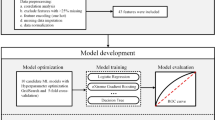
Abstract
Early diagnosis of Kawasaki disease (KD) allows timely treatment to be initiated, thereby preventing coronary artery aneurysms in children. However, it is challenging due to the subjective nature of the diagnostic criteria. This study aims to develop a machine learning prediction model using routine blood tests to distinguish children with KD from other febrile illnesses in Chinese children within the first five days of fever onset. The retrospective cross-sectional data for this study was collected from the records of Guangzhou Women and Children’s Medical Center, spanning January 1, 2020, to April 30, 2024. A retrospective analysis was performed using three machine learning models and five ensemble models based on this dataset. This study included 1,089 children with KD (mean age 32.8 ± 27.0 months; 34.5% female) and a control group of 81,697 children without KD (mean age 45.3 ± 33.6 months; 42.8% female). The supervised method, Xtreme Gradient Boosting (XGBoost), was applied. It was tested without feature selection, achieved an area under the ROC curve (AUC) of 0.9999, sensitivity of 0.9982, specificity of 0.9975, F1 score of 0.9979, accuracy of 0.9979, positive predictive value (PPV) of 0.9975, and negative predictive value (NPV) of 0.9982. The SHapley Additive exPlanations (SHAP) summary plot identified the top five significant features, which were the percentage of eosinophils (EO%), hematocrit (HCT), platelet crit (PCT), gender, and absolute basophil count (BA#). This study demonstrates that the application of the machine learning model, XGBoost, on routine blood test results can predict KD.
Similar content being viewed by others
Introduction
Kawasaki disease (KD) is an acute systemic pediatric vasculitis characterized by fever, mainly affecting medium-sized arteries in children aged 6 months to 5 years, with an unknown etiology1. Without treatment, 25% of pediatric patients with KD are at risk of developing coronary artery aneurysms, which can result in myocardial infarction and death2. Treating KD with intravenous immunoglobulin (IVIG) within the first 10 days reduces the incidence of coronary artery aneurysms from 25 to 3% and lowers mortality3. Moreover, treating children with KD using IVIG on or before the fifth day of fever can lead to improved coronary outcomes and shorten the duration of clinical symptoms4,5. Therefore, early detection of KD is crucial for allowing timely IVIG therapy, avoiding possible unnecessary antibiotics use, and reducing severe complications, mortality, as well as burden to the healthcare system.
The clinical diagnostic criteria for KD primarily rely on observable clinical manifestations6. The key criterion is a persistent fever exceeding five days, alongside at least four out of five specific physical symptoms7. However, this requirement poses a challenge, as it is difficult to administer IVIG within the first five days of fever onset without a concrete diagnosis. Additionally, some cases of KD do not present with classic symptoms, and signs that are overlooked may disappear quickly. Therefore, it is crucial to develop an early diagnostic tool that can quantitatively assess and predict KD, providing insights into its onset and underlying mechanisms.
The currently proposed rapid methods for KD diagnosis using machine learning are not yet being implemented into routine clinical practice. A recent study developed KD-CNN, a deep convolutional neural network that diagnoses KD by analyzing clinical photographs with high accuracy8. Tsai et al.. created a machine learning model using the supervised eXtreme Gradient Boosting (XGBoost) to differentiate children with KD from other febrile illnesses in children presenting to emergency departments, demonstrating high sensitivity and specificity based on objective lab tests9. Another study presented a machine learning nomogram that effectively distinguishes KD from sepsis using clinical and biomarker data10. However, conducting extensive tests beyond routine blood tests is uncommon and impractical for febrile children within the first five days in a clinic, emphasizing on fever within this timeframe.
The rapid advancements in artificial intelligence have accelerated the transition of machine learning from academic research to clinical applications11. Machine learning found widespread application within clinical big data frameworks, effectively uncovering nonlinear relationships to assist in clinical diagnosis, treatment, and surveillance12. Its utility in various medical domains, particularly in binary classification tasks, such as in KD, has been extensively explored and has demonstrated promising results8,9,10. Hence, it offers an opportunity to explore and develop a practical tool for rapidly detecting KD using only routine blood tests.
The aim of this study is to develop an algorithmic model to predict KD in febrile Chinese children within five days of symptom onset. The model relies on a small set of routinely collected laboratory parameters that are easily accessible in primary care or emergency room settings. To select key features for the model, we employed three methods: Lasso L1 regularization, t-tests/Chi-square tests, and a no-selection approach. Additionally, SHapley Additive exPlanations (SHAP) analysis was used to identify risk factors associated with biomarkers, offering insights into potential underlying pathophysiological mechanisms.
