Abstract
The cultivation and trade of underutilized fruits are gaining prominence worldwide, largely on account of their capacity to contribute to a nutritious diet. Manila tamarind [Pithecellobium dulce (Roxb.) Benth] is a fast-growing, nitrogen-fixing tree, with a fairly high tolerance to abiotic stresses. Despite significant potential in terms of food, fodder, timber, and medicine, it has largely remained an orphan crop. There is a dearth of systematic research on the exploration, conservation, and genetic improvement of Manila tamarind. Our study aimed to assess the genetic variability for commercially important fruit, aril, and leaf attributes in 22 diverse accessions of Manila tamarind comprising both white and red aril genotypes. Precise characterization of the existing genetic resources is a requisite for the commercial cultivation of Manila tamarind. The study was conducted with 15 white and 7 red accessions of P. dulce, which were planted in a square system of planting between and within row distances of 5 m each. One of the major contributions of the present study was that we examined genotypic variations in biochemical attributes, such as TSS, acidity, TSS: acidity ratio, total sugars, ascorbic acid, protein, mineral contents, and bioactive compounds; these factors significantly improve the nutritional value and eating quality of Manila tamarind arils. Most of the traits examined by us differed remarkably (p < 0.001) among the accessions. Some economically relevant traits, such as pulp weight, aril weight, aril total phenols, aril flavonoids, aril total antioxidant activity, and leaf flavonoids exhibited a high degree of variability, indicating the scope for the selection of elite genotypes and divergent parents for future hybridization programs. The highly variable values of total soluble solids (17.33–26.46 °Brix), acidity (0.54–1.07%), ascorbic acid (82.54-138.49 mg 100 g− 1), total sugars (12.45–18.81%), and aril protein (3.15–6.32%) recorded in this study broadly meet fresh consumption and aril processing standards for Manila tamarind. A significant finding was that Manila tamarind accessions differed greatly in aril mineral contents (mg/100 g FW), including potassium (220.44-334.33), phosphorus (21.63–62.34), and calcium (14.06–39.12). Overall, two red aril genotypes (CHESM-27 and CHESM-33), and three white aril genotypes (CHESM-4, CHESM-20, and CHESM-24) were found to be particularly promising in terms of pod and aril quality attributes. Our findings are expected to pay the way for commercial cultivation of elite Manila tamarind genotypes, and their applications in pharmaceutical applications. Future studies should aim to elucidate the molecular basis of genetic diversity and relationships in Manila tamarind.
Similar content being viewed by others
Introduction
The production and trade of underutilized fruits are gaining importance globally, mainly in recognition of their contribution to a healthy diet24,35,56,58,69and significant medicinal potential14,17,23,59,61. Compared with many other commercially grown fruit crops, the cultivation of underutilized fruit crops, such as Manila tamarind [Pithecellobium dulce(Roxb.) Benth; family Leguminosae; subfamily Mimosoideae], has numerous advantages in terms of ease of management, hardiness, and resilience to climatic change35,63,64. Manila tamarind is a fast-growing, evergreen, nitrogen-fixing, and salt-tolerant tree or shrub found throughout the plains of India and in the Andaman Islands5,53,66. It can tolerate degraded soil conditions and is resistant to fire and drought8,48. It is also widely distributed in many semi-arid regions of tropical Africa and America9,28. The P. dulce tree has multiple uses as a source of timber, firewood, honey, food, fodder, and medicinal ingredients12,29,47,48,67. It is also recommended for soil biological restoration8.
The fruit of Manila tamarind is a pod consisting of sweet edible arils covering black seeds; depending on genotype, the arils are white or red50,73. Aril is a fleshy and sweet pulp that covers the seed partially or completely9. Pods are tightly coiled, irregularly shaped, and turn greenish brown to reddish at the time of maturity. Each pod has about 5–12 seeds which are shiny black and attached to the pods by a red funicle43,65. P. dulce aril is a good source of vitamins, minerals, dietary fiber, protein, and carbohydrates50,52,56,74, and shows antioxidant and hepatoprotective properties33,36. The level and composition of nutraceutical and physicochemical parameters in P. dulce fruit vary greatly with genotype50,56, and the stage of fruit maturity74. The aril is consumed raw, roasted, or used in the preparation of a beverage similar to lemonade7,42,53, and various value-added products like RTS, squash, and syrup37,40,56. It has numerous medicinal uses, and exhibits strong analgesic, anti-inflammatory, antibacterial, antidiarrheal, antiulcer, antioxidant, hypoglycemic, and hepatoprotective properties13,25,30,52,66,73,75.
Limited efforts have been made to methodically explore, assess, and improve the genetic resources of Manila tamarind16,19,40. Until recently, the Manila tamarind germplasm resources were mostly sourced from the landraces adapted to certain niche areas, resulting in a very narrow genetic base63. To our knowledge, some selections have been carried out only in the Philippines for big pods with tiny seeds, and red, sweet, and less astringent arils18. There is a need to identify prolific-bearing and high-yielding genotypes of Manila tamarind with large and sweet pods32. Gaining a deeper comprehension of the fruit’s physicochemical and biochemical characteristics is crucial for creating potential cultivars that are appropriate for various end uses38. Precise characterization of the existing genetic resources is a requisite for the commercial cultivation of Manila tamarind in India and elsewhere. Selecting the parents for genetic gains can be facilitated by having a thorough understanding of the gene pool that can be used in genetic improvement programs as well as morpho-genetic characterization of each genotype38. Plant trait determination and genetic characterization studies are important in establishing plant breeding programs80,81.
Until recently, genetic diversity analysis in P. dulce has mainly focused on variations in aril, leaf, and bark phytochemical traits in one or two genotypes i.e. either white aril or red aril types. To the best of our knowledge, this is the first study where a fairly large number of red-aril and white-aril P. dulce accessions were characterized using a range of economically important traits. To address the aforementioned research gaps, the present study was carried out with the objectives of examining the diversity in pod physicochemical traits, and bioactive compounds in the white and red accessions of P. dulce, and identify promising P. dulce accessions for commercial cultivation and for use as parents in the genetic improvement programs.
Materials and methods
Study site
The study was conducted at the Central Horticultural Experiment Station (CHES) of the Indian Council of Agricultural Research- Central Institute for Arid Horticulture located in Vejalpur, Panchmahal, Gujarat, India (22°41′N, 73°33′E with an altitude of 113 m above sea level) during 2023 and 2024 fruiting seasons. The experimental ___location has a hot, semi-arid climate with an average annual precipitation of about 750 mm. The soils of the experimental farm are mostly shallow, and sandy loam in texture. The soil pH is approximately 6.65, and the organic carbon content ranges from 0.35 to 0.45%. The formal identification of the samples was performed by Dr. Mishra. A voucher specimen of this material has been deposited in the publicly available herbarium of ICAR-Central Horticultural Experiment Station, Vejalpur with deposition number PD-3425.
Experimental material
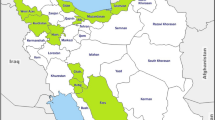
The study was conducted with 15 white and 7 red accessions of P. dulce planted in the field gene repository at CHES, Panchmahal, Gujarat, India (Table 1; Fig. 1). The trees of each P. dulce accession, aged 8–9 years, were planted in a square system of planting with between and within row distances of 5 m each. Recommended crop management practices were adopted for healthy tree growth.
