Abstract
This study was conducted to develop and validate a novel deep reinforcement learning (DRL) algorithm incorporating the segment anything model (SAM) to enhance the accuracy of automatic contouring organs at risk during radiotherapy for cervical cancer patients. CT images were collected from 150 cervical cancer patients treated at our hospital between 2021 and 2023. Among these images, 122 CT images were used as a training set for the algorithm training of the DRL model based on the SAM model, and 28 CT images were used for the test set. The model’s performance was evaluated by comparing its segmentation results with the ground truth (manual contouring) obtained through manual contouring by expert clinicians. The test results were compared with the contouring results of commercial automatic contouring software based on the deep learning (DL) algorithm model. The Dice similarity coefficient (DSC), 95th percentile Hausdorff distance, average symmetric surface distance (ASSD), and relative absolute volume difference (RAVD) were used to quantitatively assess the contouring accuracy from different perspectives, enabling the contouring results to be comprehensively and objectively evaluated. The DRL model outperformed the DL model across all evaluated metrics. DRL achieved higher median DSC values, such as 0.97 versus 0.96 for the left kidney (P < 0.001), and demonstrated better boundary accuracy with lower HD95 values, e.g., 14.30 mm versus 17.24 mm for the rectum (P < 0.001). Moreover, DRL exhibited superior spatial agreement (median ASSD: 1.55 mm vs. 1.80 mm for the rectum, P < 0.001) and volume prediction accuracy (median RAVD: 10.25 vs. 10.64 for the duodenum, P < 0.001). These findings indicate that integrating SAM with RL (reinforcement learning) enhances segmentation accuracy and consistency compared to conventional DL methods. The proposed approach introduces a novel training strategy that improves performance without increasing model complexity, demonstrating its potential applicability in clinical practice.
Similar content being viewed by others
Introduction
Cervical cancer is the fourth most common cancer among female persons worldwide and the second most common cancer in female persons aged 15–44 years1,2,3. Radiotherapy is used as primary or adjuvant therapy in the curative treatment of patients with cervical cancer4. When administering radiotherapy for cervical cancer, delineating the organs at risk (OARs) is time-consuming and laborious; While inter-observer variability in contouring OARs is a recognized challenge in clinical practice, for the purposes of this study, the contours drawn by experienced physicians were considered the reference standard. In recent years, scholars worldwide have studied the automatic delineation of OAR via atlas-based autosegmentation (ABAS) algorithms, which are widely used for contouring OARs5,6,7,8. ABAS algorithms use deformable image registration to transform OARs contours from the atlas into a new image. Deformable registration algorithms are uncertain, particularly when intensity-based algorithms are used9,10. Deep learning (DL) approaches have been recently applied to various medical imaging modalities to increase organ segmentation accuracy, reproducibility, and consistency11,12,13,14,15. Various DL algorithms, including recurrent neural networks (RNNs), restricted residual networks (ResNets), encoder decoders, and convolutional neural networks (CNNs), have been devised to address the typical challenges encountered in medical imaging research15,16,17,18,19. CNN models in particular have been used for image registration, autosegmentation, and classification20,21. Several studies have used CNNs to autocontour regions, such as head and neck, and lung cancer regions22,23. Although DL-based methods have demonstrated promising segmentation performance, they still face challenges in handling complex anatomical structures and inter-observer variability. DL algorithms have been applied in various medical imaging modalities to increase the precision, repeatability, and consistency of organ segmentation and achieve certain delineation effects. However, physicians must still manually modify the results before using them in clinical practice.
For machine learning (ML), manual adjustments made by physicians essentially constitute a feedback loop that serves as a reward mechanism. For the portions of the OARs delineation produced by ML algorithms that are well executed, physicians may need to make minimal or no changes, which can be viewed as a reward; conversely, areas that are poorly delineated may require significant manual correction, which can be viewed as a punishment. Based on this, researchers have attempted to develop a deep reinforcement learning (DRL) algorithm model that combines DL algorithms with reinforcement learning (RL) algorithms. DL is used to extract graphic features from high-dimensional raw CT data to automatically delineate OARs, while RL is employed to learn the optimal strategy through interactions with the results of manual adjustments made by physicians, thereby maximizing the cumulative rewards. Unlike traditional DL-based segmentation models that rely solely on static labeled datasets, DRL incorporates an adaptive learning process, enabling continuous improvement based on physician feedback. The integration of these two approaches in DRL utilizes neural networks as agents, learning to interact and allocate rewards from the state-action pairs of delineation and thereby addressing the perceptual and decision-control issues of automatic organ delineation and manual adjustments in radiation therapy practice.