Materials and methods
Study design
The retrospective cross-sectional data for this study was retrieved from the records of Guangzhou Women and Children’s Medical Center, covering the period from January 1, 2020, to April 30, 2024. Ethical approval for the study was obtained from the Ethics Committee of Guangzhou Women and Children’s Medical Center in July 2024. The need for informed consent was waived by the committee, owing to the retrospective nature of this study as well as the fact that patient contact was not required. All procedures and methods were conducted in compliance with Chinese national legislation, institutional guidelines, and the Declaration of Helsinki.
Participants and data collection
Children under 15 years of age who had routine blood tests within five days of fever onset at the Guangzhou Women and Children’s Medical Center were identified. Both inpatient and outpatient cases were included. Our analysis focused solely on the first routine blood tests performed within five days of the fever’s start, considering only patients with a definitive diagnosis for that fever episode. The exclusion criteria were blood tests taken more than five days after fever onset, cases with unclear or absent diagnoses, and diagnoses unrelated to the fever episode.
Clinical data documented for analysis encompassed gender, age, final diagnosis outcomes, and results from the initial blood tests conducted during the fever episode. Data extraction was conducted by two independent reviewers who retrieved relevant data elements from the hospital systems for all eligible patients. The dataset, provided by the Department of Pediatrics at Guangzhou Women and Children’s Medical Center, included 87,868 cases. Data collected comprised participant age, gender, routine blood test parameters, and final diagnosis. Patients with known malignancies, chronic inflammatory diseases, or other infections that could influence blood test parameters were excluded after a thorough review of their medical histories.
Demographic characteristics
Participants reported sociodemographic information, including age and gender.
Kawasaki disease
Currently, the clinical diagnosis of KD in China follows the 2004 American Heart Association guidelines13. KD is diagnosed based on a fever lasting at least five days and at least four of the following five clinical features: changes in extremities, polymorphous exanthema, bilateral bulbar conjunctival injection without exudates, changes in lips and oral cavity, and cervical lymphadenopathy. Extremity changes include erythema of the palms and soles and edema of the hands and feet during the acute phase, and periungual peeling of the fingers and toes in the subacute phase. Polymorphous exanthema presents as a diffuse maculopapular rash, urticarial rash, erythroderma, or erythema-multiforme-like rash, but excludes vesicular or bullous rashes. Changes in the lips and oral cavity involve erythema, cracked lips, strawberry tongue, and diffuse injection of the oral and pharyngeal mucosa. Cervical lymphadenopathy is characterized by a lymph node diameter greater than 1.5 cm, usually unilateral. To confirm a KD diagnosis, it is crucial to exclude other diseases with similar symptoms, such as scarlet fever, viral infections like measles and adenovirus, Stevens-Johnson syndrome, and toxic shock syndrome.
Routine blood tests
The first routine blood tests obtained within five days of the fever’s onset were used for analysis. They comprised of 25 laboratory parameters, including absolute basophil count (BA#), basophil percentage (BA%), absolute eosinophil count (EO#), eosinophil percentage (EO%), hematocrit (HCT), hemoglobin (HB), absolute lymphocyte count (LYMPH#) and lymphocyte percentage (LYMPH%), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), mean corpuscular volume (MCV), absolute monocyte count (MONO#), monocyte percentage (MONO%), mean platelet volume (MPV), platelet large cell ratio (P-LCR), platelet crit (PCT), platelet distribution width (PDW), platelet count (PLT), red blood cell count (RBC), red cell distribution width (RDW), white blood cell count (WBC), absolute neutrophil count (NEUT#), neutrophil percentage (NEUT%) C-reactive protein (CRP).
Feature selection
Three strategies were used for feature selection: Lasso L1 regularization, t-test/Chi-square test, and no selection. Lasso L1 regularization14 enhances model accuracy by penalizing the absolute size of regression coefficients, effectively shrinking less important coefficients to zero. The penalty coefficient was set to 1.0, based on cross-validation to balance model complexity and prediction accuracy, and implemented this method using Scikit-learn in Python. The t-test or Chi-squared test were used to evaluate the independence between features and the target variable, retaining features with p-values below 0.05 as statistically significant predictors. This method is conducted using the scipy.stats library in Python, helps identify the most relevant predictors. Additionally, models were trained without any feature selection as a baseline to compare the impact of our feature selection methods on model performance, providing a reference point for evaluating their effectiveness.
