Abstract
Tuberculosis, caused by Mycobacterium tuberculosis, is a major global health problem. In Mexico, the State of Nuevo Leon is among the top ten in tuberculosis morbidity. Information about transmission patterns and case clustering for tuberculosis in Nuevo Leon is limited. The spoligotypes of 151 isolates from newly diagnosed pulmonary tuberculosis patients were obtained and its phenotypic drug susceptibility pattern for streptomycin, isoniazid, rifampin, ethambutol, and pyrazinamide was determined by using the Mycobacteria Growth Indicator Tube fluorometric method. Geographical data of isolates were mapped using geographic information systems. Nineteen M. tuberculosis sublineages were identified. The most frequent lineages were: T1 at 35% (n = 53), X1 at 19.2% (n = 29), and LAM at 10.2% (n = 15). Additionally, we identified the Beijing lineage (3.3%, n = 5) and orphan strains (9.9%, n = 15). Drug resistant strains were 25 (16.55%) DR-TB, 15 (9.93%) MDR/RR-TB and 6 (3.97%) Hr-TB. Regarding TB comorbidities, diabetes mellitus II affected 21.85% of patients, while four patients were HIV-positive (2.65%). Despite the large number of tuberculosis cases in Monterrey, no definitive correlation with clusters and comorbidities was found. However, our results suggest a potential TB transmission hotspot for the T1 lineage within the Monterrey metropolitan area.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis. It remains a major public health problem worldwide, particularly in low- and middle-income countries. In 2022, the global incidence of newly diagnosed TB cases reached 7.5 million. the highest figure since the inception of the World Health Organization’s (WHO) global TB monitoring in 1995. This number surpassed the pre-COVID-19 baseline of 7.1 million in 2019, representing an increase from 5.8 million in 2020 to 6.4 million in 20221. TB ranks among the top 10 causes of death worldwide and is the second leading cause of death from a single infectious agent after COVID-19. Furthermore, it is estimated that a quarter of the world’s population (two billion people) has latent TB infections and five to ten% of them will develop active TB disease during their lives1.
TB incidence in Mexico is influenced by factors such as healthcare resources, access to medical care and public health interventions. In 2023 Mexico reported more than 28,000 new cases of TB of all forms; however, pulmonary tuberculosis (PTB) accounted for more than 90% of those cases. The national PTB incidence rate was 14.9 cases per 100,000 inhabitants. The federal entities with the highest incidence rates were Baja California (49.8), Sonora, Sinaloa (33.2), Tamaulipas (32.0) and Tabasco (28.2). Nuevo Leon is among the top ten, with an incidence of 26.8 cases per 100,000 inhabitants2. Each year, the Secretary of Health of the State of Nuevo Leon (SHSNL), Mexico performs more than 2,000 microbial cultures, of which more than 1,300 cultures test positive for new TB cases each year. These cultures account for 80–90% of the total TB clinical samples in the state of Nuevo Leon3.
Comorbidities such as HIV coinfection and diabetes4 and the emergence of drug-resistant (DR-TB) and multidrug-resistant (MDR-TB) strains complicate treatment and increase the number of deaths caused by TB5.
Spoligotyping is a PCR-based molecular technique that uses the reverse hybridization principle of the highly polymorphic direct repeat (DR) genome region for the genotypic characterization of M. tuberculosis isolates6,7. The use of spoligotyping data combined with drug resistance analysis and geospatial information system (GIS) mapping provides a tool for examining the spatial distribution of TB patients8, risk factors9,10 and drug resistance. Additionally, we can analyze TB transmission dynamics11,12, and cluster formation13,14 and identify highly virulent and drug resistant lineages such as the Beijing lineage15. The present study aimed to analyze the molecular epidemiology and drug resistance patterns of M. tuberculosis strains isolated from patients newly diagnosed with pulmonary tuberculosis. Additionally, we used GIS to analyze the distribution of tuberculosis in specific urban geographical areas of the state of Nuevo León, México.