Fruit physical properties
At the commercial maturity stage (March-April), when pods begin to exhibit distinctive colors with some noticeable splitting, ten pods from different directions of three trees of each accession were randomly collected to record various observations. The pod weight (PWt, g), aril weight (ArWt, g), peel weight (PlWt, g), and seed weight (SWt, g) were recorded using a precision balance (0.01 g accuracy). Pod width (PW, mm), seed length (SL, mm), and seed width (SW, mm) were measured using a Vernier caliper (Mitutoyo, Japan). Number of seeds/pod (NSP) were manually counted.
Fruit chemical properties
The filtered aril juice was used for determining titrable acidity (Acid, %) and total soluble solids (°Brix). Total soluble solids were estimated using an Erma Hand Refractometer (0–32 °Brix). Titrable acidity (% of citric acid) was determined using N/10 NaOH and phenolphthalein as indicators as described in AOAC1. The TSS: acidity ratio (TA) was calculated as the ratio between TSS and acidity. Ascorbic acid content (mg 100 g−1) was determined using 2,6-dichlorophenol indophenol dye1. Total sugars (%) were estimated by Lane and Eynon’s method using Fehling’s solutions as described in AOAC1. Protein determination was carried out by the micro-Kjeldahl method using the factor 6.25. The sample (0.50 g) was digested in H2SO4 (10 ml) in the presence of 1 g catalyst K2SO4 (10 g) + CuSO4 (1 g). Subsequently, the volume was made up to 100 ml. 10 ml of sodium hydroxide (NaOH, 40%) was added to 10 ml of digest, followed by steam distillation; the distillate was collected in 10 ml of boric acid (4%). Then nitrogen content was determined by using titration with 0.005 N H2SO4 (Bharghava and Raghupathi, 1993). All results are expressed on a fresh weight basis (FW).
Bioactive compounds and antioxidants
Leaf biochemical parameters were determined on recently mature leaves. Total phenols (TP) present in the arils (ArTP) and leaf extracts (LTP), expressed as mg of gallic acid equivalent per g (mg GAE g−1), were estimated following the Folin-Ciocalteu reagent4,78. The reaction mixture containing an appropriate aliquot of ethanolic extract, 0.5 ml of 1 N phenol reagent (Folin- Ciocalteu), and 6% sodium carbonate solution (2 ml) (w/v) was incubated at a boiling water bath for 2 min. Then, the tubes were kept at room temperature to cool the solution and the absorbance was recorded at 650 nm against a reagent blank. The total flavonoid content in leaves (LFlav) and arils (ArFlav) was determined by the aluminum chloride-based colorimetric method79. A volume of extracts (0.50 ml) was mixed with 0.3 ml each of 5% NaNO2 and 10% AlCl3 and 3.4 ml of 1 M NaOH. The resultant reaction mixtures were incubated for 15 min at room temperature and the OD was measured at 510 nm against the reagent blank. The total flavonoid content was expressed as mg of catechol equivalent per g (mg CE g−1). In the case of total antioxidant activity (TAA), the reducing capacity of P. dulce aril (ArTAA) and leaf (LTAA) extracts were assayed by the DPPH method3. Different concentrations (100 to 500 µg/ml) of aqueous methanolic extract of aril and leaves of P. dulce (100 µl) were allowed to react with 2.9 ml of 0.006% methanolic DPPH for 10 min under dark conditions. A control was also run simultaneously with 100 µl distilled water instead of extract and absorbance was taken at 450 nm using a UV‑VIS spectrophotometer (UV‑2550, SHIMADZU). Ascorbic acid was used as a reference standard (10–50 µg) and the results were expressed as mg of ascorbic acid equivalent per g (mg AAE g−1).
Aril mineral contents
The calcium (ArCa), phosphorus (ArP), and potassium (ArK) contents in arils were estimated on a fresh weight basis using diacid-digested samples38. One gram of fresh aril was digested in 25 ml of diacid mixture (concentrated nitric acid and perchloric acid in a 9:4 ratio), and distilled water was added to achieve the final volume (100 ml). ArCa was determined by EDTA titration38, ArP was estimated colorimetrically using an atomic absorption spectrophotometer (ELICO SL 164, India), and ArK using a flame photometer (ELICO CL 361, India)5.
Statistical analyses
The experiment was laid out in a randomized complete block design. Observations related to pod (fruit) physical parameters were recorded from randomly sampled trees (n = 3) of each accession. Fruit physical and chemical properties, biochemical, antioxidants, and mineral contents were determined on randomly sampled firm-ripe fruits (n = 10). One-way Analysis of Variance (ANOVA) was used to test the significant differences (p< 0.05) (JASP v. 0.17.3). Means were compared using the Tukey test. Data are presented as mean ± standard deviation. Principal Component Analysis (PCA) was carried out to discern the key trends in data. Pearson’s bivariate correlations between variables and the associated significance levels were calculated. Hierarchical cluster analysis was carried out using the Ward’s distance20.
Results
Genotypic differences for pod and peel-related traits
The results for the analysis of variance (ANOVA) and mean comparisons revealed strong differences among the studied genotypes for most of the pod and peel-related traits. The highly significant F values for all the traits (p < 0.001) suggested that genotype accounted for a significant proportion of variance in the dependent variables (Table 2). The pod weight (PWt) varied between 10.22 g (CHESM-29) and 32.92 g (CHESM-6), while pod width (PW) ranged between 12.24 (CHESM-30) and 19.65 mm (CHESM-10). Compared with the average value of 19.27 g, the PWt was considerably higher in genotypes CHESM-6 and CHESM-24. The peel weight (PlWt) varied between 1.26 g (CHESM-31) and 5.12 g (CHESM-24), while peel percent (PlP) ranged between 7.05% (CHESM-6) and 34.32% (CHESM-30). Notably, genotypes CHESM-6 and CHESM-31 had very low peel content (< 10.0%) when compared with most other genotypes. Of these traits, PWt was the most variable with a coefficient of variation of ~ 20.0% (Table 2).
Genotypic differences for seed-related traits
Table 3 shows the variation among the tested genotypes for seed physical properties. The F values were highly significant for all the traits (p < 0.001) except seed width (SW) (F = 2.40, p = 0.039) and number of seeds per pod (NSP) (F = 3.98, p = 0.003). It was interesting to note that both lower (1.19 g) and higher (3.40 g) values of seed weight (SWt) values were recorded in red aril genotypes (CHESM-31 and CHESM-33, respectively) while it ranged between 1.66 g (CHESM-10) and 2.62 g (CHESM-24) in white aril genotypes. White aril genotype CHESM-6 recorded the lowest seed percent (SP) (6.04%) while red aril genotype CHESM-30 recorded the highest SP (24.85%). The seed length (SL) ranged between 10.13 mm (CHESM-28) and 12.96 mm (CHESM-33), SW between 6.57 mm (CHESM-22) and 9.68 mm (CHESM-20), and NSP between 6.66 (CHESM-31) and 14.0 (CHESM-30). In general, seed-related traits varied more among red aril genotypes than among white aril genotypes (Table 3).