Machine learning models
Three machine learning algorithms and five ensemble models were selected, all of which are known for their effectiveness in classification tasks. Logistic regression15 is a fundamental linear model used for binary classification tasks due to its simplicity and interpretability. Support vector machine16 excels in high-dimensional and complex settings, providing robust classification performance. Multilayer perceptron17 is a neural network model that effectively identifies complex patterns through its layered structure.
Ensemble models enhance prediction performance by combining multiple models to mitigate overfitting and improve generalization. Random forest18 is esteemed for its high accuracy and ability to mitigate overfitting through ensemble learning, aggregating the predictions of decision trees via a majority vote to enhance predictive strength. LightGBM19 is a highly efficient gradient boosting framework that uses a histogram-based method to bin continuous features, accelerating training speed, optimizing memory usage, and excelling in processing large-scale datasets with remarkable speed and effectiveness. AdaBoost20 prioritizes difficult scenarios, enhancing classification precision by iteratively adjusting weights to improve the model. XGBoost21, an advanced gradient boosting system developed by Chen, iteratively refines models by splitting tree nodes and fitting residuals, demonstrating exceptional scalability and superior performance across diverse applications. CatBoost22, introduced in 2018, is a cutting-edge gradient boosting algorithm known for its exceptional handling of categorical features, reduced training times, and the use of a greedy strategy to pinpoint optimal tree splits, thereby enhancing prediction accuracy.
Statistical analysis
Statistical analyses were conducted using Python (Version 3.11.5) in the Microsoft Visual Studio Code environment. Incomplete data samples were excluded, and categorical variables were encoded into numerical values using Scikit-learn’s LabelEncoder. Descriptive statistics were used to summarize categorical variables (frequencies) and continuous variables (means and standard deviations). Chi-square tests and unpaired t-tests were applied where necessary. All features were normalized using Scikit-learn’s StandardScaler, and imbalanced data were addressed with the RandomOverSampler function.
To explore variable relationships, correlation heatmaps were created. Feature selection was performed using three methods: Lasso L1 regularization, t-test/chi-square test, and no selection. The dataset was split into training (80%) and testing (20%) sets, with 5-fold cross-validation. Eight machine learning models were trained: Logistic Regression, Support Vector Machine, Multilayer Perceptron, Random Forest, LightGBM, AdaBoost, XGBoost, and CatBoost. Model performance was evaluated using metrics such as AUC, sensitivity, specificity, F1 score, accuracy, positive predictive value (PPV), and negative predictive value (NPV).
Log loss versus epoch curves were used to monitor performance and detect overfitting. Precision-recall curves evaluated the balance between precision and recall, especially for imbalanced datasets. Learning curves illustrated model performance on training and validation sets as the sample size increased, helping diagnose underfitting or overfitting. Feature importance was analyzed using SHAP values, which also assessed each feature’s contribution to the models. A two-sided p-value of < 0.05 was considered statistically significant. The Python environment included essential libraries such as pandas 2.1.4, numpy 1.24.3, scikit-learn 1.3.0, scipy 1.11.4, catboost 1.2, lightgbm 4.1.0, seaborn 0.12.2, SHAP 0.42.1, and matplotlib 3.8.0.
Results.
Background and clinical characteristics
The study flowchart illustrating patient identification, inclusion and exclusion is shown in Fig. 1A. The number of patients with missing data in the KD and the non-KD groups is shown in Fig. 1B. Among the 1,169 KD patients, notable missing data were observed for CRP (58, 4.962%) and for MPV, P-LCR, PCT, and PDW (each 24, 2.053%). In contrast, among the 85,530 patients without KD, the highest percentages of missing data were for BA% (862, 1.008%), MPV (2875, 3.361%), P-LCR (2874, 3.360%), PCT (2879, 3.366%), and PDW (2874, 3.360%). Most of the remaining variables had negligible missing data in both groups, often less than 0.01%. The gender distribution amongst the study participants is shown in Fig. 1C. In the KD group (N = 1,089), 714 were male and 376 were female. In the group without KD (N = 81,697), 46,729 were male and 34,968 were female, indicating a higher proportion of males in both groups. The mean age of KD patients was significantly younger at 32.833 months compared to 45.310 months in non-KD patients (p < 0.001; Fig. 2). Significant differences were observed in various blood parameters: KD patients had higher mean values for BA#, BA%, CRP, EO#, EO%, LYMPH#, LYMPH%, MCHC, and PLT, while lower values were noted for HCT, hemoglobin (HGB), MCV, MONO#, MONO%, NEUT#, NEUT%, and RBC (all p-values < 0.001). No significant differences were found in MPV, P-LCR, and PDW (p > 0.05). These findings highlight the distinct clinical and hematological profiles of KD patients.