Materials and methods
Study population
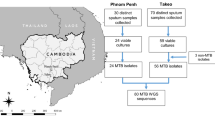
One hundred and fifty-one patients with three serial sputum samples were confirmed to be acid-fast bacillus positive and culture positive. The patients (41 females and 110 males) were attending the Secretary of Health of the State of Nuevo León (SHSNL), México. The samples were collected from June 2019 to June 2020. Epidemiological and clinical information, including age, HIV infection status and diabetes status, was obtained from the clinical records of each patient. The age range was 18–96 years. This study included only patients who were diagnosed with pulmonary TB for the first time and had not received any prior TB treatment. Patients with extrapulmonary TB, and underaged and inmate patients were excluded from this study.
Ethic statement
All methods were carried out in accordance with the institutional, national (Mexican General Health Law and Regulations of the General Health Law on Health Research) and international (Declaration of Helsinki) guidelines and regulations. Informed consent was obtained from all subjects and/or their legal guardian(s). This study was approved by the ethics committee of the Facultad de Enfermería, Universidad Autónoma de Nuevo León, with registration number FAEN-E-1496.
Isolation and purification of M. tuberculosis DNA
Sputum samples were decontaminated using Petroff´s modified method16. Briefly, 2 mL of the sample was combined with 2 mL of 4% NaOH in a sterile conical-bottom tube. The mixture was vortexed for 30 s to ensure homogenization. The samples were subsequently centrifuged at 3000 rpm for 15 min, after which the supernatant was discarded. HCl was used to neutralize the pH. Primary isolation was performed in Lowenstein-Jensen medium. After 5–8 weeks of incubation at 37 °C, a loop of each culture was resuspended in 2 mL of phosphate-buffered saline (PBS) (pH ~ 7.4) and inactivated at 95 °C for 60 min, and 50 µL of lysozyme (50 mg/ml) was added and incubated overnight at 37 °C. The Precellys® VK05 Lysing Kit (Bertin Technologies, France) was used for DNA purification. The purified DNA was dissolved in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH = 8.0) and quantified with a Nanodrop™ 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, USA). The DNA samples were stored at – 20 °C until use.
Spoligotyping
Spoligotyping assays were performed using the Spoligotyping Diagnostic Kit (Mapmygenome India Limited, Madhapur, India). The direct repeat region of the M. tuberculosis genome was amplified using the DRa forward (5′-GGTTTTGGGTCTGACGAC-3′ biotinylated) and DRb reverse (5′-CCGAGAGGGGACGGAAAC-3′) primers17. The chromosomal DNA of the M. tuberculosis H37Rv strain and Mycobacterium bovis BCG P3 strain were used as positive controls, whereas molecular grade water (Corning, New York, NY, USA) was used as a negative control. The amplified products were hybridized on a membrane precoated with the 43 spacer oligonucleotides of the direct repeat (DR) region (Ocimum Biosolutions, Hyderabad, India). The spoligotype patterns were visualized on X-ray films after incubation with streptavidin‐peroxidase and enhanced chemiluminescence (ECL) detection (Santa Cruz Biotechnology, USA). The obtained spoligotypes were entered into the SITVIT2 WEB database (Institut Pasteur de la Guadeloupe) (http://www.pasteur-guadeloupe.fr:8081/SITVIT2/), and the octal method was used to determine the M. tuberculosis sublineage and families.
Drug susceptibility testing
Phenotypic drug susceptibility testing for the first-line drugs streptomycin (S), isoniazid (H), rifampin (R), ethambutol (E), and pyrazinamide (Z) was performed using the Mycobacteria Growth Indicator Tube fluorometric method (MGIT 960, Becton–Dickinson, New Jersey, United States) at standard concentrations (H > 0.1 µg/mL, R 1.0 µg/mL, E 5.0 µg/mL and S 1.0 µg/mL). The Z resistance determination was performed using the BACTEC™ MGIT™ 960 PZA kit 100 µg/mL. Classification of resistant strains was made according to the WHO operational handbook on tuberculosis18.