Genotypic differences for aril-related traits
The genotypic differences for aril physical attributes were invariably highly significant (p < 0.001) except for ArWt (F = 4.47, p = 0.002). Both aril weight (ArWt) and aril percent (ArP) were the highest in CHESM-6 (28.45 g and 86.31%, respectively). Comparably, CHESM-29 recorded the lowest ArWt (5.60 g) and CHESM-30 recorded the lowest ArP (40.59%). Genotype CHESM-6 had the highest aril length (ArL) (25.20 mm) and aril width (ArW) (28.45 mm), while these two traits were the lowest (14.20 and 10.60 mm, respectively) in CHESM-22. In comparison with other traits, ArP exhibited a very high degree of variability (CV = 69.29%) (Table 4).
Genotypic differences for fruit and leaf biochemical attributes
Table 5 shows the genotypic differences for fruit biochemical parameters. The F values were mostly highly significant (p < 0.001), except for aril protein (ArPt) (F = 2.87, p = 0.002). The TSS ranged between 17.33 °Brix (CHESM-10) and 26.46 °Brix (CHESM-28) with an average of 21.32 °Brix. Both acidity [0.54 (CHESM-6) and 1.07 (CHESM-24)] and TSS: acidity ratio (TA) differed remarkably among genotypes [20.65 (CHESM-1) and 42.79 (CHESM-29)], and were found to be more variable in terms of coefficient of variation (≥ 20.0%) when compared with TSS (CV = 12.51%). The ascorbic acid (AA) content ranged between 82.54 mg/100 g (CHESM-22) and 138.49 mg/100 g (CHESM-4) with an average of 107.26 mg/100 g. Of the tested genotypes, CHESM-28 exhibited the highest content of total sugars (TS) (18.81%) while it was the lowest (12.45%) in CHESM-10. Similarly, ArPt ranged between 3.15% (CHESM-24) and 6.32% (CHESM-27). There were highly significant differences among the genotypes for aril minerals as well as aril and leaf bioactive compounds (Table 6). The aril total phenols (ArTP) were the lowest (2.13 GAE mg g−1 FW) in CHESM-22 and the highest (11.18 GAE mg g−1 FW) in CHESM-29. Compared with the average value of 0.53 CE mg g−1 FW, some genotypes, including CHESM-29, CHESM-32, CHESM-33, and CHESM-34 had much higher aril flavonoids (ArFlav) contents (> 0.80 CE mg g−1 FW) while it was considerably lower (< 0.20 CE mg g−1 FW) in some genotypes, such as CHESM-2, CHESM-6, and CHESM-7.
The tested genotypes differed remarkably from one another in aril total antioxidant activity (ArTAA) with CHESM-6 exhibiting its lowest value (4.11 AAE mg g−1) while CHESM-27 showed the highest ArTAA (27.09 AAE mg g−1). Considerable genotypic differences were also noted for the aril minerals, including K (220.44–334.33 mg 100 g−1 FW), P (21.63–62.34 mg 100 g−1 FW), and Ca (14.06–39.74 mg 100 g−1 FW). However, aril P content was found to be more variable (CV = 34.18%) than both K (CV = 13.86%) and Ca (CV = 25.63%) (Table 6). The leaf total phenols (LTP) ranged between 7.25 GAE mg g−1 FW (CHESM-2), and 16.95 GAE mg g−1 FW (CHESM-27). Similarly, the leaf flavonoid (LFlav) content varied between 0.34 CE mg g−1 FW (CHESM-33), and 1.21 CE mg g−1 FW (CHESM-27) and LTAA differed between 8.61 AAE mg g−1 (CHESM-31)−30.54 AAE mg g−1 (CHESM-27). In general leaf samples had more bioactive compounds than aril and amongst white and red genotypes, red accessions contained a higher quantity in leaf and aril both (Table 6).
Principal component analysis
The results of principal component analysis (PCA) are shown in Supplementary 1. The first four Principal Components with Eigenvalue > 1.0 accounted for 62.30% of the cumulative variance in data. The PC1 accounted for 27.60% of the total variation; it was largely a linear combination of ArTP, ArFlav, ArTAA, LFlav, and LTP. The PC2 explained 14.90% of the total variance in data and was loaded heavily on PWt, Arwt, ArL, ArW, TSS, AA, TS, ArK, and ArCa. The PC3 summarized about 12.20% of the variance in data and was mainly a construct of PlWt, SWt, SL, and NSP. As expected, the Eigenvalues and the proportion of variance explained by the subsequent Principal Components declined with PC4 accounting for 7.70% of the total variation in data (Supplementary 1). The variable loadings on the first two principal components are illustrated as the PCA biplot in Fig. 2. It was found that the PC1 separated most of the pod edible components (aril physical attributes) from non-edible components (PlP and SP) as well as the nutritional and bioactive compounds. Given its strong association with the traits accounting for antioxidant value (e.g., ArTP, ArTAA, ArFlav, LFlav, and LTP), the PC1 was labeled as the ‘bioactive component’. The PC2 was well represented by the mixed variables (e.g., PWt, ArL, ArWt, SW, AA, TSS, TS, ArK, and ArCa) that directly or indirectly influence the pod edible quality and mineral contents; it may thus be termed as the ‘pod quality component’ (Fig. 2).
Correlation of variables in a 2-dimension plot of Pithecellobium dulce genotypes. Abbreviations: PWt, pod weight (g); PW, pod width (mm); PlWt, peel weight (g); PlP, peel percentage (%); SWt, seed weight (g); SP, seed percentage (%); SL, seed length (mm); SW, seed width (mm); NSP, number of seed/pod; ArWt, aril weight (g); ArL, aril length (mm); ArW, aril width (mm); ArP, aril percentage (%); TSS, total soluble solids (°Brix); Acid, acidity (%); AA, ascorbic acid (mg/100 g); TS, total sugars (%); ArPt, aril protein (%); ArTP, aril total phenols (GAE mg g−1); ArFlav, aril flavonoids (CE mg g−1); TAA, aril total antioxidant activity (AAE mg g−1); ArK, aril potassium (mg g−1); ArP.1, aril phosphorus (mg g−1); ArCa, aril calcium (mg g−1); LTP, leaf total phenols (GAE mg g−1); LFlav leaf flavonoids (CE mg g−1); LTAA, and leaf total antioxidant activity (AAE mg g−1).