Feature selection using Lasso L1 regularization
A heatmap of all features was generated to show significant positive or negative associations among features (Fig. 3). The relationship between the inverse of regularization strength and the coefficients for all features in Lasso regularization (L1) is shown in Fig. 4A. These coefficient values were ranked (Fig. 4B), indicating the impact of each feature on the model’s predictions. Positive coefficients, such as for CRP (1.162), HCT (4.422), and PLT (2.377), suggest these features positively influence the outcome. Conversely, negative coefficients such as those for LYMPH% (-3.366), MCH (-4.071), and RBC (-6.013) indicate a negative association. Features such as NEUT# and NEUT% with coefficients of 0.000 and − 4.847 respectively, suggest they have minimal or negative impact. Age in months (0.967) shows a moderate positive influence. These coefficients collectively inform the model’s predictive performance under Lasso regularization and suggest the exclusion of features like NEUT# and PCT.
Model performances using 5-fold cross validation
The performance of the various models using different feature selection strategies was evaluated (Fig. 5). The evaluated metrics include AUC, Sensitivity, Specificity, F1 Score, Accuracy, PPV, and NPV. The metrics for different models using Lasso L1 regularization are shown in Fig. 5A. The XGBoost model achieved the highest AUC of 0.9999 and an accuracy of 0.9978, along with high sensitivity (0.9981) and specificity (0.9976). Similarly, the Multilayer Perceptron, LightGBM, and Random Forest models also demonstrated exceptional performance, with AUC values close to 1.0. In contrast, the logistic regression model, while performing well, had relatively lower metrics compared to the other models.
The metrics for different models using the t-test/Chi-squared test for feature selection are shown in Fig. 5B. The XGBoost model again exhibited high performance, with an AUC of 0.9999 and an accuracy of 0.9978. The Multilayer Perceptron and LightGBM models also maintained strong performance, with only slight variations compared to their Lasso regularization results. Overall, the models demonstrated robustness across different feature selection methods. The metrics for different models without any feature selection are shown in Fig. 5C. The results were consistent with those in Fig. 5A and B. The XGBoost model continued to excel, with an AUC of 0.9999 and an accuracy of 0.9979. The logistic regression model again showed strong but slightly worse performance metrics compared to the more complex models. The metrics of the XGBoost model using Lasso L1 selection, t-test/Chi-square test, and no selection were then compared (Fig. 5D). The performance metrics were nearly identical across all three strategies, with accuracy of 0.9978 (Lasso), 0.9978 (t-test/Chi-squared), and 0.9979 (no selection). This consistency suggests that the XGBoost model is highly effective regardless of the feature selection method used.
Metrics for the three strategies for feature selection of the eight models using a five-fold cross validation. (A) Metrics using Lasso regularization (L1). (B) Metrics using t-test/Chi-squared test. (C) Metrics without feature selection. (D) Metrics for the three feature selection strategies using the XGBoost model.
Performance of the XGBoost model
The performance of the XGBoost model was then evaluated (Fig. 6). The log loss versus epoch curves for both the training and test sets are shown in Fig. 6A, demonstrating the model’s convergence as epochs increase. The precision-recall curve shows the high precision and recall achieved by the model, with an AUC of 1.00 (Fig. 6B). The learning curve shows the model’s accuracy on cross-validation sets increase without starting to decline as the number of training samples increases, indicating strong performance stability and no signs of overfitting (Fig. 6C). SHAP values show the impact of each feature on the model’s output, with features such as EO%, HCT, and PCT significantly influencing the predictions (Fig. 6D).
Performance evaluation of the XGBoost model. (A) Log loss versus epoch curves. These curves illustrate the model’s performance on the training and test sets as the number of epochs increases. The convergence and stabilization of log loss for both sets suggest that the model is neither underfitting nor overfitting and demonstrates good generalization. (B) Precision-Recall curve. This curve evaluates the trade-off between precision (the proportion of true positive predictions among positive predictions) and recall (the proportion of true positives identified out of all actual positives). A higher area under the Precision-Recall curve indicates strong model performance, particularly for imbalanced datasets. (C) Learning curve. This plot shows how model accuracy changes with increasing training data. The decreasing gap between training and cross-validation accuracy as sample size increases indicates that the model generalizes well to unseen data. (D) SHAP values plot. This visualization highlights the contribution of individual features to the model’s predictions. Positive SHAP values increase the likelihood of a positive prediction, while negative values decrease it, providing interpretability to the model’s decisions.
