Abstract
We investigated whether baseline levels of biomarkers related to endotheliopathy, thromboinflammation, and fibrosis were associated with clinical outcomes in hospitalized COVID-19 patients. We analyzed the associations between baseline levels of 21 biomarkers and time to hospital discharge and change in NEWS-2 score in patients from DisCoVeRy trial. We fitted multivariate models adjusted for baseline ISARIC 4C score, disease severity, D-dimer values, and treatment regimen. Between March 22 and June 29, 2020, 603 participants were randomized; 454 had a sample collected at baseline and analyzed. The backward selection of multivariate models showed that higher baseline levels of soluble suppressor of tumorigenicity 2 (sST2) and nucleosomes were statistically associated with a lower chance of hospital discharge before day 29 (sST2: aHR 0.24, 95% CI [0.15–0.38], p < 10−9; nucleosomes: aHR 0.62, 95% CI [0.48–0.81], p < 10−3). Likewise, higher levels of baseline sST2 were statistically associated with lower changes in the NEWS-2 score between baseline and day 15 (adjusted beta 4.47, 95% CI [2.65–6.28], p < 10−5). Moreover, we evaluated sST2 involvement in a confirmation cohort (SARCODO study, 103 patients) and found that elevated baseline sST2 levels were significantly associated with lower rates of hospital discharge before day 29 and a higher model performance (AUC at day 29 of 92%) compared to models without sST2. sST2 emerged as an independent predictor of clinical outcomes in two large cohort of hospitalized COVID-19 patients, warranting further investigation to elucidate its role in disease progression and potential as a therapeutic target.
Similar content being viewed by others
Introduction
The emergence of SARS-CoV-2 sparked a global COVID-19 crisis, with the case fatality ratio dropping from 2% in February 2020 to below 0.3% in 20221. Beyond pulmonary entanglements culminating in acute respiratory distress syndrome (ARDS)2, severe COVID-19 cases exhibit an array of extrapulmonary manifestations. These include acute kidney injury, acute cardiac damage, coagulation dysregulation, thromboembolic episodes in particularly pulmonary embolism, and disseminated microthrombi observed in several autopsy studies3,4. To date, biomarker research has primarily focused on unraveling pathophysiology, with limited applications in predicting outcomes, severity, and in-hospital mortality. However, studies on inflammation, coagulopathy, and endotheliopathy suggest potential biomarkers for disease severity5,6. These include hypercoagulation, evidenced by the presence of fibrin degradation products (D-dimer), which are associated markers with COVID-19 severity and outcomes7. Direct endothelial infection by SARS-CoV-2 remains uncertain8. Endotheliopathy-driven dysfunction could fuel hypercoagulation by increasing the levels of von Willebrand factor (VWF)9. Beyond thrombotic events, recent studies have highlighted the role of pathological angiogenesis3,5 which could give rise to subsequent fibrotic remodeling10,11.
The objective of our study was to examine the associations between baseline levels of various circulating biomarkers linked to thromboinflammation, endothelial damage, angiogenesis, or fibrosis, and clinical outcomes in hospitalized COVID-19 patients participating in the European randomized controlled study DisCoVeRy12. The biomarker identified in the primary analysis was tested in an independent confirmation cohort of hospitalized COVID-19 patients (SARCODO study)9.
Material and methods
Participants
This protocol is based on the protocol produced by the National Institute of Health for the World Health Organization (WHO), version of 03/03/2020, which further led to the Solidarity protocol of WHO. This study supports the integration of the Solidarity WHO trial worldwide. Hospitalized patients with a laboratory-confirmed SARS-CoV-2 infection were enrolled in the DisCoVeRy trial13, sponsored by the Institut national de la santé et de la recherche médicale (Inserm, France). The aim of this trial was to compare the effectiveness of standard of care (SoC) plus lopinavir/ritonavir (SoC + Lopi/Rito), SoC plus lopinavir/ritonavir-interferon (IFN)-β-1a (SoC + Lopi/Rito + IFB), or SoC plus hydroxychloroquine (Soc + HCQ) to SoC alone, in hospitalized patients during the first wave of the COVID-19 pandemic. Written informed consent was obtained from all participants or their legal representatives if they were unable to consent. From March 22 to June 29, 2020, a total of 603 individuals were randomly assigned to different treatment groups at 30 locations in France and two in Luxembourg. The study included adults aged 18 and older who were hospitalized due to a PCR-confirmed SARS-CoV-2 infection, exhibiting pulmonary rales or crackles, and having a peripheral oxygen saturation of 94% or lower, or who required supplemental oxygen. Patients were randomly allocated in a 1:1:1:1 ratio to receive SoC alone, SoC + Lopi/Rito, SoC + Lopi/Rito + IFB, or SoC + HCQ as previously described12,13. The trial was conducted in accordance with the Declaration of Helsinki and national laws and regulations, approved by the Ethics Committee (CPP Ile-de-France-III, approval #20.03.06.51 744), and declared on the clinicaltrials.gov registry (NCT04315948, first posted on 20/03/2020).