DRL, as a significant branch of the DL field, has been increasingly applied and developed in the medical ___domain since 201624. However, research on DRL-based automatic delineation of OARs in cervical cancer radiotherapy remains scarce. To our knowledge, limited research has been conducted on the automatic delineation of OARs in cervical cancer radiotherapy planning using DRL. Furthermore, no prior studies have integrated the Segment Anything Model (SAM) with RL to enhance segmentation performance. This study is primarily conducted to develop and validate a DRL algorithm incorporating SAM, leveraging its robust feature extraction capability alongside RL’s decision-making process. To the best of our knowledge, this is the first study that combines SAM with RL for medical image segmentation, specifically in the context of OARs delineation in cervical cancer radiotherapy. By harnessing SAM’s advanced feature extraction capabilities and the iterative decision-making process of reinforcement learning (RL), our approach seeks to improve segmentation accuracy and robustness relative to traditional DL-based methods, without increasing model complexity.
Materials and methods
Clinical data
The CT images of 150 Asian cervical cancer patients, collected between 2021 and 2023, were used in this study. All the data were 512 × 512 pixels (pixel spacing: 0.3 × 0.3 mm), 5 mm thick, and acquired with a large-aperture CT simulator (version Discovery CT590, GE, Wisconsin, USA). The scanning range encompassed the pelvic lymphatic drainage area from the lower edge of the second lumbar vertebra to the ischial tuberosity and pelvis, with the field of view (FOV) selected for the maximum range. The CT images were anonymized, and the standard DICOM files were transferred to the MonacoV5.11 (Elekta AB, Stockholm, Sweden) treatment planning system. A senior radiation oncologist with less than 10 years of experience then delineated the OARs, which included the left kidney (kidney L), right kidney (kidney R), liver, bladder, spleen, duodenum, bone marrow, pancreas, stomach, and rectum25,26. To ensure the accuracy of delineation, the accuracy of the delineation should be reviewed by an expert radiation oncologist with more than 15 years of experience. The OARs were delineated based on the consensus of experts in the cervical cancer target area and OAR delineation fields27. The inclusion criteria for patients were as follows: a pathologically confirmed diagnosis of cervical cancer, no contraindications to radiation therapy, and a Karnofsky Performance Score (KPS) greater than 70. The exclusion criteria for patients were as follows: contraindications to radiation therapy and having previously undergone radiation therapy, chemotherapy, or other anticancer treatments. The human CT images used in this study were anonymized, and informed consent was obtained from all participants. This study was approved by the Ethics Institutional Review Board of Zhejiang Provincial People’s Hospital (QT2024040), and it was conducted in accordance with the ethical standards of the Declaration of Helsinki.
DRL algorithm model training
The architectural framework is centered around the synergy between the SAM and a RL model, illustrated in Fig. 1. When operating under the prompt mode paradigm, the SAM model demonstrates remarkable adaptability by embracing a dual-point directive—a foreground point nestled within the confines of the object of interest (target mask) and a contrasting background point situated external to it. This dual input, coined as a ‘point prompt’, serves as the catalyst for the SAM model to meticulously delineate and produce an exacting target mask that encapsulates the intended segmentation area with precision. The RL component, a cornerstone of this integrated design, assumes a pivotal role in advancing the autonomous functionality of the system. By harnessing the collective power of the immediate visual context (the image), a nuanced understanding encapsulated in the probability map, and the distilled knowledge from prior segmentation attempts (previous mask), the RL model executes a sophisticated predictive analysis. The probability map is generated using a hint point as the central reference. Specifically, a 9 × 9 Gaussian kernel with a sigma value of 2.5 is constructed. The Gaussian kernel is defined as:
where \(\sigma\) is the standard deviation (set to 2.5 in our case), and \(x,\;y\) represent the coordinates relative to the kernel center.
This Gaussian kernel is then superimposed onto an initially blank image, with the hint point serving as the center of the kernel. The resulting probability map reflects the spatial distribution of the Gaussian function around the hint point, emphasizing its importance for subsequent segmentation tasks. It ingeniously forecasts the most efficacious subsequent point prompt, thereby orchestrating a sequence of informed decisions aimed at optimizing segmentation outcomes and enhancing the model’s iterative learning and adaptation process. This harmonious interplay between the SAM and RL models not only bolsters the system’s segmentation accuracy but also imbues it with an enhanced capacity for intelligent decision-making, navigating complex visual scenes with heightened efficiency and precision.