Discussion
KD is a common systemic vasculitis affecting young children, predisposing to the coronary artery aneurysms, and is the leading cause of acquired heart disease in this age group23. Currently, its diagnosis relies on clinical manifestations without quantitative methods. Some cases of KD do not present with classic symptoms, and the signs may pass unnoticed. Thus, developing new methods to facilitate the clinical diagnosis of KD is crucial. Despite the potential of machine learning, few studies or decision-support systems have been developed to classify and diagnose diseases like KD. Our study is the first to explore the possibility of diagnosing KD using only routinely collected blood tests yet achieving high accuracy with machine learning. A key highlight is addressing the clinical challenge of conducting extensive tests within the first five days of fever, which our study successfully overcomes.
This study utilized 25 routine blood test parameters. Previous research has highlighted that Kawasaki Disease (KD) exhibits distinct patterns in routine blood tests, particularly in inflammatory markers. However, these patterns are often non-linear and do not always follow a consistent trend, presenting both challenges and opportunities for deeper analysis using machine learning techniques. In KD, CRP levels are increased, which are mainly driven by systemic inflammation and vasculitis. Whilst EO# and eosinophil percentage (EO%) usually remain normal, they may rise during allergic reactions or episodes of inflammation. HCT and HGB levels can decrease as a result of anemia. During the acute phase, both LYMPH# and LYMPH% are often low, but these levels increase as the body begins to heal24,25. MCH and MCHC levels tend to remain stable, although MCH may fluctuate in cases of anemia. For individuals with anemia, MCV can be normal or abnormal. In most cases, inflammation or illness leads to elevated MONO# and MONO% levels. Increased platelet production can result in a higher MPV. The onset of inflammation triggers a rise in neutrophil numbers. Significant changes in platelets may cause P-LCR to increase substantially. High levels of PCT can indicate severe systemic inflammation and a potential bacterial infection. A high PDW value suggests greater variation in platelet size. The intermediate and late stages of KD significantly elevate PLT levels. Anemia often results in a decreased RBC, with RDW and RDW-1 reflecting variations in red blood cell size. Over time, the immune response and systemic inflammation lead to increased WBC, NEUT#, and NEUT%.
Our study found similar performance of three feature selection methods (Lasso, t-test/Chi-squared, and no-selection) and identified EO%, HCT, PCT, gender, and BA# as the top five significant features. The similar model performance observed across these methods can be attributed to the specificity of KD-related changes in routine blood tests. For example, inflammatory markers such as CRP, WBC, and PLT typically increase during the acute phase of KD, while hemoglobin levels may decrease in later stages. These features exhibit a strong linear relationship with the target variable, leading to overlapping feature sets and comparable performance across methods. Furthermore, distinct yet sometimes inconsistent patterns in KD-related inflammatory markers, suggesting opportunities for further investigation using advanced machine learning techniques. Liu et al. highlighted eosinophilia’s critical role in distinguishing KD from other febrile infections with high predictive accuracy26. Similarly, HCT and PCT have been evaluated in KD23, although they were not statistically significant in another study27. The male-to-female ratio averaged 1.47:128 across these studies, underscoring their relevance to KD. These findings underscore the efficacy of using SHAP summary plots to explore risk factors, alongside supportive evidence from log loss versus epoch curves, precision-recall curves, and learning curves, all confirming XGBoost as an effective model.
In our study, XGBoost demonstrated the highest accuracy (0.9979) and AUC (0.9999) in diagnosing KD without feature selection, aligning with previous research findings. The work by Tsai et al.. incorporated urinalysis results and liver enzymes, and found that XGBoost achieved a sensitivity of 92.5%, specificity of 97.3%, PPV of 34.5%, NPV of 99.9%, and a positive likelihood ratio of 34.029. Similarly, Habgood-Coote et al.. used a multi-class supervised machine-learning approach, incorporating the clinical consequences of misdiagnosis as a “cost” weighting, on a whole-blood transcriptomic microarray dataset with 1,212 children across 18 infectious or inflammatory diseases30. Lam et al. developed a machine-learning algorithm utilizing patient age, five classic signs of KD, and 17 laboratory parameters to diagnose multisystem inflammatory syndrome and KD, achieving an accuracy rate of 94-96%10. Although these studies demonstrated excellent performance, they involved complex tests and examinations, making them impractical for routine clinical use, especially in the primary care setting or in the emergency room. However, our method, based on limited laboratory parameters, proves to be more effective, accurate, and simple-to-use. Whilst other studies employed different feature selection strategies to explore the relationship between clinical data and KD, our results indicate that interestingly no feature selection yields better outcomes.