Spatial analysis
To identify hotspots and cluster formation, we created maps of the TB lineage and strain clusters using ArcGIS® (version 10.2) software. These maps incorporated the geographic ___location of each patient. To maintain confidentiality, we used the intersections of each block and the municipality of each patient instead of their exact addresses. We constructed maps displaying the distribution of all the tuberculosis cases in the neighborhoods to identify spatial point patterns.
Statistical analysis
To investigate associations between epidemiological variables (such as age, sex, risk factors and drug resistance) and a specific SIT or cluster, odd ratios were calculated. A p value < 0.05 was considered to indicate statistical significance. Tests were performed using GraphPad Prism (V 9) software.
Results
The patient distribution revealed an approximate 3:1 sex ratio, with ages ranging from 18 to 96 years and a mean age of 40–44 years (± 12) (Fig. 1). Among the patients, 32 had type II diabetes mellitus (21.2%) and 4 were HIV positive (2.65%). Other risk factors, such as smoking (more than 10 cigarettes per week) (1.32%) and the use of drugs (2.64%), were also reported (Table 1). The statistical analysis showed no significant association between the different variables considered (age, sex, risk factors and drug resistance) with respect to any lineage.
Spoligotyping
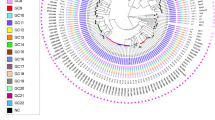
The spoligotyping patterns of the 151 samples corresponding to nineteen sublineages are shown in Table 2. The two most frequent clades were T1 with 53 (35.1%) isolates and X1 with 29 (19.2%) isolates. Lam family (including clades 1, 2, 4, 5 and 9) had 15 (9.93%) isolates. Clade H1 had 1 (0.67%) isolate and clade H3 had 4 (2.60%) isolates, while MANILLA had 3 (1.98%) isolates. Clades U had 4 (2.6%), X2 had 5 (3.3%), X3 had 4 (2.6%), S and T had 3 (1.98%) isolates. Additionally, we identified 5 (3.3%) isolates belonging to the Beijing lineage and 15 (9.9%) classified as orphan strains. The spoligotype patterns of the 151 samples are shown in Supplementary Table 1 (sTable 1).
Drug resistance
Among the 151 clinical isolates, 25 (16.55%) were resistant to at least one drug (DR-TB), 15 isolates (9.93%) were MDR/RR-TB, and 6 isolates (3.97%) were Hr-TB. The resistance to each drug, regardless of whether the strain was resistant to one or multiple drugs, was 27 (17.9%) streptomycin-resistant isolates, 24 (15.9%) isoniazid-resistant isolates, 15 (9.9%) rifampicin-resistant isolates, 7 (4.6%) ethambutol-resistant isolates, and 21 (13.9%) pyrazinamide-resistant isolates. Table 3 shows the resistance to first-line drugs for each strain. Additionally, 105 (69.1%) clinical isolates were susceptible to all the drugs tested.
Spatial distribution
The approximate address of each patient at the time of diagnosis was geocoded by ArcGIS®. Among the municipalities, Monterrey had the greatest number of cases (42), followed by Juarez with 25 and Gudalupe with 24. Apodaca accounted for 20 cases. Outside the metropolitan area, Montemorelos had 9 cases, whereas Linares and Cadereyta had 3 cases each (Fig. 2). Figure 3 shows the spoligotype distribution in the metropolitan area of Monterrey; the drug-resistant and multidrug-resistant strains are highlighted (orange and red circles, respectively). Clades X and T had the most drug-resistant and multidrug-resistant strains, with 18 and 17 respectively.
Number and spatial distribution of pulmonary tuberculosis cases. The green area delinates the Monterrey metropolitan area. Colored dots with letters represent spoligotypes. Regarding drug resistance: blue cricled dots represent DR-TB, red circled dots represent Hr-TB, and black circled dots represent MDR/RR-TB.
Hotspots
The spoligotype T1 had the greatest number of cases in the metropolitan area, with 54 isolates. Most of them were located near the border limits of the Guadalupe and Juarez municipalities. These data suggest a TB transmission hotspot within the metropolitan area (Fig. 4).