Initialization
In the initialization phase, the state space and action space are first defined. The state is characterized by the image to be segmented, the current segmentation status, and the Gaussian probability map generated by the hint points. Actions are defined as the coordinates of the hint points. The algorithm model needs to provide two hint points each time, one representing the foreground area and the other representing the background area. The output of the Agent algorithm model is thus the mean and standard deviation of the two hint points. The specific action points for each iteration are obtained by sampling based on these mean and standard deviation values. ResNet-5028, with two additional fully connected layers, is employed as the backbone network for the Agent model. Pretrained ResNet-50 weights are utilized. Additionally, the authors incorporate an attention module to calculate the weight of each pixel. This module, composed of a convolutional layer and a fully connected layer, takes the feature vector of each pixel as input and computes a weight vector for each pixel. The original feature map is then weighted and summed using these weight vectors to produce a weighted feature map. Incorporating the attention mechanism into the CNN algorithm model enables surgeons to better focus on key areas of the image, thereby enhancing segmentation accuracy.
Provide cue points
A Gaussian probability distribution map is generated for each cue point based on its coordinate ___location. During the probability map generation, the coordinates of each cue point are set as the mean of the Gaussian distribution and the standard deviation of the action is set as the standard deviation of the Gaussian distribution. Thus, each Gaussian probability distribution map produced represents the degree of influence of that cue point on the segmentation result. These maps are subsequently input into a DL algorithm model along with the original image. This allows the algorithm model to better focus on these key areas, improving the segmentation effect. To evaluate the impact of the cue points on the segmentation results, the Dice similarity coefficient (DSC) is calculated by comparing the current segmentation state with the labels, establishing an initial DSC benchmark. After each output of the action points by the algorithm model, the input is fed into the SAM model, generating the next segmentation state. This state is subsequently compared with the labels to calculate the DSC value. The difference between the DSC value and the DSC benchmark is the reward value for this action.
Training algorithm model
During the training phase, DL algorithm models must be integrated with RL algorithms. The ratio of the training set to the test set is 122:28. The researchers first employed the proximal policy optimization (PPO) algorithm to train the model, enabling it to select the optimal action based on the current state. PPO is an optimization algorithm designed to address RL challenges. It operates within the actor-critic framework by minimizing the policy loss function to train the model. PPO primarily limits the divergence between new and old policies during updates, ensuring the stability of the strategy. Unlike the Monte Carlo method, which uses the entire dataset for training, PPO utilizes only the latest batch of trajectories, enhancing the training efficiency. To further improve training stability, the researchers also implemented the generalized advantage estimation (GAE) method. This approach normalizes the advantage function estimates and introduces an importance sampling strategy, which mitigates the instability commonly associated with traditional advantage function estimation methods based on the temporal difference error (TDE). This approach avoids the “excessive advantages” that can arise when a neural network is used to estimate the value of state–action pairs. The DRL model was implemented using PyTorch (version 2.0.1) as the primary framework. The training was conducted on a system equipped with an Intel i7-10700 CPU and an NVIDIA RTX 3090 GPU with 24 GB memory. The key training parameters were set as follows: the learning rate was 1e-4, and the Adam optimizer was used. The batch size was 128, and the total number of iterations reached 30,000. The experience replay buffer size was 2048, while the reward discount factor (gamma) was set to 0.99. Additional parameters included an entropy coefficient (lambda) of 0.005 and an advantage clipping threshold of 0.2.
Deep learning algorithm model
The AccuContour (version 3.2, Manufactured by MANTEIA, XiaMen, China), a high-performance DL algorithm model, was developed by Manteia Technologies Co., Ltd., and generated using their advanced DL platform. The images are preprocessed using an adaptive approach based on the image intensity range and distribution characteristics to standardize and resample the images. Data augmentation is first performed on the loaded images, followed by balanced cropping according to the label area, to generate data that fit the input size of the algorithm model. The U-Net29 network architecture is chosen for algorithm model training. This architecture utilizes an adaptive network structure adjustment strategy based on gradient feedback and a loss function to adapt to the features of the training dataset. During data collection, the principles of multicenter, multiregional, and multi-disease data types were applied to increase the diversity of the training data, thereby enhancing the accuracy and generalizability of the DL algorithm model. During manual data annotation, multiple people annotate a single data instance. After examining the differences between the annotations of the individuals and obtaining a certain degree of consistency, the data are added to the training library, thereby improving the quality of the training data annotation. Therefore, the automatic segmentation algorithm model integrated into AccuContour exhibits strong accuracy and stability.
