8), 1374–1378 (2012).
Tarhini, Z. et al. The effect of metformin on the survival of colorectal cancer patients with type 2 diabetes mellitus. Sci. Rep. 12(1), 12374 (2022).
Vekic, J. et al. Diabetes and colorectal cancer risk: A new look at molecular mechanisms and potential role of novel antidiabetic agents. Int. J. Mol. Sci. 22(22), 12409 (2021).
Scheper, A. F., Schofield, J., Bohara, R., Ritter, T. & Pandit, A. Understanding glycosylation: Regulation through the metabolic flux of precursor pathways. Biotechnol. Adv. 67, 108184 (2023).
Vasconcelos-Dos-Santos, A. et al. Biosynthetic machinery involved in aberrant glycosylation: Promising targets for developing of drugs against cancer. Front. Oncol. 5, 138 (2015).
Loponte, H. F. et al. Hyperglycemia alters N-glycans on colon cancer cells through increased production of activated monosaccharides. Glycoconj J. 39(5), 663–675 (2022).
Singh, J. P., Zhang, K., Wu, J. & Yang, X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 356(21), 244–250 (2015).
Drury, J. et al. Overexpression of fatty acid synthase upregulates glutamine-fructose-6-phosphate transaminase 1 and O-Linked N-acetylglucosamine transferase to increase O-GlcNAc protein glycosylation and promote colorectal cancer growth. Int. J. Mol. Sci. 25(9), 4883 (2024).
Jiang, M., Xu, B., Li, X., et al. O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit [published correction appears in Oncogene. 2019 Jul;38(28):5744–5745. https://doi.org/10.1038/s41388-019-0834-2]. Oncogene 38(3), 301–316 (2019).
Wang, Y. et al. O-GlcNAcylation destabilizes the active tetrameric PKM2 to promote the Warburg effect. Proc. Natl. Acad. Sci. USA 114(52), 13732–13737 (2017).
Very, N. et al. Thymidylate synthase O-GlcNAcylation: A molecular mechanism of 5-FU sensitization in colorectal cancer. Oncogene 41(5), 745–756 (2022).
Farook, V. S., Bogardus, C. & Prochazka, M. Analysis of MGEA5 on 10q241–q243 encoding the beta-O-linked N-acetylglucosaminidase as a candidate gene for type 2 diabetes mellitus in Pima Indians. Mol. Genet. Metab. 77(1–2), 189–193 (2002).
Masaki, N. et al. O-GlcNAcylation mediates glucose-induced alterations in endothelial cell phenotype in human diabetes mellitus. J. Am. Heart Assoc. 9(12), e014046 (2020).
Chu, B. et al. Integrative analysis of single-cell and bulk RNA seq to reveal the prognostic model and tumor microenvironment remodeling mechanisms of cuproptosis-related genes in colorectal cancer. Aging (Albany NY) 15(23), 14422–14444 (2023).
Olsson, A. H. et al. Decreased expression of genes involved in oxidative phosphorylation in human pancreatic islets from patients with type 2 diabetes. Eur. J. Endocrinol. 165(4), 589–595 (2011).
Carr, H. S., George, G. N. & Winge, D. R. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J. Biol. Chem. 277(34), 31237–31242 (2002).
Prohaska, J. R. & Gybina, A. A. Intracellular copper transport in mammals. J. Nutr. 134(5), 1003–1006 (2004).
Xue, Q. et al. Copper metabolism in cell death and autophagy. Autophagy. 19(8), 2175–2195 (2023).
Bode, M. et al. Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase. Mol. Biol Cell 26(13), 2385–2401 (2015).
Wiedmann, M. W. & Caca, K. Molecularly targeted therapy for gastrointestinal cancer. Curr. Cancer Drug Targets 5(3), 171–193 (2005).
Di Gennaro, E. et al. Modulation of thymidilate synthase and p53 expression by HDAC inhibitor vorinostat resulted in synergistic antitumor effect in combination with 5FU or raltitrexed. Cancer Biol. Ther. 8(9), 782–791 (2009).