Correlation analysis
The Pearson’s bivariate correlations and the associated p-values between the measured traits are shown in Supplementary 2 and Fig. 3. Pod physical traits of P. dulce showed significant positive correlations with each other; PWt mostly exhibited strong positive correlations with ArWt (r = 0.950, p = 0.000), ArW (r = 0.782, p = 0.000), PW (r = 0.699, p = 0.000), ArL (r = 0.647, p = 0.000) and ArP (r = 0.491, p = 0.000). Similarly, PWt had significant negative correlations with PlP (r = −0.384, p = 0.001), ArTP (r = −0.359, p = 0.003), ArPt (r = −0.328, p = 0.007), ArTAA (r = −0.282 p = 0.022), and SP (r = −0.535, p = 0.000). The PW had strong positive correlations with ArL (r = 0.736, p = 0.000), ArWt (r = 0.734, p = 0.071), ArW (r = 0.677, p = 0.000), ArP (r = 0.634, p = 0.000), and strong negative correlations with SP (r = −0.712, p = 0.000), PlP (r = −0.475, p = 0.000) and ArTAA (r = −331, p = 0.000). The PlWt was correlated positively with PlP (r = 0.604, p = 0.000), SWt (r = 0.530, p = 0.000), NSP (r = 0.500, p = 0.000), and inversely with ArP (r = −0.437, p = 0.000) (Fig. 3). The PlP and SP were strongly positively correlated (r = 0.626, p = 0.000), and had inverse relationships with edible components, such as ArWt, ArL and ArW (p = 0.000). However, both of them had positive correlations with most of the bioactive components of aril (ArFlav and ArTAA) and leaves (LFlav and LTAA). The SWt showed strong positive correlations with NSP, SP, SL, and SW, and a strong negative correlation with ArP (r = −0.488, p = 0.000). The SP had significant negative correlations with fruit aril physical, including ArP, ArWt, ArW, and ArL (p = 0.000). The ArWt had strong positive correlations with ArP, ArL, and ArW (p = 0.000), and moderate negative correlations with ArFlav, ArTAA, and ArTP. The TSS showed remarkably strong positive correlations with TS (r = 0.963, p = 0.000), and AA (r = 0.630, p = 0.000). Likewise, ArTP had strong positive correlations with ArTAA (r = 0.831, p = 0.000), ArFlav (r = 0.632, p = 0.000), and ArK (r = 0.656, p = 0.000), and ArTAA exhibited strong positive correlations with LTP (r = 0.611, p = 0.000), and ArK (r = 0.574, p = 0.000). There were strong positive correlations between AA and TS (r = 0.613, p = 0.000), and ArFlav and ArTAA (r = 0.738, p = 0.000) (Supplementary 2; Fig. 3).
Correlation plot showing Pearson’s bivariate correlations between the measured traits in Pithecellobium dulce genotypes. Ellipse size and color reflect the strength and direction (positive or negative) of the correlation. Individual cells marked with a cross (X) denote non-significant correlations. For abbreviations, see Fig. 2.
Hierarchical cluster analysis
The hierarchical cluster analysis using Ward’s linkage method grouped the P. dulce genotypes into two broad clusters (Fig. 4). The first cluster (cluster I) was further divided into subclusters IA and IB, both comprising five genotypes each. The sub-cluster IA had three white (CHESM-12, CHESM-26, and CHESM-29) and two red genotypes (CHESM-27 and CHESM-30). Similarly, sub-cluster IB had three red (CHESM-32, CHESM-33, and CHESM-34), and two white genotypes (CHESM-2 and CHESM-22). The Cluster II was also divided into sub-clusters IIA and IIB. While IIA consisted exclusively of 7 white genotypes (CHESM-10, CHESM-6, CHESM-24, CHESM-5, CHESM-4, CHESM-3, and CHESM-7), cluster IIB comprised two red aril (CHESM-28 and CHESM-31) and three white aril genotypes (CHESM-17, CHESM-20, and CHESM-1). A perusal of the heat map also revealed that genotypes in Cluster I were mostly low in pod width (PW) and aril-related attributes, such as ArP, ArL, ArW, and ArWt. Likewise, the genotypes in Cluster II generally exhibited low values of seed-related (e.g., SWt, SP, and NSP) and leaf bioactive parameters. The genotypes, such as CHESM-30 (NSP and PIP), CHESM-29 (SP), CHESM-33 (SL and SWt), CHESM-6 (ArL, ArW, ArWt, PWt, ArP, and PW), and CHESM-24 (Acid and PlWt) had positive value(s) of certain traits, suggesting that these traits distinguished these genotypes from others and may be utilized in genetic studies and genetic improvement programs. Based on the hierarchical clustering heatmap, three white aril types (CHESM-4, CHESM-6, and CHESM-20) and two red aril types (CHESM-27, CHESM-31, and CHESM-33) were adjudged to be promising in terms of desirable traits (Fig. 4).
Hierarchical clustering heat map showing the clustering of genotypes and traits of Pithecellobium dulce genotypes based on Ward’s linkage method. Data were scaled before clustering, and the values were normalized for each trait by Z-Fisher transformation. Yellow and blue represent high and low trait values, respectively. For abbreviations, see Fig. 2.
Discussion
Although Manila tamarind could be a promising alternative crop for enhanced food production from marginal lands, there have been no systematic attempts to properly assess its genetic resources for the development of improved cultivars. Because Manila tamarind continues to be grown in certain niche tribal and marginalized areas as a subsistence crop16, adequate emphasis on its genetic improvement is a prerequisite for promoting its commercial cultivation. An improved understanding of the fruit’s physicochemical, mineral, and biochemical characteristics is crucial for developing Manila tamarind cultivars that are appropriate for various end uses38. There is a growing worldwide interest in non-conventional plant foods to meet the daily needs for vital nutrients and health-promoting bioactive chemicals. The global food industry is also increasingly exploring locally adapted and underutilized natural products that contain a blend of vital nutrients and bioactive compounds74. To our knowledge, there is a lack of comprehensive understanding regarding the genetic diversity for fruit and leaf nutritional and medicinal properties of Manila tamarind; our study seeks to address this knowledge gap5.
In this study, we characterized 15 white and 7 red aril P. dulce accessions using pod physical attributes (13), aril chemical composition (6), minerals (3), bioactive compounds (3), and leaf bioactive compounds (3) to assess the extent of genotypic variability and to identify the promising genotypes. Our findings demonstrated highly significant (p< 0.001) genotypic differences for almost all the traits examined, suggesting the scope for further selection. Hitherto, our understanding of the fruit quality attributes in Manila tamarind comes largely from the investigations on a few non-descript landraces from countries, such as the Philippines18, Thailand24, Mexico50, Pakistan23, and India6,16,27. Interestingly, one of the major shortcomings in the majority of these studies is that they examined nutritional and bioactive compound profiles exclusively in fruit50,74, or bark/leaf samples22,27. They are thus not comprehensive because only a small number of genotypes were screened, overlooking several desirable characteristics, which makes it challenging to draw reasonable conclusions. Our study addresses these gaps, at least partly, by investigating horticultural, nutritional, and antioxidant traits in fruits of 22 P. dulce accessions, and establishing their relationships with leaf bioactive compounds. The majority of the economically significant attributes in our study showed a high degree of variation, indicating ample scope for the selection of elite genotypes16.