Discussion
Knowledge of the M. tuberculosis strains circulating in a specific region may aid in the implementation of health programs to control tuberculosis. The State of Nuevo León, which is located in northeast Mexico and has a population of over five million inhabitants, boasts a well-developed commercial and industrial infrastructure that attracts immigrants from other regions of the country. While a few studies have investigated the genetic variability of M. tuberculosis strains circulating in urban areas of the State of Nuevo León, Mexico, our present study provides an update on the TB lineages occurring in the state and their drug resistance profiles. Additionally, by using GIS, we were able to geolocate the cases, linking them with their corresponding lineages and drug sensitivities to predict potential TB transmission hotspots.
Several studies performed in Mexico have reported a wide range of genetic diversity of M. tuberculosis strains depending on the geographical region of the country. We found that the predominant spoligotype group was T1 (SIT53), as previously reported by Molina Torres19,20 and Zenteno Cuevas21,22. However, we could not establish a relationship between clades and age groups. Another common spoligotype found is “LAM”, which is a global spoligotype and is common in Mexico23.
An important lineage, which is globally distributed, corresponds to Beijing strains. It has been associated with increased virulence and a drug resistance phenotype. Beijing strains cause up to 50% of TB cases in East-Asia24. In recent years, this strain has been reported in México25,26,27. A recent study reported that 14 Beijing strains among 406 TB isolates from 2008 to 2015 in Nuevo Leon were classified as modern strains because of the deletion of the TbD1 region28. In this work, we identified 5 strains (3.3% of TB cases) belonging to the Beijing lineage in patients newly diagnosed with TB. Only one of them was resistant to four first line antibiotics (Table 3), whereas the other four Beijing isolates were drug sensitive. Although the number of strains found is still low, identification of these strains is important for molecular epidemiology, determining transmission routes and determining whether migration phenomena occur, which we were unable to do owing to the small number of Beijing isolates. Efforts to control the spread of Beijing family include identifying and treating infected individuals and implementing infection control measures in healthcare settings.
Our results showed that sublineage X1 is the lineage most strongly associated with the drug- resistant phenotype. Similarly, the X3 sublineage is strongly associated with MDR-TB29. We are concerned that, as the 62.06% of X1 strains are drug resistant and constitute the second largest group of isolates, their spread could represent an increase in TB cases; adequate measures need to be taken to control it. Additionally, a larger number of samples need to be analyzed to determine the importance of these strains in the region. However, we found that 46 out of 151 strains were resistant to one or more drugs. Becerril et al. reported a high prevalence of MDR-TB among previously treated patients in Monterrey30. We detected MDR-TB strains in newly diagnosed patients, which may indicate the transmission of drug-resistant strains into the TB naive population. The detection of drug-resistant strains and the ___location of those cases by GIS may explain the spread of those strains with the potential formation of local foci, increasing the transmission risk31,32,33. Poojar et al.33 explored the spatiotemporal aggregation and proved the usefulness of this method. They geolocated the precise address of each single TB case to avoid a selection bias due to misinformation in routine records. This also limited the aggregation bias to construct heat maps based on the occurrence of an event. Poojar reported that the sample size used in that work (larger than 1000) was adequate for the spatial heterogeneity analysis. We observed similar spatial heterogeneity in our results; the geographic distribution of the TB lineages and resistant strains were randomly dispersed.
GIS are very useful tools for creating maps to visualize the geographic distribution of TB cases to analyze the distribution and transmission of M. tuberculosis in a specific region11. The association of spoligotyping information with GIS analysis allows us to track the spread of specific strains in the community and understand their transmission dynamics34. By using GIS, we geolocated the highest rates of TB cases within the municipality of Monterrey (the most populated city of the state). Similar results were reported in other cities in Mexico with large populations. Those cities, as expected, also have the highest rates of TB transmission. The studies were performed in several states, such as Veracruz35, the state of Mexico36, Mexico City37 and Jalisco25.