Bálintová, L., Matúšková, M. & Gábelová, A. The evaluation of the efficacy and potential genotoxic hazard of combined SAHA and 5-FU treatment in the chemoresistant colorectal cancer cell lines. Mutat Res. Genet. Toxicol. Environ. Mutagen 874–875, 503445 (2022).
Liang, X. L. et al. Histone deacetylase inhibitor pracinostat suppresses colorectal cancer by inducing CDK5-Drp1 signaling-mediated peripheral mitofission. J. Pharm Anal. 13(10), 1168–1182 (2023).
Li, Q. & Chen, H. Silencing of Wnt5a during colon cancer metastasis involves histone modifications. Epigenetics 7(6), 551–558 (2012).
Kim, S. L. et al. A novel HDAC1 inhibitor, CBUD-1001, exerts anticancer effects by modulating the apoptosis and EMT of colorectal cancer cells. Int. J. Oncol. 57(4), 1027–1038 (2020).
Ye, C. et al. Inhibition of histone deacetylase 7 reverses concentrative nucleoside transporter 2 repression in colorectal cancer by up-regulating histone acetylation state. Br. J. Pharmacol. 175(22), 4209–4217 (2018).
Zeng, Z. et al. Three single nucleotide variants of the HDAC gene are associated with type 2 diabetes mellitus in a Chinese population: a community-based case-control study. Gene 533(1), 427–433 (2014).
Khan, S. & Jena, G. The role of butyrate, a histone deacetylase inhibitor in diabetes mellitus: Experimental evidence for therapeutic intervention. Epigenomics 7(4), 669–680 (2015).
Daneshpajooh, M. et al. MC1568 improves insulin secretion in islets from type 2 diabetes patients and rescues β-cell dysfunction caused by Hdac7 upregulation. Acta Diabetol. 55(12), 1231–1235 (2018).
Hu, Y. et al. Sodium butyrate mitigates type 2 diabetes by inhibiting PERK-CHOP pathway of endoplasmic reticulum stress. Environ. Toxicol. Pharmacol. 64, 112–121 (2018).
Lee, T. I. et al. HDAC inhibition modulates cardiac PPARs and fatty acid metabolism in diabetic cardiomyopathy. PPAR Res. 2016, 5938740 (2016).
Yang, M. et al. Association between insulin resistance related indicators with the prognosis of patients with colorectal cancer. Cancer Epidemiol. 87, 102478 (2023).
Acknowledgements
We thank the investigators who made their sequencing data publicly available in the public database and the subjects who provided samples.
Funding
This research was funded by Fund for development of Science and Technology of Hospital of Chengdu University of TCM, grant number 23YY36.
Author information
Authors and Affiliations
Contributions
SL: Conceptualization, Visualization, Supervision, and Writing— original draft. YZ: Investigation, Formal analysis, Data curation, and Writing— original draft. ZG: Methodology and Software, and Writing— original draft. XF: Validation, and Writing— original draft. CZ: Project administration, and Writing— original draft. FY: Project administration, Resources and Writing—review and editing. XL: Project administration, Funding acquisition and Writing—review and editing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Institutional review board statement and informed consent statement
This study used sequencing data from public databases for analysis. The dataset involved in this study provides ethical approval in the corresponding database, so no additional ethical approval and informed consent are required for this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Luo, S., Zhu, Y., Guo, Z. et al. Exploring biomarkers and molecular mechanisms of Type 2 diabetes mellitus promotes colorectal cancer progression based on transcriptomics. Sci Rep 15, 4086 (2025). https://doi.org/10.1038/s41598-025-88520-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-88520-4
Keywords
This article is cited by
-
Bridging diabetes and cancer: harnessing biomarkers as dual sentinels for diagnosis, prognosis, and therapeutic advancements
Naunyn-Schmiedeberg's Archives of Pharmacology (2025)