Higher fruit weight and aril yield are two crucial factors in selecting the superior cultivars of arillate fruits39,59,62. The weight of the fruit and arils may have varied across the Manila tamarind accessions due to the varying translocation of photosynthates from the leaves to developing pods and seeds74which is also influenced by the genetic makeup of the accessions31,41,71,72and different growing environments2,68. In our case, considerable variability was recorded for PWt (10.22–32.92 g), PW (12.24–19.65 mm), and PlWt (1.21–5.12 g). Although such variability has not previously been documented in P. dulce, similar results have been reported in tamarind26,34,55. In contrast to the findings of Narayan46 for a lower pod width (9.33–13.60 mm), our results suggest a greater variability in this attribute. In comparison with white aril genotypes (1.66–2.62 g), we observed that red aril genotypes showed comparatively more diversity in seed weight (1.19–3.40 g), suggesting that red aril types may provide greater possibilities for the selection for lower seed weight. Pio-Leon et al.50 also reported lower seed weight in ‘white’ and higher seed weight in ‘red’ aril P. dulce genotypes. Our results for NSP are slightly higher than those reported in Goyal et al.16, and Narayan46. Overall, the results of this study for seed physical properties revealed rich genetic diversity, and agree with the previous findings in P. dulce16,50and tamarind26,34,55. We recorded higher ArWt in white aril genotypes while it was lower in red aril genotypes. The aril physical properties in our study are more variable than values previously reported in P. dulce by Pio-Leon et al.50.
One of the major contributions of the present study was that we examined genotypic variations in biochemical attributes, such as TSS, acidity, TSS: acidity ratio, total sugars, ascorbic acid, and protein contents; these factors significantly improve the nutritional value and eating quality of Manila tamarind arils. Such variations have also been reported in other fruit crops34,38,77. The highly variable values of TSS (17.33–26.46 °Brix), acidity (0.54–1.07%), TA (21.24–36.33), AA (82.54–138.49 mg 100 g−1), TS (12.45–18.81%), and ArPt (3.15–6.32%) recorded by us on the whole meet fresh consumption and aril processing standards9,24,49,53,74. Our results agree with the findings of Pio-Leon et al.50, we also found that white aril genotypes had greater levels of ascorbic acid; nevertheless, in contrast to them, we found that white aril types had higher TSS whereas red aril types had higher ArPt and TS. Such differences are reasonable on account of a comparatively large number of P. dulce accessions assessed by us. Because their high sugar content provides a quick energy boost, the consumption of Manila tamarind arils is considered an alternate food source for the marginalized people in the semi-arid tropics. Similarly, in addition to antioxidant properties, higher ascorbic acid levels can also alleviate vitamin C deficiencies9,70.
P. dulce accessions varied appreciably with each other in aril mineral content and bioactive compounds. The variability in ArK (220.44–334.33 mg 100 g−1 FW), ArP (21.63–62.34 mg 100 g−1 FW), and ArCa (14.06–39.12 mg 100 g−1 FW) observed in this study are supported by the findings of Pio-Leon et al.50 in Mexican P. dulce and Yadav et al.76 in Indian P. dulce. It is also reported that red aril types had higher ArCa and ArK contents than white aril types50,53. The range for ArTP content (2.13–11.18 GAE mg g−1FW) in our study conforms the findings of Kubola et al.24 in Thai P. dulce. However, Recuenco et al.54 recorded low ArTP content in Philippines P. dulce accessions whereas Rao et al.53 recorded a fairly high ArTP in white types; supporting our findings. Our results for total phenols are comparable to the values reported for Mexican P. dulce accession50,74, but higher than those reported for Thai Manila tamarind57. The range for ArFlav content (0.12–0.93 CE mg g−1FW) in our study is consistent with the earlier findings10,50,74. We recorded a high variability for ArTAA (4.11–27.09 AAE mg g−1), which has previously been substantiated in P. dulce from other countries, including Thailand24, India6,51, and the Philippines54. Leaf nutraceutical parameters, including LTP (7.25–16.95 GAE mg g−1FW), LFlav (0.34–1.21 CE mg g−1FW), and LTAA (9.08–30.54 AAE mg g−1) also varied remarkably among Manila tamarind genotypes in the present study. In general, leaves were found to have more bioactive compounds than arils. Likewise, red aril types were observed to contain more bioactive compounds in both leaves and arils when compared with white aril accessions. It is known that plants belonging to the Fabaceae family are frequently rich in secondary metabolites, such as tannins, flavonoids, and other phenolic compounds11. Several factors, including genetic, agronomic, and environmental aspects, can alter the nutraceutical levels in the red and white aril varieties38,74.
Highly significant correlations between PWt on one hand and ArW, PW, ArL, and PlWt on the other indicate that increases in pod weight covary with the increases in other pod components26. The inverse relationships of PWt with PlP and SP suggest that selection for a higher pod weight can result in significant reductions in non-edible components of the pod21,34. We observed significant negative correlations of SP with aril physical attributes, such as ArP and ArWt. Kanupriya et al.21reported that the seed percent had significant negative effects on pulp mass and pulp percentage in tamarind and that this could be useful in selecting the genotypes with a higher pulp weight. The ArWt had strong positive correlations with ArP, ArL, and ArW, and negative correlations with ArTP. A negative relationship between pulp weight and tannin content has previously been demonstrated in tamarind34 and may be useful in selecting less acrid and more palatable P. dulce genotypes. TSS showed strong positive correlations with TS, AA, ArTP, LTP, and ArTAA, substantiated by previous reports in red-pulped guava38and tamarind34. In our study, ArTP showed strong positive correlations with ArTAA, ArFlav, ArK, and LTP which is supported by the findings of Nagmoti et al.44. They demonstrated the better antioxidant and free radical scavenging potential of P. dulce seeds extract powder via an in-vitro assay compared with standard antioxidants, and attributed high antioxidant activity in P. dulce to its high phenolic content. ArTAA exhibited strong positive correlations with LTP, ArK, and ArFlav. Such positive correlations between various phenolic and antioxidant compounds have previously been demonstrated22,57. These results indicate that the aril and leaf extracts of this plant are an important source of natural antioxidants22,73. Principal Component Analysis (PCA) and cluster analysis highlighted significant variations among P. dulce genotypes. The first three Principal Components accounted for over half of the cumulative variance in data suggesting that PCA efficiently reduced the dimensionality and summarized the major patterns in data. These results are largely agreed with the previous reports on tamarind21,26and guava38. Hierarchical cluster analysis revealed both the similarities and differences among the Manila tamarind genotypes in terms of various physical, biochemical, and bioactive compound attributes26,45. The identification of specific traits deriving clustering can inform breeding strategies aimed at improving crop performance and sustainability15.
Conclusions
Our findings demonstrated a high degree of diversity among 22 P. dulce accessions for several traits of interest. Our results assume significance because these 22 accessions represent an unexplored gene pool of P. dulce. The genetic divergence for pod physical and chemical quality traits along with leaf and aril bioactive components is expected to assist in selecting the suitable genotypes for various end uses. Significant variations were observed across all accessions for pod quality and phytochemical attributes. Among the white aril genotypes, CHESM-6 exhibited superior pod physical characteristics, including a higher pod weight (32.92 g), aril weight (28.45 g), and aril percentage (86.31%). However, among red aril genotypes, CHESM-27 recorded the higher pod weight (22.21 g), aril weight (15.72 g), aril protein (6.32%), and total antioxidant activity (27.09 AAE mg g−1); CHESM-28 recorded the maximum TSS (26.46 °Brix) and total sugars (18.81%), while CHESM-31 showed the higher aril percentage (81.04%). Bioactive compounds varied significantly, with strong correlations between key traits, suggesting these genotypes as promising candidates for both nutritional and bioactive value. Some promising accessions identified (CHESM-4, CHESM-6, CHESM-24, CHESM-27, CHESM-31, CHESM-33) these genotypes stand out as excellent candidates for future breeding programs targeting both yield and quality improvements. A fairly high variability in ascorbic acid, minerals, and antioxidant compounds in P. dulce accessions also indicates their suitability for fresh consumption, and for use in processing industries.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
AOAC. Official Method of Analysis 18th edn (Association of Officiating Analytical Chemists, 2005).