Quantitative evaluation metrics
In this study, CT scans from 150 cervical cancer patients were utilized, with 122 of these scans serving as the training set for the DRL algorithm model based on the SAM model. The other 28 CT scans served as the test set for comparing the performances of the DRL and DL algorithm models in delineating cervical cancer OARs. Although various delineation accuracy evaluation standards have been used for automatic delineation assessment, due to the varying sizes, shapes, and locations of different organs, four different evaluation metrics were used in this study: the DSC, 95th percentile Hausdorff distance (95HD), average symmetric surface distance (ASSD), and relative absolute volume difference (RAVD). These standards, each with a different focus, were used to comprehensively and objectively evaluate the delineation results of OARs using different algorithm models.
-
1.
The DSC is the volume of overlap between the predicted and manual annotations (ground truth), given by:
$${\text{DSC (A,B) = }}\frac{{{\text{2|A }} \cap {\text{ B|}}}}{{\text{(|A| + |B|)}}}$$(1)where A and B are the volumes of voxels in the predicted segmentation and ground truth, respectively, and A ∩ B is the volume of voxels that are consistent between the two methods. The DSC scores range between 0 and 1, with higher values indicating better segmentation performance.
-
2.
HD95 is a distance-based metric used to measure the boundary distance between the predicted segmentation and ground truth. A smaller HD95 value indicates better learning of edge information. To calculate the 95th percentile of HD, HD95 is obtained as follows:
$$HD 95(A,B) = max (h(a,b),h(a,b))*95\%$$(2)$$h(a,b) = min||a - b||,a \in A,b\in B$$(3)where a and b are the elements in A and B, respectively.
-
3.
The ASSD calculates the minimum Euclidean distance from all points in a predicted surface point set to a reference surface point set and then computes the average of these distances. A smaller ASSD value indicates better smoothness and consistency in the delineation. The ASSD is given by:
$$ASSD (A,B) = mean(h (A,B),h (A,B))$$(4)$$h(A,B) = min ||a - b||,a \in A,b \in B$$(5) -
4.
The RAVD is used to calculate the relative coefficient of the nonoverlapping regions between the predicted volume and the reference volume. A smaller RAVD value indicates a smaller relative difference between the nonoverlapping regions.
The RAVD is calculated as follows:
$$RAVD = \frac{{\left| {{\text{A }}{-}{\text{ B}}} \right|{ }}}{{\text{B}}}$$(5)In this formula, \(|A - B|\) represents the absolute difference between the reference volume and the predicted volume.
Statistical analysis
All the data underwent statistical analysis using SPSS (v20, SPSS, Inc., Chicago, IL, USA) software. The data that did not follow the normal distribution were represented by M (Q1, Q3), and nonparametric rank-sum tests were used for group comparisons. A p value < 0.05 was considered indicative of statistical significance. To visualize the data, Prism software (GraphPad Prism 5, GraphPad Software, Inc.) was used to graph the data.
Ethics approval and consent to participate
The human CT images used in this study were anonymized, and informed consent was obtained from all participants. This study was approved by the Ethics Institutional Review Board of Zhejiang Provincial People’s Hospital (QT2024040), and it was conducted in accordance with the ethical standards of the Declaration of Helsinki.
Results
The training time of our DRL model was approximately 12 h, reflecting the computational effort required to achieve optimal performance. Once trained, the model demonstrated rapid prediction capabilities, with the creation of OARs contours taking approximately 30 s per case. This is consistent with the performance of AccuContour, suggesting that our approach is both efficient and practical for clinical implementation. Examples of manual and predicted contours from the DRL and DL models for the OAR are shown in Fig. 2. The authors employed nonparametric rank-sum tests to compare the delineations of all OARs by the DRL and DL algorithm models using metrics such as the DSC, HD95, ASSD, and RAVD. The results revealed that the median DSC scores of the DRL were greater than 0.9 for the kidney L, kidney R, liver, bladder, spleen, and stomach. For the bone marrow, pancreas, and rectum, the median DSC scores of the DRL were greater than 0.8, and the median DSC for the duodenum was 0.77. As shown in Table 1, evaluating the DSC, HD95, and ASSD scores of the DRL and DL delineation results reveals that the delineation effects of all OARs obtained by DRL are superior to those obtained by DL. The differences between the two groups were statistically significant (P < 0.05) for all OARs except for the ASSD of the pancreas. A separate evaluation of the RAVD of the two algorithm models revealed that the differences between the delineations of the liver, pancreas, and stomach of the two models were not statistically significant (P > 0.05); however, the differences between the two were statistically significant (P < 0.05) for other OARs. A box plot comparing the delineation results of OARs using DRL and DL is shown in Fig. 3.