The geographic ___location of Nuevo León, bordering Texas, makes it a transit route for migrants seeking to enter the US. Official data highlight the increased number of foreign individuals seeking temporary or permanent residence in Nuevo León, placing it among the top ten states in Mexico with the highest number of foreigners. The National Institute of Statistics and Geography (INEGI) reported over 19,000 foreigners in the state in 2000, which increased to 49,500 in 202038. Foreign residents in Nuevo León mainly come from the US (46.9%), Venezuela (12.1%), and Honduras (9.0%). The high number of U.S. residents is easily explained by the commercial and cultural exchange between Texas and Nuevo León, while people from Venezuela and Honduras come to Mexico seeking better living conditions or reaching the U.S. Moreover, the Mexican Migration Policy Unit, Registration and Identity of Persons (UPMRIP, 2021) reported that 2,674 temporary residence cards were issued in 2021, mainly to citizens from South Korea (16%), Colombia (11%), Venezuela (9%), Honduras (6%), and Brazil (5%)39.
The M. tuberculosis lineages with low incidence in this study may represent introduced lineages associated with recent migratory movements between countries40, as migration phenomena play an important role in tuberculosis epidemiology41. Additionally, the geographical proximity of Nuevo León to Texas in the United States underscores the importance of exploring spatial-temporal relationships in densely populated regions for understanding tuberculosis transmission dynamics. Agarwal, et al.42 described the temporal and geospatial characteristics of TB in Harris County, Texas, which includes the Houston metropolitan area, the third most populous county in the United States and has one of the highest TB rates in the United States of America. They analyzed active TB transmission, identified hot and cold spots and related them to geographic areas with high and low TB incidence. A similar trend was observed in the Monterrey metropolitan area, indicating a correlation between population size and TB incidence. This city requires constant surveillance as a potential emerging hot spot for TB transmission.
In Montemorelos, a small city located 80 km away from Monterrey (still with easy access and constant communication due to business), the transmission dynamics exhibited intriguing patterns. The identification of four distinct lineages (LAM, X1, Manilla, and T1), alongside an orphan strain, suggested random transmission from urban to less populated areas. The identification of areas where TB is prevalent and has active transmission could lead to the initiation of improvements in TB control programs.
Figure 4 shows the spatial distribution of spoligotype T1, which had the highest incidence in the metropolitan area, particularly in the municipalities of Guadalupe, Juarez, and Monterrey. The darkest colored areas have the highest number of TB cases.
These data suggest a TB transmission hotspot formation area on the border line between Gudalupe and Juarez. Geospatial tuberculosis hotspots serve as hubs of TB transmission both within and across community groups12. Confirmation of these hotspots involves several challenges that complicate accurate results: the interpretation of spatial clustering with low TB case reporting rates, the number of analyzed strains, short-term analyses (less than one year), and the chosen spatial analysis method. For example, kernel density estimation, which is based on the ___location of TB cases may differentiate between regions with a high number of diagnosed TB cases and those with high disease density in studies with 2000–5000 patients with a statistically normal distribution43. Alternatively, Zelner et al. utilized a nonparametric distance-based mapping (DBM) approach to construct hotspot maps. Compared with kernel density estimation, DBM is less sensitive to asymmetric spatial patterns of cases and controls, thus providing an attractive alternative for detecting high-risk areas in real communities with long-term follow up44. Although, we found a potential hot spot, it may not reflect actual TB transmission, as we used coordinates close to real residential addresses, which provide approximate neighborhoods; additionally, the number of patients was relatively low, and this was a short-term study. Additionally, it must be considered that transmission could have occurred in other areas where patients work, commute, or socialize.
Significant spatial heterogeneity was observed. Geospatial techniques can identify spatial variability in reported TB case rates and provide critical information for program planning. It remains unclear whether the observed heterogeneity in reported TB case rates is due to underlying differences in disease burden or barriers to accessing TB medical services14. High-resolution genomic data are required to establish real transmission links between individual cases and spatial locations and to explore the possibility that specific clones possess high reproductive fitness. Future analyses should integrate geographic and genetic information with dynamic models to highlight individual-level heterogeneity in TB transmission45. Additionally, further geographic analyses and mathematical modeling studies are essential to evaluate the potential of such strategies to curtail transmission.