Our study focuses on universally accessible features in most healthcare settings, utilizing only routine blood tests. This approach does not require additional clinical evaluation, and indeed it does not require primary care physicians to be familiar with the signs of KD. Liu et al.. developed a nomogram using objective blood test results to differentiate KD from other febrile illnesses, achieving sensitivity between 78% and 84% in validation, but it potentially missed 16–22% of KD cases26, limiting its clinical utility. Tsai et al.. incorporated urinalysis results, including pyuria, achieving an AUC of 0.980 (95% CI, 0.974–0.987)29. In contrast, our study uses only essential and common tests (routine blood tests) to achieve the highest accuracy (0.9979) and AUC (0.9999) in children with fever within five days.
We employed a machine learning approach to diagnose KD with the goal of reducing the healthcare burden and expediting the diagnostic process. When children present with unexplained fever, first-line doctors often recommend extensive and costly tests, which can be time-consuming and, in some cases, unnecessary. In situations where the fever cannot be linked to common viral or bacterial infections, some physicians turn to antibiotic or steroid therapies to alleviate symptoms and address parental concerns. However, this approach does not assist in diagnosing KD or ensuring timely intervention to prevent serious complications such as coronary artery aneurysms. Moreover, complete blood count results in KD can be affected by infections or social factors, further complicating diagnosis. The dataset used in this study reflects the real-world rarity of KD, resulting in a significant class imbalance that presents challenges for model training. This imbalance may bias the model toward the majority class, reducing its sensitivity in detecting KD cases. To address this, we applied the RandomOverSampler method to balance the dataset, allowing the model to better recognize KD-specific patterns. We evaluated the model’s performance using metrics such as sensitivity, specificity, and AUC to ensure its effectiveness under imbalanced conditions. Although class imbalance may limit the model’s clinical applicability, we propose its use as a supportive diagnostic tool in conjunction with clinical judgment. Future research should focus on refining the model through strategies such as cost-sensitive learning, weighted loss functions, and ensemble methods. Additionally, prospective validation with larger and more balanced datasets is essential to further enhance the model’s robustness and reliability.
Our study has several limitations. First, the dataset was limited to pediatric patients from a single center, which may constrain the generalizability of our model to other geographic regions, ethnic groups, and populations. For example, KD incidence in the United States is estimated at 18 to 25 cases per 100,000 children under the age of five, while it is 10 to 30 times higher in Northeast Asian countries31. This difference highlights not only the regional variation in KD prevalence but also the rarity of the disease, even in high-incidence areas, which limits the size of patient populations in individual hospitals. Our dataset was obtained from Guangzhou Women’s and Children’s Medical Center, a national platform consisting of three Grade A tertiary hospitals, including a specialized children’s hospital with a large number of KD cases. This makes it a valuable resource for KD research. However, the regional characteristics of the dataset may limit its ability to reflect KD prevalence or clinical patterns in areas with different geographic, ethnic, or cultural profiles. To enhance the generalizability and applicability of the model, future studies should incorporate larger and more diverse datasets from multiple centers across various regions to include a broader range of populations. Additionally, understanding how geographic, cultural, and socioeconomic factors influence KD incidence will be critical to refining the model, addressing regional variability, and ensuring its utility across diverse settings. Second, the study only considered data readily available in routine clinical practice. While this approach enhances the model’s practicality, it excludes potentially relevant social and environmental factors, such as family income, residential status, and physical activity, which may influence the development or presentation of KD. Incorporating these factors in future research could provide a more comprehensive understanding of KD and improve the predictive power of the model. Finally, while we addressed data imbalances in the study groups using the RandomOverSampler method, prospective validation with larger sample sizes remains necessary to ensure the robustness and reliability of our XGBoost model. Larger, multi-center studies with prospective designs will be crucial to confirm the model’s performance, especially in real-world clinical settings where the prevalence of KD may differ.
Conclusion
Routine blood tests have the potential to predict KD. Additionally, the XGBoost model demonstrates excellent accuracy, offering a practical, non-invasive diagnostic tool for primary care clinics or emergency rooms.
Data availability
The datasets utilized and/or analyzed in this study are available from the corresponding author upon reasonable request.