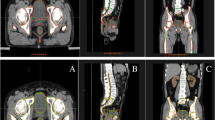
Automatic delineation results of OARs in transverse, coronal, sagittal planes and 3D visualization of OARs for a patient using two models (a kidney L, kidney R and duodenum, b bladder, rectum, bone marrow, c liver, stomach, pancreas, spleen). DRL represents deep reinforcement learning, and DL represents deep learning.
Discussion
The SAM is a robust foundational segmentation algorithm trained on more than a billion annotations, primarily targeting natural images and designed for interactive segmentation based on user-defined objects. While its performance on natural images is impressive, its application to medical imaging requires specialized anatomical knowledge. In this study, we introduced a DRL model based on the SAM algorithm to address these challenges. Our results demonstrate that the DRL-based approach successfully adapts the SAM model to medical imaging datasets, significantly improving segmentation accuracy. Specifically, the DRL model outperforms traditional DL algorithms in terms of generalizability, edge preservation, smoothness, and consistency, providing a reliable solution for the automatic delineation of OARs in medical images.
Previous studies have shown that automated treatment planning for pelvic radiotherapy is well-established and often outperforms manual planning, particularly in terms of efficiency, reducing human intervention, and enhancing plan consistency. For example, Schmidt et al.30 demonstrated that automated planning tools improved dose homogeneity and reduced the dose to OARs compared to manual planning. Prunaretty et al.31 further validated a DL-based automated treatment planning system, which not only produced dosimetrically competitive plans but also excelled in treatment machine deliverability. Automated treatment planning, compared to traditional manual planning, stands out for its significant advantages in improving workflow efficiency and reducing planning time. Buschmann et al.32 found that automated volumetric modulated arc therapy (VMAT) planning not only improved organ sparing but also decreased planning time and workload, further supporting the clinical applicability of automated planning systems. In this study, we focused particularly on the automatic delineation of OARs, where our DRL-based method demonstrated higher DSC values compared to manual contouring. Goddard et al.33 evaluated multiple AI-based auto-contouring solutions and reported that MIM achieved average DSC values of 0.76 for the bladder and 0.79 for the rectum. Similarly, a comparative study by Chen et al.34 found that RayStation, using an atlas-based approach, achieved DSC values of 0.45 for the bladder and 0.44 for the rectum. In contrast, our DRL-based method achieved median DSC values of 0.92 for the bladder and 0.82 for the rectum, demonstrating superior performance relative to these commercial systems. This result underscores the potential of automated contouring techniques in enhancing accuracy and providing a reliable foundation for automated treatment planning. By enabling automated contouring, we pave the way for an end-to-end automated planning workflow, significantly reducing patient waiting times and improving the overall efficiency of the radiotherapy process.
The authors comprehensively evaluated the accuracy and consistency of the automatic delineation of OARs in cervical cancer radiotherapy patients using a DRL algorithm based on the SAM model and a DL algorithm. DRL improved the accuracy and consistency of manual delineation across all abdominal OARs. As shown in Table 1, the DSC values for OARs delineated by DRL were significantly higher than those for DL. The duodenum had the lowest DSC (0.77), with the remaining OARs exceeding 0.8. The HD95 values showed significant differences between DRL and DL, with DRL demonstrating improved delineation, particularly at the edges. ASSD values were higher for DRL in all OARs except the pancreas, reflecting better smoothness and consistency. RAVD values for the liver and pancreas were lower for DL, but the differences were not statistically significant. Therefore, DRL outperforms DL in OAR delineation, offering greater accuracy, consistency, and adaptability to complex structures, making it a promising approach for improving radiotherapy planning in cervical cancer.