Biomarker measurements
Blood samples were collected just after inclusion/randomization and before administration of an investigational drug (baseline) in ethylenediaminetetraacetic acid (EDTA). Platelet-poor plasma (PPP) was obtained after centrifugation at 2500 × g for 15 min and stored at − 80 °C until analysis. The D-dimer level was measured with the immunoturbidimetric STA®—Liatest® D-Di PLUS (STAGO Asnières sur Seine, France). The concentrations of nucleosomes, LDH, citrullinated Histone H3 (H3cit), CRP, and P-selectin were determined using ELISA respectively from Sigma-Aldrich (nucleosomes and LDH), Cayman Chemical (H3cit) and R&D Systems (CRP and P-selectin). The plasma concentrations of angiopoietin-1 and -2, CA 15-3/MUC-1, CD40 Ligand/TNFSF5, Ferritin, FGF-2, IL-6, PDGF-BB, PIGF, soluble Suppressor of tumorigenicity 2 (ST2; receptor of soluble IL33), VEGF-A, ICAM1/CD54, Syndecan-1; VWF, D-dimers, E-selectin, VCAM-1/CD106 and Troponin I were quantified in PPP using a Human Magnetic Luminex Assay (R&D Systems, Minneapolis, MN). Data were assessed with the Bio-Plex 200 using the Bio-Plex Manager 5.0 software (Bio-Rad, Marnes-la-Coquette, France). Cell-free DNA was measured by spectrofluorimetry at 520 nm after excitation at 480 nm with Quant-iT PicoGreen dsDNA assay (Thermo Fischer Scientific, Waltham, MA) on a SAFAS spectrophotometer (Monaco, France).
Outcomes
The two main outcomes considered were the time to hospital discharge before day 29, and the change in National Early Warning Score (NEWS-2 score) between baseline and day 15.
-
The time to hospital discharge before day 29 was computed as the number of days between the randomization date, and the date of discharge from the index hospitalization, censored at day 29. Death before day 29 was treated as a competing risk of hospital discharge.
-
The NEWS-2 score is a standardized clinical scoring system developed for improved detection of deterioration in acutely ill patients that has demonstrated good performance in COVID-19 patients14,15,16.
The National Early Warning Score (NEWS) was introduced in 2012 to facilitate the early detection of clinical deterioration in acutely ill patients within NHS acute medical units17. Its primary aim was to establish a standardized system for tracking, scoring, and responding to physiological changes. The system is based on the principle that early recognition and prompt intervention significantly impact patient outcomes. Due to its effectiveness, NEWS has been widely adopted across the NHS and internationally. NEWS functions as a cumulative scoring system that assigns numerical values to routine physiological measurements recorded during patient monitoring. In 2017, an updated version, NEWS 2, was introduced, aligning its structure with the Resuscitation Council UK’s ABCDE framework18. The system evaluates six key physiological parameters: respiratory rate, oxygen saturation, systolic blood pressure, pulse rate, temperature, and level of consciousness or new confusion. Each parameter is assigned a score from 0 to 3, with higher scores indicating greater deviation from normal values and an increased risk of clinical deterioration18. The change in NEWS-2 score between baseline and day 15 was computed as the difference between the scores assessed at day 15 and at baseline. For participants who died before day 15, the NEWS-2 score was imputed to worst possible value (i.e. NEWS-2 score of 20). The proportion of patients that had died by day 29 was analyzed as a post-hoc secondary outcome.