Bender, J. Impact of environmental factors on the nutrient content of fruits. Hortic. Sci. 50 (6), 872–882 (2015).
Berwal, M. K., Chugh, L. K., Goyal, P. & Kumar, R. Total antioxidant potential of pearl millet genotypes: Inbreds and designated b-lines. Indian J. Agric. Chem. 29, 201–204 (2016).
Berwal, M. K. et al. Calligonum polygonoides L. as novel source of bioactive compounds in hot arid regions: evaluation of phyto-chemical composition and antioxidant activity. Plants 10, 1156. https://doi.org/10.3390/plants10061156 (2021).
Bhargava, R., Shukla, A. K., Chuahan, N., Vashishtha, B. B. & Dhandar, D. G. Impact of hybridity on flavonoid spectrum of ber (Ziziphus mauritiana Lamk). Environ. Exp. Bot. 53, 135–138. https://doi.org/10.1016/j.envexpbot.2004.03.008 (2005).
Bhati, D. & Jain, S. Nutrition potential of uncultivated fruits grown in Udaipur district of Rajasthan. Bioscan 11 (1), 15–18 (2016).
Brewbaker, J. L. Pithecellobium dulce - sweet and thorny. NFT Highlights 92 – 01. 2, 5 (1992).
Ceccon, E., Sanchez, I. & Powers, J. S. Biological potential of four indigenous tree species from seasonally dry tropical forest for soil restoration. Agroforest Syst. 89, 455–467. https://doi.org/10.1007/s10457-014-9782-6 (2015).
Chapparo-Santiago, A., Osuna-Fernandez, H. R., Aguillon-Arenas, J. & Osuna-Fernandez, A. M. Nutritional composition of Pithecellobium dulce, Guamuchil aril. Pakistan J. Nutr. 14 (9), 611–613 (2015).
Cheema, J., Yadav, K., Sharma, N., Saini, I. & Aggarwal, A. Nutritional quality characteristics of different wild and underutilized fruits of Terai Region, Uttarakhand (India). Int. J. Fruit Sci. 17, 72–81. https://doi.org/10.1080/15538362.2016.1160271 (2017).
Conti, M. V. et al. Bioactive compounds in legumes: implications for sustainable nutrition and health in the elderly population. Trends Food Sci. Technol. 117, 139–147. https://doi.org/10.1016/j.tifs.2021.02.072 (2021).
Crane, E., Walker, P. & Day, R. Directory of Important World Honey Sourcespp. 384 (International Bee Research Organization (IBRA), 1984).
Dhanisha, S., Drishya, S., Mony, R. & Guruvayoorappan, C. Polyphenolic rich fraction of Pithecellobium dulce attenuates methotrexate induced oxidative stress and associated tissue injury by regulating the TNF α, IL 1β and IL 6 pro inflammatory cytokines. Int. J. Funct. Nutr. 2 (3), 1–17. https://doi.org/10.3892/ijfn.2021.17 (2021).
Elhewehy, A. A., Mohsen, E., El-Fishawy, A. M. & Fayed, M. A. A. Traditional, phytochemical, nutritional and biological importance of Pithecellobium dulce. (Roxib) Benth Yuzuncu Yil Univ. J. Agril Sci. 34 (2), 354–380. https://doi.org/10.29133/yyutbd.1329407 (2024).
Frankham, R., Ballou, J. D. & Briscoe, D. A. Introduction to Conservation Genetics1–618 (Cambridge University Press, 2010).
Goyal, P., Jain, R., Kachhwaha, S. & Kothari, S. L. Assessment of genetic diversity in Pithecellobium dulce (Roxb.) Benth. Germplasm using RAPD and ISSR markers. Trees 29, 637–653. https://doi.org/10.1007/s00468-014-1141-8 (2015).
Heber, D. Vegetables, fruits and phytoestrogens in the prevention of diseases. J. Postgrad. Med. 50, 145–149 (2004).
Hendro Sunarjono, H. & Coronel, R. E. Pithecellobium dulce. In: (eds Verheij, E. W. M. & Coronel, R. E.) Plant Resources of South-East Asia No. 2. Edible Fruits and Nuts. Wageningen, Netherlands: Pudoc, 256–257 (1991).
Hughes, C. E. & Styles, B. T. Exploration and seed collection of multiple-purpose dry zone trees in Central America. Int. Tree Crops J. 3 (1), 1–31 (1984).
Julkowska, M. M. et al. MVApp-Multivariate analysis application for streamlined data analysis and curation. Plant. Physiol. 180, 1261–1276 (2019).
Kanupriya, C. et al. Phenotypic diversity in Tamarindus indica L. sourced from different provenances in India. Agrofores Syst. 98, 477–490. https://doi.org/10.1007/s10457-023-00925-0 (2023).
Katekhaye, S. D. & Kale, M. S. Antioxidant and free radical scavenging activity of Pithecellobium dulce (Roxb.) Benth wood bark and leaves. Free Rad Antiox. 2 (3), 47–57 (2012).
Khanzada, S. K., Khanzada, A. K., Shaikh, W. & Ali, S. A. Phytochemical studies on Pithecellobium dulce Benth. A medicinal plant of Sindh. Pakistan Pak J. Bot. 45 (2), 557–561 (2013).
Kubola, J., Siriamornpun, S. & Meeso, N. Phytochemicals, vitamin C and sugar content of Thai wild fruits. Food Chem. 126, 972–981. https://doi.org/10.1016/j.foodchem.2010.11.104 (2011).
Kulkarni, K. V. & Jamakhandi, V. R. Medicinal uses of Pithecellobium dulce and its health benefits. J. Pharmacogn Phytochem. 7 (2), 700–704 (2018).
Kumar, R., Dalve, P. D., Palande, A. L. & Choudhary, S. M. Multivariate diversity analysis of tamarind (Tamarindus indica L.) genotypes under the arid condition of western Maharashtra. Indian J. Ecol. 50 (3), 700–705 (2023).
Kumari, S. Evaluation of phytochemical analysis and antioxidant and antifungal activity of Pithecellobium dulce leaves’ extract. Asian J. Pharm. Clin. Res. 10 (1), 370–375. https://doi.org/10.22159/ajpcr.2017.v10i1.15576 (2017).
Lal, N. & Nath, V. Sweet tamarind [Pithecellobium dulce (Roxb.) Benth.]. In, (ed Gosh, S. N.) Minor Fruits: Nutraceutical Importance and Cultivation. Narendra Publ. House, New Delhi, 901–912. (2017).
Leon, J. Central American and west Indian species of Inga (Leguminosae). Ann. Missouri Bot. Gard. 53, 265–359 (1966).
Lopez-Angulo, G. et al. Anthocyanins of Pithecellobium dulce (Roxb.) Benth. Fruit associated with high antioxidant and α-Glucosidase inhibitory activities. Plant. Foods Hum. Nutr. 73, 308–313. https://doi.org/10.1007/s11130-018-0693-y (2018).