Perhaps the main limitations of this study were the limited number of isolates and the lack of use of other genotypic methods, such as the mycobacterium interspersed repetitive unit-variable number of tandem repeats (MIRU-VNTR), the lack of knowledge of the natural residence of the patients and follow up data. The latter would be complicated as these patients were newly diagnosed. Overall, GIS can provide valuable insights into the distribution and transmission of TB, as well as guide the development and implementation of effective TB control programs. Further studies are needed to better understand the dynamics of TB circulation in Mexico. The implementation of more robust molecular surveillance methods is key for the proper monitoring of lineages, their introduction into the area and the development of drug resistance. This study revealed the diversity of genotypes and drug resistant strains circulating in the State of Nuevo Leon, Mexico and represents a data update on surveillance.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Global Tuberculosis Report (2023, accessed 3 Jul 2024). https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2023.
Catálogo Nacional de Indicadores (2024, accessed 8 Jul 2024). https://www.snieg.mx/cni/escenario.aspx?idOrden=1.1&ind=6300000063&gen=190&d=n.
Salud, S. Boletín epidemiológico sistema nacional de vigilancia epidemiológica sistema Único de Información 2021 [Internet]. gob.mx. (2021, accessed 17 Oct 2024). http://www.gob.mx/salud/documentos/boletinepidemiologico-sistema-nacional-de-vigilancia-epidemiologica-sistema-unico-de-informacion-2021.
Tulu, B. et al. Diabetes mellitus and HIV infection among active tuberculosis patients in Northwest Ethiopia: health facility-based cross-sectional study. Trop. Med. Health 49, 68 (2021).
Seung, K. J., Keshavjee, S. & Rich, M. L. Multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis. Cold Spring Harb. Perspect. Med. 5, a017863. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4561400/ (2015).
Gori, A. et al. Spoligotyping and Mycobacterium tuberculosis. Emerg. Infect. Dis. 11, 1242–1248 (2005). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3320497/.
Driscoll, J. R. Spoligotyping for molecular epidemiology of the Mycobacterium tuberculosis complex. Methods Mol. Biol. 551, 117–128 (2009).
Readhead, A. et al. Spatial distribution of tuberculosis incidence in Los Angeles County. BMC Public Health 20, 1434. https://doi.org/10.1186/s12889-020-09523-6 (2020).
Tiwari, N., Adhikari, C., Tewari, A. & Kandpal, V. Investigation of geo-spatial hotspots for the occurrence of tuberculosis in Almora district, India, using GIS and spatial scan statistic. Int. J. Health Geogr. 5, 33. https://doi.org/10.1186/1476-072X-5-33 (2024).
Abdul Rasam, A. R., Mohd Shariff, N., Dony, J. F. & Othman, F. Spatial and statistics for profiling risk factors of diseases: a case study of tuberculosis in Malaysia. IOP Conf. Ser.: Earth Environ. Sci. 385, 012037. https://doi.org/10.1088/1755-1315/385/1/012037 (2019).
Moonan, P. K. et al. Using GIS technology to identify areas of tuberculosis transmission and incidence. Int. J. Health Geogr. 3, 23 (2004). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC529461/.
Shaweno, D., Trauer, J. M., Denholm, J. T. & McBryde, E. S. The role of geospatial hotspots in the spatial spread of tuberculosis in rural Ethiopia: a mathematical model. R. Soc. Open Sci. 5, 180887 (2018). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6170575/.
Izumi, K. et al. Detection of tuberculosis infection hotspots using activity spaces based spatial approach in an urban Tokyo, from 2003 to 2011. PLOS ONE 10, e0138831. https://doi.org/10.1371/journal.pone.0138831 (2015).
Brooks, M. B. et al. Mapping local hot spots with routine tuberculosis data: a pragmatic approach to identify spatial variability. PLOS ONE 17, e0265826. https://doi.org/10.1371/journal.pone.0265826 (2022).
Cerezo-Cortés, M. I., Rodríguez-Castillo, J. G., Hernández-Pando, R. & Murcia, M. I. Circulation of M. Tuberculosis Beijing genotype in Latin America and the Caribbean. Pathog. Glob. Health 113, 336–351 (2019).