Figure 3 illustrates that the delineation stability of the bladder and duodenum by the DRL was inferior compared to that of the other OARs. This could be attributed to several factors. The bladder’s filling state can significantly vary, affecting its size and shape and thereby increasing the delineation complexity. The anatomical ___location of the duodenum is intricate, it is surrounded by complex structures, and it suffers from poor image contrast, which makes boundary recognition challenging. These factors contribute to lower delineation accuracy and stability. Additionally, the instability could be due to an insufficient volume of training samples. Substantial discrepancies were also observed between the automatic and manual delineation of certain layers for individual patients, decreasing the stability.
The CT images of test patients reveal that the soft tissues of the bladder, rectum, stomach, and duodenum of different patients were adjacent to each other; furthermore, the shape and volume of these tissues were susceptible to alterations due to varying volumes of urine and intestinal gases. In this study, RL and DL methodologies are combined to enhance OARs identification, particularly when the boundaries of the OARs are indistinct. The median DSC values for the bladder, rectum, stomach, and duodenum, as obtained by the DRL method, were 0.92, 0.82, 0.92, and 0.77, respectively, surpassing the DSC values achieved by the DL method. This can be attributed to the reward mechanism of the DRL method, which enables the algorithm model to focus more on the boundary features of the OARs and effectively handle noise information, and thereby yielding more accurate delineation results. While these results demonstrate the effectiveness of the DRL-based method in improving segmentation performance, it is important to highlight that the success of our approach stems not only from its architecture but also from the specific training framework utilized. Distinct from the investigation into DL architectures, this paper primarily focuses on enhancing the DRL training framework to optimize segmentation performance. The DRL model in this study employs a relatively straightforward architecture, which typically comprises no more than eight convolutional or fully connected layers. Notably, we do not use normalization layers in our model. Despite this simplicity, the DRL model achieves its objectives efficiently, demonstrating that the performance improvement is largely attributed to the effective training framework rather than the complexity of the model architecture. The researcher believes that the combination of an efficient training strategy and a simpler model architecture contributes to the superior performance observed with the DRL approach. In a typical clinical setting, it takes approximately 1.5 h for a physician to delineate the OARs of a patient. This time estimation is based on the practical experience of most physicians at our institution and specifically refers to the delineation of OARs. It reflects the average time required for manual contouring of OARs in cervical cancer radiotherapy. However, the DRL algorithm model developed in this study can automatically delineate all OARs within 30 s. Clinical physicians can then manually adjust the delineations before implementing them in a clinical setting, significantly reducing the time required for manual delineation by physicians.
Most related research evaluates the automatic delineation effects using one or a few evaluation indicators, such as the DSC and HD9535,36,37. To assess the merits of OAR delineation results under different algorithm models more comprehensively and objectively, four evaluation indicators are employed to holistically evaluate the feasibility of the DRL algorithm model for delineating cervical cancer OARs. This multi-indicator evaluation aids in obtaining a deeper understanding of the strengths and limitations of the performance of the algorithm and provides a clear direction for future improvements and optimizations. The proposed model is specifically focused on the automatic segmentation of OARs and does not include the delineation of target volumes, such as the gross tumor volume (GTV), clinical target volume (CTV), or planning target volume (PTV). Furthermore, the segmentation of the small intestine and sigmoid colon, which are critical structures in gynecological radiotherapy planning, was not addressed in this study. This omission is primarily due to the challenges posed by their high variability in shape and position, as well as the limited availability of high-quality, multi-center datasets for training and analysis. In the future our goal is to improve the accuracy of automatic contouring for these organs by optimizing DRL models and leveraging larger, high-quality, multi-center datasets. This approach aims to enhance both the precision and generalizability of the model, enabling it to better adapt to diverse clinical scenarios and ultimately improve the efficiency and effectiveness of radiotherapy planning.
Conclusion
In this study, a DRL algorithm based on SAM is presented. This algorithm yields accurate OARs contouring results and exhibits high clinical acceptability. Furthermore, using DRL can enhance physician efficiency and improve the precision and consistency of OARs in cervical cancer patients, without increasing model complexity.
Data availability
The datasets from the current study are not publicly available because the data contain information that may compromise patients, but are available from the corresponding author upon reasonable request.
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. https://doi.org/10.3322/caac.21492 (2018).
Ishida, S., McCormick, F., Smith-McCune, K. & Hanahan, D. Enhancing tumor-specific uptake of the anticancer drug cisplatin with a copper chelator. Cancer Cell 17, 574–583. https://doi.org/10.1016/j.ccr.2010.04.011 (2010).