Abbreviations
- AUC:
-
Area under the ROC curve
- BA#:
-
Absolute basophil count
- BA%:
-
Basophil percentage
- CRP:
-
C-reactive protein
- EO#:
-
Absolute eosinophil count
- EO%:
-
Eosinophil percentage
- HCT:
-
Hematocrit
- HGB:
-
Hemoglobin
- IVIG:
-
Intravenous immunoglobulin
- KD:
-
Kawasaki disease
- LYMPH#:
-
Absolute lymphocyte count
- LYMPH%:
-
Lymphocyte percentage
- MCH:
-
Mean corpuscular hemoglobin
- MCHC:
-
Mean corpuscular hemoglobin concentration
- MCV:
-
Mean corpuscular volume
- MONO#:
-
Absolute monocyte count
- MONO%:
-
Monocyte percentage
- MPV:
-
Mean platelet volume
- NEUT#:
-
Absolute neutrophil count
- NEUT%:
-
Neutrophil percentage
- NPV:
-
Negative predictive value
- PCT:
-
Platelet crit
- PDW:
-
Platelet distribution Width
- P-LCR:
-
Platelet large cell ratio
- PLT:
-
Platelet count
- RBC:
-
Red blood cell count
- RDW:
-
Red cell distribution width
- SHAP:
-
SHapley Additive exPlanations
- WBC:
-
White blood cell count
- XGBoost:
-
Supervised eXtreme Gradient Boosting
References
Cohen, E. & Sundel, R. Kawasaki disease at 50 years. JAMA Pediatr. 170, 1093–1099. https://doi.org/10.1001/jamapediatrics.2016.1446 (2016).
La Vecchia, A. et al. Risk factors and scores for prediction of coronary artery aneurysms in Kawasaki disease: a European monocentric study. BMC Pediatr. 24, 139. https://doi.org/10.1186/s12887-024-04623-3 (2024).
Newburger, J. W. et al. A single intravenous infusion of gamma Globulin as compared with four infusions in the treatment of acute Kawasaki syndrome. N. Engl. J. Med. 324, 1633–1639. https://doi.org/10.1056/NEJM199106063242305 (1991).
Tse, S. M. L., Silverman, E. D., McCrindle, B. W. & Yeung, R. S. M. Early treatment with intravenous Immunoglobulin in patients with Kawasaki disease. J. Pediatr. 140, 450–455. https://doi.org/10.1067/mpd.2002.122469 (2002).
Broderick, C., Kobayashi, S., Suto, M., Ito, S. & Kobayashi, T. Intravenous Immunoglobulin for the treatment of Kawasaki disease. Cochrane Database Syst. Reviews. https://doi.org/10.1002/14651858.CD014884.pub2 (2023).
Faye, A. Kawasaki disease: a new Understanding of the clinical spectrum. Lancet Child. Adolesc. Health. 7, 672–673. https://doi.org/10.1016/S2352-4642(23)00191-8 (2023).
Wang, H. et al. Subgroups of children with Kawasaki disease: a data-driven cluster analysis. Lancet Child. Adolesc. Health. 7, 697–707. https://doi.org/10.1016/S2352-4642(23)00166-9 (2023).
Xu, E., Nemati, S. & Tremoulet, A. H. A deep convolutional neural network for Kawasaki disease diagnosis. Sci. Rep. 12, 11438. https://doi.org/10.1038/s41598-022-15495-x (2022).
Tsai, C. M. et al. Use of machine learning to differentiate children with Kawasaki disease from other febrile children in a pediatric emergency department. JAMA Netw. Open. 6, e237489–e237489. https://doi.org/10.1001/jamanetworkopen.2023.7489 (2023).
Lam, J. Y. et al. A machine-learning algorithm for diagnosis of multisystem inflammatory syndrome in children and Kawasaki disease in the USA: a retrospective model development and validation study. Lancet Digit. Health. 4, e717–e726. https://doi.org/10.1016/s2589-7500(22)00149-2 (2022).
Theodosiou, A. A. & Read, R. C. Artificial intelligence, machine learning and deep learning: potential resources for the infection clinician. J. Infect. 87, 287–294. https://doi.org/10.1016/j.jinf.2023.07.006 (2023).
Swanson, K., Wu, E., Zhang, A., Alizadeh, A. A. & Zou, J. From patterns to patients: advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell 186, 1772–1791. https://doi.org/10.1016/j.cell.2023.01.035 (2023).
Newburger, J. W. et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the committee on rheumatic fever, endocarditis and Kawasaki disease, Council on cardiovascular disease in the young, American heart association. Circulation 110, 2747–2771 (2004).
Fonti, V. & Belitser, E. Feature selection using Lasso. VU Amsterdam Res. Paper Bus. Analytics. 30, 1–25 (2017).
Nusinovici, S. et al. Logistic regression was as good as machine learning for predicting major chronic diseases. J. Clin. Epidemiol. 122, 56–69. https://doi.org/10.1016/j.jclinepi.2020.03.002 (2020).
Suthaharan, S. & Suthaharan, S. Support vector machine. In Machine Learning Models and Algorithms for Big Data Classification: Thinking with Examples for Effective Learning. 207–235 (2016).