Covariates
All analyses were adjusted on four baseline covariates: (1) the ISARIC 4C mortality score, (2) the disease severity at baseline during randomization (moderate: WHO 7-point ordinal scale 3 or 4; or severe: WHO 7-point ordinal scale 5 or 6), (3) the log10 D-dimer value at baseline, and (4) the treatment regimen: SoC, Soc + HCQ, SoC + Lopi/Rito, and SoC + Lopi/Rito + IFB. The D-dimer levels at baseline have been shown to be associated with clinical outcomes of hospitalized COVID-19 patients7. The disease severity at randomization and the ISARIC 4C mortality score have been selected as they are key markers of clinical severity of patients at baseline. The ISARIC 4C mortality score is a risk stratification score that predicts in-hospital mortality for hospitalized COVID-19 patients, produced by the ISARIC 4C consortium19,20. Developed by the International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC), this score integrates clinical, demographic, and laboratory parameters to assess disease severity. It is calculated using eight variables: age, sex, number of comorbidities (0, 1, 2 or more), respiratory rate, oxygen saturation, Glasgow Coma Scale score, blood urea level, and C-reactive protein level. Increased BMI is considered as a comorbidity. Each parameter is assigned a numerical score based on its deviation from the normal range, resulting in a total score between 0 and 21, where higher values indicate an increased risk of mortality. The stratification system categorizes patients into different risk groups to aid clinical decision-making. The classification system defines risk groups based on the total score. A low-risk classification applies to patients scoring 0 to 3, corresponding to an estimated 1.2% mortality risk. The intermediate-risk category includes scores from 4 to 8, where mortality risk increases to 9.9%. The high-risk category consists of patients with scores 9 or above, who have an estimated 48.2% mortality rate. These thresholds provide essential guidance for clinicians in triaging COVID-19 patients and optimizing medical resource allocation. The ISARIC 4C Mortality Score has been externally validated and remains one of the most reliable predictors of mortality in hospitalized COVID-19 patients. When one of the variables needed to compute the ISARIC 4C score was missing, the missing 4C score was imputed by a single multivariate imputation using a predictor matrix comprised of the outcome variables (i.e. NEWS-2 score, time to hospital discharge, event of discharge before day 29, event of death before day 29), and the 8 variables used to compute the score. Missing values for other variables were not imputed.
Statistical analysis
The baseline concentration of the biomarkers, and other continuous variables, were described by their median value and interquartile range. Categorical variables were described by the absolute and relative frequencies. For some descriptive analyses, the ISARIC 4C score was considered categorical, using the categories defined by the ISARIC consortium: low mortality risk [0–3], intermediate mortality risk (3,8], high mortality risk (8,14], very high mortality risk (14,21]. First, associations between the log10 baseline values of the biomarkers and disease severity at baseline during randomization (moderate or severe) were tested with univariate logistic regression, and the p-values, q-values (according to Storey’s procedure), as well as the area under receiver operating characteristic (ROC) curves (AUC) were reported. Given that disease severity at randomization was a stratification factor in the DisCoVeRy study, and to maintain consistency with previous work, associations between baseline covariates and disease severity at randomization were evaluated using Wilcoxon rank-sum tests, Pearson’s chi-squared tests, or Fisher’s exact tests, as appropriate. The proportions of patients discharged from hospital by day 29, considering the competing risk of death before day 29, were estimated using the Aalen-Johansen estimator (equivalent of the Kaplan–Meier method in the competing risk setting), and compared according to disease severity at randomization using a normal approximation. Correlations coefficients between each pair of biomarkers were computed on log10 values and tested using Pearson test. To assess the relationships between the four baseline covariates and clinical outcomes, we first fitted multivariate models that included only the covariates. The time to hospital discharge before day 29 was analyzed using a multivariate proportional-hazards model with a Fine and Gray approach considering the competing risk of death before day 29 (R package “survival”). The change in NEWS-2 score between baseline and day 15 was analyzed using a multivariate linear model. Then, to assess the individual association between the log10 baseline concentration of the biomarkers and the two outcomes, we included each marker individually in the models along with the rest of the covariates. Finally, we fitted models to assess the association between a combination of biomarkers and the two outcomes. To do so, biomarkers individually associated with the outcomes with a p-value less than or equal to 0.20 were selected using a backward selection process, with the covariates forced in the final model. The goodness of fit of proportional-hazards models were assessed using cumulative/dynamic time-dependent AUC. The goodness of fit of the different models were compared statistically using the time-dependent AUC evaluated at t = 29 (R package “timeROC”). The confidence intervals and p-values were computed using the jackknife approach. In a secondary, post-hoc analysis, the associations between the log10 biomarker concentrations and the mortality by day 29 was assessed by multivariate logistic regressions, with the same approach as the other outcomes. All analyses were done using the R software, version 4.3.1.