Petroff, S. A. A new and rapid method for the isolation and cultivation of tubercle bacilli directly from the sputum and feces. J. Exp. Med. 21, 38–42 (1915). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2125265/.
Kamerbeek, J. et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35, 907–914 (1997).
WHO Operational Handbook on Tuberculosis. Module 4: Treatment—Drug-Resistant Tuberculosis Treatment, 2022 Update 1st edn (World Health Organization, 2022).
Molina-Torres, C. A. et al. Mycobacterium tuberculosis spoligotypes in Monterrey, Mexico. J. Clin. Microbiol. 48, 448–455 (2010).
Molina-Torres, C. A. et al. Genetic diversity of Mycobacterium tuberculosis isolates from an Amerindian population in Chiapas, México. Front. Cell Infect. Microbiol. 12, 875909 (2022). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9326120/.
Zenteno-Cuevas, R. et al. Mutations conferring resistance to first- and second-line drugs in multidrug-resistant Mycobacterium tuberculosis clinical isolates in southeast Mexico. Int. J. Antimicrob. Agents 45, 671–673 (2015). Available from: https://www.sciencedirect.com/science/article/pii/S0924857915000771.
Zenteno-Cuevas, R. et al. Genetic diversity and drug susceptibility of Mycobacterium tuberculosis in a city with a high prevalence of drug resistant tuberculosis from Southeast of Mexico. BMC Infect. Dis. 21, 1202 (2021). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8630842/.
Martinez-Guarneros, A. et al. Genetic diversity among multidrug-resistant Mycobacterium tuberculosis strains in Mexico. Infect. Genet. Evol. 14, 434–443 (2013). https://www.sciencedirect.com/science/article/pii/S1567134813000099.
Parwati, I., van Crevel, R. & van Soolingen, D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 10, 103–111 (2010).
Flores-Treviño, S. et al. Genetic diversity of Mycobacterium tuberculosis from Guadalajara, Mexico and identification of a rare multidrug resistant Beijing genotype. PLOS ONE 10, e0118095. https://doi.org/10.1371/journal.pone.0118095 (2015).
Martinez-Gamboa, A. et al. Molecular analysis of Mycobacterium tuberculosis strains with an Intact pks15/1 gene in a rural community of Mexico. Arch. Med. Res. 39, 809–814 (2008). https://www.sciencedirect.com/science/article/pii/S0188440908002221.
López-Rocha, E. et al. Genetic diversity of the Mycobacterium tuberculosis complex in San Luis Potosí, México. BMC Res. Notes 6, 172 (2013).
Catana-Botello, N. et al. Mycobacterium tuberculosis Beijing in the State of Nuevo Leon, Mexico. Rev. Argentina Microbiol. (2024). http://www.elsevier.es/en-revista-revista-argentina-microbiologia-372-articulo-mycobacterium-tuberculosis-beijing-in-state-S0325754124000464.
Munro-Rojas, D. et al. Genetic diversity of drug and multidrug-resistant Mycobacterium tuberculosis circulating in Veracruz, Mexico. PLOS ONE 13, e0193626. https://doi.org/10.1371/journal.pone.0193626 (2018).
Becerril-Montes, P. et al. A population-based study of first and second-line drug-resistant tuberculosis in a high-burden area of the Mexico/United States border. Mem. Inst. Oswaldo Cruz. 108, 160–166 (2013). https://www.scielo.br/j/mioc/a/rZ9sLwHhXqJmCpTzWrYcsgH/?lang=en.
Manjourides, J. et al. Identifying multidrug resistant tuberculosis transmission hotspots using routinely collected data. Tuberculosis (Edinb.) 92, 273–279 (2012). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3323731/.
Theron, G. et al. Data for action: collection and use of local data to end tuberculosis. Lancet 386, 2324–2333 (2015).
Poojar, B. et al. Spatiotemporal analysis of drug-resistant TB patients registered in selected districts of Karnataka, South India: a cross-sectional study. Trop. Med. Health 48, 15 (2024). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7063724/
Tulu, B. & Ameni, G. Spoligotyping based genetic diversity of Mycobacterium tuberculosis in Ethiopia: a systematic review. BMC Infect. Dis. 18, 140. https://doi.org/10.1186/s12879-018-3046-4 (2018).