Kostaras, D., Karampli, E. & Athanasakis, K. Vaccination against HPV virus: A systematic review of economic evaluation studies for developed countries. Expert Rev. Pharmacoecon. Outcomes Res. 19, 147–158. https://doi.org/10.1080/14737167.2019.1555039 (2019).
van Meir, H. et al. Impact of (chemo)radiotherapy on immune cell composition and function in cervical cancer patients. Oncoimmunology 6, e1267095. https://doi.org/10.1080/2162402X.2016.1267095 (2017).
Anders, L. C. et al. Performance of an atlas-based autosegmentation software for delineation of target volumes for radiotherapy of breast and anorectal cancer. Radiother. Oncol. 102, 68–73. https://doi.org/10.1016/j.radonc.2011.08.043 (2012).
Arabi, H. et al. Comparative study of algorithms for synthetic CT generation from MRI: Consequences for MRI-guided radiation planning in the pelvic region. Med. Phys. 45, 5218–5233. https://doi.org/10.1002/mp.13187 (2018).
Arabi, H., Koutsouvelis, N., Rouzaud, M., Miralbell, R. & Zaidi, H. Atlas-guided generation of pseudo-CT images for MRI-only and hybrid PET-MRI-guided radiotherapy treatment planning. Phys. Med. Biol. 61, 6531–6552. https://doi.org/10.1088/0031-9155/61/17/6531 (2016).
Voet, P. W. et al. Does atlas-based autosegmentation of neck levels require subsequent manual contour editing to avoid risk of severe target underdosage? A dosimetric analysis. Radiother. Oncol. 98, 373–377. https://doi.org/10.1016/j.radonc.2010.11.017 (2011).
Mohammadi, R. et al. Evaluation of deformable image registration algorithm for determination of accumulated dose for brachytherapy of cervical cancer patients. J. Contemp. Brachyther. 11, 469–478. https://doi.org/10.5114/jcb.2019.88762 (2019).
Yeo, U. J. et al. Performance of 12 DIR algorithms in low-contrast regions for mass and density conserving deformation. Med. Phys. 40, 101701. https://doi.org/10.1118/1.4819945 (2013).
Chougrad, H., Zouaki, H. & Alheyane, O. Deep convolutional neural networks for breast cancer screening. Comput. Methods Programs Biomed. 157, 19–30. https://doi.org/10.1016/j.cmpb.2018.01.011 (2018).
Im, J. H. et al. Impact of denoising on deep-learning-based automatic segmentation framework for breast cancer radiotherapy planning. Cancers (Basel) https://doi.org/10.3390/cancers14153581 (2022).
Moradi, S. et al. MFP-Unet: A novel deep learning based approach for left ventricle segmentation in echocardiography. Phys. Med. 67, 58–69. https://doi.org/10.1016/j.ejmp.2019.10.001 (2019).
Wu, J. et al. A deep Boltzmann machine-driven level set method for heart motion tracking using cine MRI images. Med. Image Anal. 47, 68–80. https://doi.org/10.1016/j.media.2018.03.015 (2018).
Guo, Z., Li, X., Huang, H., Guo, N. & Li, Q. Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans. Radiat. Plasma Med. Sci. 3, 162–169. https://doi.org/10.1109/trpms.2018.2890359 (2019).
Yu, L. et al. The applications of deep learning algorithms on in silico druggable proteins identification. J. Adv. Res. 41, 219–231. https://doi.org/10.1016/j.jare.2022.01.009 (2022).
Yu, L. et al. Systematic analysis and accurate identification of DNA N4-methylcytosine sites by deep learning. Front. Microbiol. 13, 843425. https://doi.org/10.3389/fmicb.2022.843425 (2022).
Anwar, S. M. et al. Medical image analysis using convolutional neural networks: A review. J. Med. Syst. 42, 226. https://doi.org/10.1007/s10916-018-1088-1 (2018).
Zhou, S. et al. High-resolution encoder-decoder networks for low-contrast medical image segmentation. IEEE Trans. Image Process. https://doi.org/10.1109/tip.2019.2919937 (2019).
van Dijk, L. V. et al. Improving automatic delineation for head and neck organs at risk by Deep Learning Contouring. Radiother. Oncol. 142, 115–123. https://doi.org/10.1016/j.radonc.2019.09.022 (2020).
Lamichhane, N. & Sharma, S. J. H. Development of flood warning system and flood inundation mapping using field survey and LiDAR data for the Grand River near the city of Painesville. Ohio Dent. J. 4, 24 (2017).