Post-hoc analyses in SARCODO cohort
Post-hoc analyses were performed on a confirmation cohort of hospitalized patients with a laboratory-confirmed SARS-CoV-2 infection enrolled in the SARCODO cohort (NCT04624997), sponsored by the Assistance Publique–Hôpitaux de Paris. This confirmation cohort is a monocentric cross-sectional study of adult (> 18-years old) COVID-19 hospitalized patients in European Georges Pompidou Hospital between March 13 and June 5, 2020. Covid-19 disease severity was classified according to the WHO Working Group on the Clinical Characterization and Management of Covid-19 infection as previously described9. Written informed consent was obtained from all participants or their legal representatives if they were unable to consent. 103 patients for this confirmation cohort were included as previously described9,21. At the time of hospital admission for suspected COVID-19, routine laboratory tests and sST2 measurements were conducted within the first 48 h. Venous blood samples were taken from patients and handled using standard laboratory procedures. The blood was collected in 0.129M trisodium citrate tubes (9NC BD Vacutainer, Plymouth, UK). Platelet-poor plasma (PPP) was prepared by centrifuging the blood twice at 2500 g for 15 min, and then stored at – 80 °C until further analysis. The plasma concentrations of sST2 were quantified in PPP using a Human Quantikine Elisa (R&D Systems, Minneapolis, MN). Demographic, clinical and biological characteristics, and clinical outcomes of patients included in SARCODO were described by their median value and interquartile range (continuous variables), and by the absolute and relative frequencies (categorical variables). The confirmation cohort of the SARCODO trial did not have the variables necessary to compute the NEWS-2 score, so the post-hoc analyses were performed only on the time to hospital discharge before day 29, defined as for the DisCoVeRy analysis. Moreover, as the treatment strategies used in DisCoVeRy were not used in SARCODO, and as the disease severity at baseline was not available, these variables were not included in the models. The biomarkers selected in the final models of DisCoVeRy were tested in SARCODO in models including ISARIC 4C-score and D-dimer, using the same statistical procedures as for the DisCoVeRy analysis.
Results
Baseline characteristics and outcomes description
Of the 603 participants randomized in the Discovery trial, 593 were evaluable for analysis. Of them, 454 had a sample collected at baseline and measured (Fig. 1 and supplementary Table S1).
Of the patients who had at least one baseline sample, 323 (71%) were men, with a median age of 63 years (IQR 54–71) and a median BMI of 27.7 kg/m2 (IQR 25.2–32.0). The median time from symptom onset to hospitalization was 9 days (IQR 7–12), and 309 patients (68%) had a moderate disease severity assessed at baseline, whereas 145 (32%) were severe. Among the 309 patients with moderate disease severity, 281 (91%) needed only supplemental oxygen. In contrast, of the 145 patients classified as having severe disease, 35 (24%) required non-invasive ventilation or high-flow oxygen devices, while 96 (66%) necessitated invasive ventilation or extracorporeal membrane oxygenation (ECMO). Overall, 25 patients had died by day 29 (Table 1).
The ISARIC 4C score was significantly associated with disease severity at randomization. As previously described, all biomarkers of thromboinflammation, endothelial dysfunction or fibrosis were associated with disease severity at randomization, notably D-dimer, Ang-2, syndecan-1 and soluble Suppressor of Tumorigenicity 2 (sST2) (Table 2). The two outcomes, namely time to hospital discharge before day 29 and change in NEWS-2 score between baseline and day 15, were both associated with disease severity at randomization (Table 1). The results of the multivariate models that included only the covariates showed that a higher baseline log10 concentration of D-dimer, a greater ISARIC 4C score, and a more severe disease at baseline were associated with lower rates of hospital discharge before day 29 (supplementary Figures S1A and S2). On the other hand, a greater ISARIC 4C score and a more severe disease at baseline were associated with lower changes of NEWS-2 score between baseline and day 15 (supplementary Figures S1B and S3).
Baseline soluble ST2 is the best biomarker associated with the time to hospital discharge before day 29 and NEWS-2 score, in DisCoVeRy
We then explored the association between the log10 baseline concentrations of each of the 21 biomarkers and the two outcomes in Discovery (Table 3). Fourteen and eight biomarkers had a p-value less than or equal to 0.20 for the association with the time to hospital discharge before day 29, and the change in NEWS-2 score between baseline and day 15, respectively. Figure 2A,B show the results of the final model after backward selection.