Lopez-Bucio, J., Cruz-Ramırez, A. & Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant. Biol. 6 (3), 280–287. https://doi.org/10.1016/S1369-5266(03)00035-9 (2003).
Malik, S. K., Chaudhury, R., Dhariwal, O. P. & Bhandari, D. C. Pithecellobium dulce (Manila Tamarind). Genetic Resources of Tropical Underutilized Fruits in India, p.114–119 (NBPGR, 2010).
Manna, P., Bhattacharyya, S., Das, J., Ghosh, J. & Sil, P. C. Phytomedicinal role of Pithecellobium dulce against CCl(4)-mediated hepatic oxidative impairments and necrotic cell death. Evid. Based Complement. Altern. Med. 2011, 1–17. https://doi.org/10.1093/ecam/neq065 (2011).
Mayavel, A., Amaravel, M., Bagathsingh, C., Krishnan, G. R. & Nagarajan, B. Exploring morphobiochemical diversity in tamarind (Tamarindus indica L) for advanced breeding approaches. Legume Res. https://doi.org/10.18805/LR-5295 (2024).
Meena, V. S. et al. Underutilized fruit crops of Indian arid and semi-arid regions: importance, conservation and utilization strategies. Horticulturae 8 https://doi.org/10.3390/horticulturae8020171 (2022).
Megala, J. & Geetha, A. Free radical-scavenging and H+, K+-ATPase inhibition activities of Pithecellobium dulce. Food Chem. 121 (4), 1120–1128. https://doi.org/10.1016/j.foodchem.2010.01.059 (2010).
Mishra, D. S. Enhancing income through value-addition. Indian Hortic. 63 (5), 107–109 (2018).
Mishra, D. S. et al. Phenotypic diversity for fruit quality traits and bioactive compounds in red-fleshed guava: insights from multivariate analyses and machine learning algorithms. S Afr. J. Bot. 149, 591–603. https://doi.org/10.1016/j.sajb.2022.06.043 (2022).
Mishra, D. S., Singh, S. & Saroj, P. L. Evaluation of pomegranate varieties under semi-arid environment of central Gujarat. Indian J. Arid Hort. 2 (1&2), 67–69 (2020).
Mishra, D. S., Singh, S., Singh, A. K. & Yadav, V. Future fruit crops for semi-arid conditions of western India. In, Lakhawat, S.S. (ed.) Compendium of Winter School Exploitation of underutilized fruit crops of arid and semi-arid region held at MPUAT, Udaipur from Oct. 04–24, 2016, pp. 187–192. (2016).
Mishra, D. S. et al. Assessment of genetic diversity in guava. Indian J. Hortic. 75 (3), 362–368 (2018).
Monroy, R. & Colin, H. El Guamuchil Pithecellobium dulce (Roxb.) Benth, Un ejemplo de uso multiple. Madera y Bosques. 10 (1), 35–53 (2004).
Murugesan, S., Lakshmanan, D. K., Arumugam, V. & Alexander, R. A. Nutritional andtherapeutic benefits of medicinal plant Pithecellobium dulce (Fabaceae): a review. J. Appl. Pharm. Sci. 9 (7), 130–139. https://doi.org/10.7324/JAPS.2019.90718 (2019).
Nagmoti, D. M., Khatri, D. K., Juvekar, P. R. & Juvekar, A. R. Antioxidant activity free radical-scavenging potential of Pithecellobium dulce Benth seed extracts. Free Rad Antiox. 2 (2), 37–43. https://doi.org/10.5530/ax.2012.2.2.7 (2012).
Nagar, B. L. & Fageria, M. S. Genetic divergence in lehsua (Cordia myxa Roxb). Indian J. Genet. Plant. Breed. 66 (01), 67–80 (2006).
Narayan, J. P. Exploratory studies on occurrence and potential benefits of high fruit yielding reproductive phenophasic variants of Pithecellobium dulce in augmenting fruit based semi-arid agro forestry systems. Eur. J. Med. Plants. 28 (2), 1–17. https://doi.org/10.9734/EJMP/2019/v28i230132 (2019).
Olivares-Perez, J., Aviles-Nova, F., Albarran-Portillo, B., Castelan-Ortega, O. A. & Rojas-Hernandez, S. Nutritional quality of Pithecellobium dulce and Acacia cochliacantha fruits, and its evaluation in goats. Livesto Sci. 154 (1), 74–81 (2013).
Parrota, J. .Pithecellobium dulce (Roxb.) Benth. Guamuchil, Madras Thorn. SO-ITF-SM-4, New Orleans, LA; U.S. Department of Agriculture, Forest Service, Southern Forest Experiment Station, 5 (1991).
Pandya, J. B. & Mehta, S. K. Biochemical analysis of Pithecellobium dulce (Roxb.) Bth. Fruit during its successive stages of development. Int. J. Res. Biosci. Agric. Technol. 17, 529–534 (2021).
Pio-Leon, J. F., Pazdiaz-Camacho, S., Montes-Avila, J., Lopez-Angulo, G. & Delgado-Vargas, F. Nutritional and nutraceutical characteristics of white and red Pithecellobium dulce (Roxb.) Benth fruits. Fruits 68 (5), 397–407 (2013).
Preethi, S. & Saral, M. A. GC-MS analysis of microwave assisted ethanolic extract of Pithecellobium dulce. Malaya J. Biosci. 1, 242–247 (2014).
Rao, B. G., Samyuktha, P., Ramadevi, D. & Battu, H. Review of literature: phyto pharmacological studies on Pithecellobium dulce. J. Glob Trends Pharm. Sci. 9, 4797–4807 (2018).
Rao, G. N., Nagender, A., Satyanarayana, A. & Rao, D. G. Preparation, chemical composition and storage studies of quamachil (Pithecellobium dulce L.) aril powder. J. Food Sci. Technol. 48 (1), 90–95. https://doi.org/10.1007/s13197-010-0135-9 (2011).
Recuenco, M. C., Lacsamana, M. S., Hurtada, W. A. & Sabularse, V. C. Total phenolic and total flavonoid contents of selected fruits in the Philippines. Philippine J. Sci. 145 (3), 275–281 (2016).
Reddy, A. G. K. et al. Performance of tamarind (Tamarindus indica L.) accessions under Dryland conditions. Legume Res. 46 (9), 1179–1183. https://doi.org/10.18805/LR-4810 (2023).
Roselin, C. & Parameshwari, S. A systematic review on the materialistic use of Pithecellobium dulce in food formulations. Mater. Today Proc. 66 (3), 996–1001. https://doi.org/10.1016/j.matpr.2022.04.779 (2020).
Samee, W. et al. Correlation analysis between total acid, total phenolic and ascorbic acid contents in fruit extracts and their antioxidant activities. Thai Pharm. Health Sci. J. 1, 196–203 (2006).
Seetaloo, A. D., Aumeeruddy, M. Z., Kannan, R., Mahomoodally, R. R. & M.F Potential of traditionally consumed medicinal herbs, spices, and food plants to inhibit key digestive enzymes geared towards diabetes mellitus management- A systematic review. S Afr. J. Bot. 120, 3–2. https://doi.org/10.1016/j.sajb.2018.05.015 (2019).