Stapleford, L. J. et al. Evaluation of automatic atlas-based lymph node segmentation for head-and-neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 77, 959–966. https://doi.org/10.1016/j.ijrobp.2009.09.023 (2010).
van Baardwijk, A. et al. PET-CT-based auto-contouring in non-small-cell lung cancer correlates with pathology and reduces interobserver variability in the delineation of the primary tumor and involved nodal volumes. Int. J. Radiat. Oncol. Biol. Phys. 68, 771–778. https://doi.org/10.1016/j.ijrobp.2006.12.067 (2007).
Xu, L., Zhu, S. & Wen, N. Deep reinforcement learning and its applications in medical imaging and radiation therapy: A survey. Phys. Med. Biol. https://doi.org/10.1088/1361-6560/ac9cb3 (2022).
Shi, F. et al. Deep learning empowered volume delineation of whole-body organs-at-risk for accelerated radiotherapy. Nat. Commun. 13, 6566. https://doi.org/10.1038/s41467-022-34257-x (2022).
Chino, J. et al. The ASTRO clinical practice guidelines in cervical cancer: Optimizing radiation therapy for improved outcomes. Gynecol. Oncol. 159, 607–610. https://doi.org/10.1016/j.ygyno.2020.09.015 (2020).
Lim, K. et al. Consensus guidelines for delineation of clinical target volume for intensity-modulated pelvic radiotherapy for the definitive treatment of cervix cancer. Int. J. Radiat. Oncol. Biol. Phys. 79, 348–355. https://doi.org/10.1016/j.ijrobp.2009.10.075 (2011).
Shafiq, M. & Gu, Z. Deep residual learning for image recognition: A survey. Appl. Sci. https://doi.org/10.3390/app12188972 (2022).
Ronneberger, O., Fischer, P. & Brox, T. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18, 234–241 (Springer).
Schmidt, M. C. et al. Technical report: Development and implementation of an open source template interpretation class library for automated treatment planning. Pract. Radiat. Oncol. 12, e153–e160. https://doi.org/10.1016/j.prro.2021.11.004 (2022).
Prunaretty, J. et al. Quantitative evaluation of a fully automated planning solution for prostate-only and whole-pelvic radiotherapy. Cancers https://doi.org/10.3390/cancers16223735 (2024).
Buschmann, M. et al. Automated volumetric modulated arc therapy planning for whole pelvic prostate radiotherapy. Strahlenther. Onkol. 194, 333–342. https://doi.org/10.1007/s00066-017-1246-2 (2018).
Goddard, L. et al. Evaluation of multiple-vendor AI autocontouring solutions. Radiation Oncology 19, 69 (2024).
Chen, W. et al. A comparative study of auto-contouring softwares in delineation of organs at risk in lung cancer and rectal cancer. Sci. Rep. 11, 23002 (2021).
Ju, Z. et al. Automatic segmentation of pelvic organs-at-risk using a fusion network model based on limited training samples. Acta Oncol. 59, 933–939. https://doi.org/10.1080/0284186X.2020.1775290 (2020).
Liu, Z. et al. Development and validation of a deep learning algorithm for auto-delineation of clinical target volume and organs at risk in cervical cancer radiotherapy. Radiother. Oncol. 153, 172–179. https://doi.org/10.1016/j.radonc.2020.09.060 (2020).
Liu, Z. et al. Segmentation of organs-at-risk in cervical cancer CT images with a convolutional neural network. Phys. Med. 69, 184–191. https://doi.org/10.1016/j.ejmp.2019.12.008 (2020).
Acknowledgements
Thanks to American Journal Expert for help with language editing. The authors thank Home for Researchers (www.home-for-researchers.com) for English language editing. Thanks to Manteia for technical support. We are deeply indebted to our editor and reviewers, a great part of the discussion and the development beyond the study original intent owe to their insightful comments.
Author information
Authors and Affiliations
Contributions
L YC: Manuscript drafting, editing, and statistical analysis. C WJ: Design, supervision, data interpretation, and critical review. Q LY, S KN, Z WM and: Patient surveillance and data acquisition. J YS and D JN: analyzed parts of the data, and interpreted the data. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yucheng, L., Lingyun, Q., Kainan, S. et al. Development and validation of a deep reinforcement learning algorithm for auto-delineation of organs at risk in cervical cancer radiotherapy. Sci Rep 15, 6800 (2025). https://doi.org/10.1038/s41598-025-91362-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-91362-9