Blanco-Guillot, F. et al. Genotyping and spatial analysis of pulmonary tuberculosis and diabetes cases in the state of Veracruz, Mexico. PLOS ONE 13, e0193911. https://doi.org/10.1371/journal.pone.0193911 (2018).
Zaragoza Bastida, A. et al. Spatial and temporal distribution of tuberculosis in the state of Mexico, Mexico. Sci. World J. 2012, 570278 (2012). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3417174/.
Flores-Treviño, S., Mendoza-Olazarán, S. & Garza-González, E. Drug resistance and molecular epidemiology of Mycobacterium tuberculosis in Mexico: a systematic review. Salud Públ. México 56, 63–77 (2014). https://www.saludpublica.mx/index.php/spm/article/view/7324.
Censo de Población y. Vivienda (2020, accessed 15 Aug 2024). https://www.inegi.org.mx/programas/ccpv/2020/.
Power, B. I. Report (2024, accessed 15 Aug 2024). https://app.powerbi.com/view?r=eyJrIjoiYjY4NWE3YzItYjZjZi00MWYzLWI4NzgtNTdkNDA4MTU5YTQ5IiwidCI6ImQ5MDc5M2MzLTY0ZWQtNGJkZS04ZGQ3LTE4NmI4MWFkNWIyOCIsImMiOjR9 .
Vinueza, L. C. & Arias, P. J. Distribution of Mycobacterium tuberculosis lineages in South America. Anatom. Digital 4, 34–58 (2021). https://cienciadigital.org/revistacienciadigital2/index.php/AnatomiaDigital/article/view/1755.
Flores-López, C. A. et al. Molecular epidemiology of Mycobacterium tuberculosis in Baja California, Mexico: a result of human migration? Infect. Genet. Evol. 55, 378–383 (2017).
Agarwal, S., Nguyen, D. T., Teeter, L. D. & Graviss, E. A. Spatial-temporal distribution of genotyped tuberculosis cases in a county with active transmission. BMC Infect. Dis. 17, 378 (2017).
Ogbudebe, C. et al. Identifying hot spots of tuberculosis in nigeria using an early warning outbreak recognition system: retrospective analysis of implications for active case finding interventions. JMIR Public Health Surveill. 9, e40311 (2023). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9947752/.
Zelner, J. L. et al. Identifying hotspots of multidrug-resistant tuberculosis transmission using spatial and molecular genetic data. J. Infect. Dis. 213, 287–294 (2016). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4690150/.
Ypma, R. J. F., Altes, H. K., van Soolingen, D., Wallinga, J. & van Ballegooijen, W. M. A sign of superspreading in tuberculosis: highly skewed distribution of genotypic cluster sizes. Epidemiology 24, 395–400 (2013).
Funding
This work was supported by the Support Program for Scientific and Technological Research of the UANL (Programa de Apoyo a la Investigación Cientifíca y Tecnológica; PAICYT-UANL) 2019–2020. Grant SA837-19.
Author information
Authors and Affiliations
Contributions
JLZH and JCG: Conceptualization of the work. JLZH, AFA, LGRM and JCG: Formal analysis. RSTG, PBM, LGRM and CRP: Supervision: JLZH, AFA, LGRM and JCG: Validation. JLZH, LGRM and JCG: Original draft: JLZH, RSTG, JMVG and MOC: Project administration. JLZH and CGVC: Investigation. JLZH, PBM and CGVC: Methodology. JCG, RSTG, CRP and LGRM: Editing. JLZH, JCG and LGRM: Writing, reviewing, and editing. LGRM, RSTG and CRP: Funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zacarías-Hernández, J.L., Flores-Aréchiga, A., Tamez-Guerra, R.S. et al. Geographical ___location and genotyping analysis of pulmonary tuberculosis in the state of Nuevo Leon, Mexico. Sci Rep 15, 7098 (2025). https://doi.org/10.1038/s41598-025-90579-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-90579-y