Singh, A., Mann, A., Kumar, R. & Yadav, R. K. Delineating eco-physiological traits linked to salt tolerance and fruit yield in pomegranate. Sci. Hortic. 322, 112422. https://doi.org/10.1016/j.scienta.2023.112422 (2023).
Singh, A., Mishra, D. S., Kumar, R. & Kumar, P. Physico-chemical changes in litchi cultivar rose scented during fruit development and maturation. Ind. J. Hort. 70 (3), 328–332 (2013).
Singh, A. K., Mishra, D. S. & Sharma, B. D. Seventy five years of research and development in arid and semi-arid fruit crops. Int. J. Innov. Hort. 11 (2), 214–227. https://doi.org/10.5958/2582-2527.2022.00019.7 (2022).
Singh, A. K., Mishra, D. S., Yadav, L. P. & Rane, J. Alternate fruit crops for sustainable food system. Indian Farming. 73 (6), 66–69 (2023).
Singh, S., Saroj, P. L., Mishra, D. S. & Singh, A. K. Underutilized Fruit Crops: Crop Improvement and agro-techniquespp.1–306 (KAAV Publication, 2019).
Singh, A. K. et al. Cultivation of underutilized fruit crops in hot semi-arid regions: developments and challenges-a review. Curr. Hort. 8 (1), 12–23 (2020).
Sneha, D., Prashanth, S., Kaveti, V. S. & Boggula, N. Systematic Review of Pithecellobium dulce (Roxb.) Benth.: A Traditional Medicinal Herb. J. Innov. Dev. Pharm. Tech. Sci. 3(5), 1–9. (2020).
Srinivas, G., Geeta, H. P., Shashikumar, J. N. & Champawat A review on Pithecellobium dulce: a potential medicinal tree. Int. J. Chem. Stud. 6 (2), 540–544 (2018).
Subbiah, A. et al. Studies on the influence age of rootstocks and season on grafting success in Manila tamarind (Pithecellobium dulce Roxb). Legume Res. 47 (1), 78–81. https://doi.org/10.18805/LR-5244 (2024).
Thepbandit, W. & Athinuwat, D. Rhizosphere microorganisms supply availability of soil nutrients and induce plant defense. Microorganisms 12 (3), 558. https://doi.org/10.3390/microorganisms12030558 (2024).
Tiwari, A. K., Mishra, D. S., Kumar, S. & Champathi Gunathilake, D. M. C. Exploitation of climate resilient minor tropical fruit crops for nutritional and livelihood security in Fiji Islands. Int. J. Curr. Microbiol. App Sci. 7 (11), 2135–2142 (2018).
Traver, M. & Stevens, J. Vitamin C and E beneficial effects from a mechanistic perspective. Free Radical Biol. Med. 51, 1000–1013 (2011).
Tripathi, P. C. et al. Phenotypic Diversity and Genetic Characterization of Cordia myxa L. using multivariate analysis. Flora. 2025,152673. (2025). https://doi.org/10.1016/j.flora.2025.152673
Tunc, Y. et al. Determination of genetic diversity in persimmon accessions using morphological and inter simple sequence repeat markers. Sci. Rep. 15, 2297. https://doi.org/10.1038/s41598-025-86101-z (2025).
Vargas-Madriz, A. F. et al. Phenolic profile and antioxidant capacity of Pithecellobium dulce (Roxb) Benth: a review. J. Food Sci. Technol. 57 (12), 4316–4336. https://doi.org/10.1007/s13197-020-04453-y (2020).
Wall-Medrano, A. et al. Ripening of Pithecellobium dulce (Roxb.) Benth. [Guamuchil] fruit: physicochemical, chemical and antioxidant changes. Plant. Foods Hum. Nutr. 71, 396–401. https://doi.org/10.1007/s11130-016-0575-0 (2016).
Wetchakul, P., Net-Anong, S., Goon, J. A. & Sanpinit, S. Anti-oxidative stress and gastroprotective effect of Tri-tharn-thip tea against ethanol-induced gastric ulcer in rats. S Afr. J. Bot. 170, 130–136. https://doi.org/10.1016/j.sajb.2024.05.027 (2024).
Yadav, A. et al. Manila tamarind: a multipurpose plant suitable for dryland areas. Agric. Environ. E- NewsLett. 2 (6), 92–97 (2021).
Yousaf, A. A. et al. Physico-chemical and nutraceutical characterization of selected indigenous guava (Psidium guajava L.) cultivars. Food Sci. Technol. (Campinas). 41 (1), 47–58. https://doi.org/10.1590/fst.35319 (2021).
Hussain, A. et al. Determination of total phenolic, flavonoid, carotenoid, and mineral contents in peel, flesh, and seeds of pumpkin (Cucurbita maxima). J. Food Process. Preserv. 45 (6), e15542. https://doi.org/10.1111/jfpp.15542 (2021).
Hussain, A. et al. Evaluation of leaves, Flowers, and seeds of coriander (Coriandrum sativum L.) through microwave drying and Ultrasonic-assisted extraction, for biologically active components. J. Food Process. Preserv. 2024(1), 2378604. https://doi.org/10.1155/2024/2378604 (2024).
Coşkun, O. F. & Gülşen, O. Determination of markers associated with important agronomic traits of watermelon (Citrullus lanatus L). J. Agricultural Sci. Technol. 26 (6), 1359–1371. https://doi.org/10.22034/JAST.26.6.1359 (2024).
Coşkun, Ö. F., Toprak, S. & Mavi, K. Some seed properties and molecular analysis with inter-primary binding site (iPBS) retrotranposons markers of edible-seeded watermelon genotypes. Genet. Resour. Crop Evol. 71, 3151–3162. https://doi.org/10.1007/s10722-023-01845-9 (2024).
Acknowledgements
We are indebted to Director, ICAR- Central Institute for Arid Horticulture (CIAH), Bikaner, India for providing the all types of logistic support to conduct the experiment. Mr K.V. Parmar, T.O. is admired for his technical support in managing the experiment and recording the observations.
Funding
None.
Author information
Authors and Affiliations
Contributions
DSM conceived the idea, conducted the experiments, and recorded the data. MKB and VVA helped in analyzing fruit biochemical, bioactive compounds, and mineral contents. AMS performed the statistical analyses. DSM, AMS, and PR wrote the manuscript. AMS, VY, DKS, JR, YT, and AK edited the manuscript before submission. All authors read and approved the final draft.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mishra, D.S., Singh, A., Rao, V.V.A. et al. Comparative evaluation of red and white aril genotypes of Manila tamarind for fruit physicochemical and bioactive attributes. Sci Rep 15, 6865 (2025). https://doi.org/10.1038/s41598-025-90683-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-90683-z
Keywords
This article is cited by
-
Characterization and evaluation of tamarind (Tamarindus indica L.) germplasm: implications for tree improvement strategies
BMC Plant Biology (2025)
-
Taxonomic significance of morphological and molecular variation in Egyptian Malvaceae species
BMC Plant Biology (2025)
-
Determination of gene association analysis of some leaf traits in mulberries using simple sequence repeat (SSR) markers
Genetic Resources and Crop Evolution (2025)