Tang, J., Deng, C. & Huang, G. B. Extreme learning machine for multilayer perceptron. IEEE Trans. Neural Networks Learn. Syst. 27, 809–821 (2015).
Segal, M. R. Machine Learning Benchmarks and Random Forest Regression. (2004).
Shehadeh, A., Alshboul, O., Al Mamlook, R. E. & Hamedat, O. Machine learning models for predicting the residual value of heavy construction equipment: an evaluation of modified decision tree, LightGBM, and XGBoost regression. Autom. Constr. 129, 103827 (2021).
Zhang, P. B. & Yang, Z. X. A novel adaboost framework with robust threshold and structural optimization. IEEE Trans. Cybern. 48, 64–76. https://doi.org/10.1109/tcyb.2016.2623900 (2018).
Chen, T. & Guestrin, C. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining.. 785–794.
Hancock, J. T. & Khoshgoftaar, T. M. CatBoost for big data: an interdisciplinary review. J. Big Data. 7, 1–45 (2020).
Noval Rivas, M., Kocatürk, B., Franklin, B. S. & Arditi, M. Platelets in Kawasaki disease: mediators of vascular inflammation. Nat. Rev. Rheumatol. https://doi.org/10.1038/s41584-024-01119-3 (2024).
Tremoulet, A. H. et al. Novel data-mining approach identifies biomarkers for diagnosis of Kawasaki disease. Pediatr. Res. 78, 547–553. https://doi.org/10.1038/pr.2015.137 (2015).
Ye, Q. et al. A comprehensive assessment of the value of laboratory indices in diagnosing Kawasaki disease. Arthritis Rheumatol. 67, 1943–1950. https://doi.org/10.1002/art.39112 (2015).
Liu, X. P. et al. A nomogram model identifies eosinophilic frequencies to powerfully discriminate Kawasaki disease from febrile infections. Front. Pediatr. 8, 559389. https://doi.org/10.3389/fped.2020.559389 (2020).
Zheng, X., Wu, W., Zhang, Y. & Wu, G. Changes in and significance of platelet function and parameters in Kawasaki disease. Sci. Rep. 9, 17641. https://doi.org/10.1038/s41598-019-54113-1 (2019).
Wu, M. H. et al. Prevalence and the long-term coronary risks of patients with Kawasaki disease in a general population < 40 years: a National database study. Circ. Cardiovasc. Qual. Outcomes. 5, 566–570. https://doi.org/10.1161/circoutcomes.112.965194 (2012).
Tsai, C. M. et al. Use of machine learning to differentiate children with Kawasaki disease from other febrile children in a pediatric emergency department. JAMA Netw. Open. 6, e237489. https://doi.org/10.1001/jamanetworkopen.2023.7489 (2023).
Habgood-Coote, D. et al. Diagnosis of childhood febrile illness using a multi-class blood RNA molecular signature. Med 4, 635–654e635. https://doi.org/10.1016/j.medj.2023.06.007 (2023).
Elakabawi, K., Lin, J., Jiao, F., Guo, N. & Yuan, Z. Kawasaki disease: global burden and genetic background. Cardiol. Res. 11, 9–14. https://doi.org/10.14740/cr993 (2020).
Funding
GT is supported by a Research Impact Fund from the Hong Kong Metropolitan University (RIF/2022/2.2). This research was also supported by grants from the Natural Science Foundation of Guangdong Province (Grant No. 2021A1515011308) and the Guangzhou Municipal Science and Technology Bureau (Grant No. 202102010237).
Author information
Authors and Affiliations
Contributions
W.Z. contributed to the conceptualization of the study, formal analysis, drafting of the original manuscript, and review and editing of the manuscript. S.Z. was responsible for the conceptualization, methodology, software development, validation, formal analysis, investigation, drafting of the manuscript, review and editing, and visualization. X.W. contributed to the conceptualization, investigation, resource management, data curation, and drafting of the manuscript. C.C. and Z.Z. were involved in resource management, data curation, and validation. Y.X. and X.M. provided support with resources and data curation. G.T. and X.L. contributed to the review and editing of the manuscript, as well as project administration and supervision. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Ethical approval was granted by the Ethics Committee of Guangzhou Women and Children’s Medical Center in July 2024, waiving the need for informed patient consent. All procedures and methods were conducted in compliance with Chinese national legislation, institutional guidelines, and the Declaration of Helsinki.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zheng, W., Zhu, S., Wang, X. et al. Machine learning for early diagnosis of Kawasaki disease in acute febrile children: retrospective cross-sectional study in China. Sci Rep 15, 6799 (2025). https://doi.org/10.1038/s41598-025-90919-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-90919-y