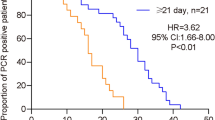
Among the 14 biomarkers included in the selection of the model of time to hospital discharge before day 29, only sST2 (aHR 0.25; 95% CI [0.16–0.39]; p < 10−8) and the nucleosomes (aHR 0.63; 95% CI [0.48–0.81]; p < 10−3) remained in the model. Higher baseline levels of these markers were significantly associated with lower rates of hospital discharge before day 29, after having adjusted for the ISARIC 4C score, the disease severity at baseline, the treatment regimen, and the baseline D-dimer level (Figs. 2A, 3A and S4).
Regarding the goodness of fit, the final model had a time-dependent AUC at t = 29 of 86% (95% CI [82–89]), while the model containing only the covariates had a time-dependent AUC at t = 29 of 83% (95% CI [79–87]), which was statistically significantly lower (p-value = 0.047) (Fig. 4A).
Among the 8 biomarkers included in the model of the change in NEWS-2 score between baseline and day 15, only sST2 remained, higher baseline levels of the marker being significantly associated with lower changes of NEWS-2 score between baseline and day 15 (adjusted beta 4.75; 95% CI [2.92–6.57]; p < 10−6) (Figs. 2B and 3B). The final model had a goodness of fit (adjusted R-squared) of 0.09 (95% CI [0.04–0.14]), while the model with only the covariates had a goodness of fit of 0.037 (95% CI [0.004–0.072]). The post-hoc analysis of mortality by day 29 showed that the association with the baseline concentrations of sST2 was statistically significant when adjusted for the other baseline covariates (p-value < 10−2, supplementary Figure S6). Scatterplots were drawn to visualize the statistically significant (p-value < 0.05) correlations between ST2/IL-33R and the other biomarkers in DisCoVeRy (Fig. 5). Interestingly, sST2 was correlated with circulating DNA, angiogenic biomarker PlGF or endotheliopathy markers Syndecan-1 and VCAM-1 (Pearson’s coefficients of 0.46 for DNA, 0.41 for PlGF, 0.40 for Syndecan-1 and 0.37 for VCAM-1).
Association between sST2 and time to hospital discharge before day 29 in SARCODO
Demographic, clinical and biological characteristics, and clinical outcomes of the 103 SARCODO participants are presented in Supplementary Table S2. As the nucleosomes’ baseline concentrations and the NEWS-2 score were not available in the SARCODO data, we only explored the association between sST2 and time to hospital discharge before day 29. The median value of sST2 at baseline was 41,918 pg/mL (IQR [22,321–106,059]). Our findings indicated that elevated sST2 levels were significantly associated with lower rates of hospital discharge before day 29, even after adjusting for the ISARIC 4C score and baseline D-dimer levels: aHR 0.13 (95% CI [0.06–0.27], p-value < 10−7) (Fig. 2C).
Regarding the goodness of fit of the model, the SARCODO model with sST2 had a time-dependent AUC at t = 29 of 92% (95% CI [86–97]), while the SARCODO model with only ISARIC and D-dimer had a time-dependent AUC at t = 29 of 79% (95% CI [68–89]), which was statistically significantly lower (p-value = 0.011) (Fig. 4B). Even a model with sST2 alone had a better time-dependent AUC at t = 29 than a model with ISARIC + D-dimer, with 90% (95% CI [85–96], p-value = 0.041).
Discussion
In this study, we address the relationship between SARS-CoV-2-mediated thromboinflammation, endotheliopathy, angiogenesis and fibrosis at hospital admission and the severity of COVID-19 in a large cohort of well-characterized patients. Among the top five biomarkers associated with outcomes, a consistent pathophysiological significance is observed across both primary endpoints. DNA, particularly its association with nucleosome levels, underscores the role of Damage-Associated Molecular Patterns (DAMPs) in COVID-19 pathology22,23. DAMPs, including nucleosomes and extracellular DNA, contribute to thromboinflammation and endotheliopathy, two key mechanisms driving severe disease progression5,24. The release of DNA and nucleosomes into circulation, often due to cell death and immune activation, can trigger inflammatory cascades and coagulation disturbances, exacerbating vascular dysfunction and organ injury in severe COVID-19 cases22,23,24. Furthermore, VCAM-1 and von Willebrand factor (vWF) have emerged as critical biomarkers of endothelial dysfunction, reinforcing the connection between endotheliopathy and disease severity5,9,25,26. Recent scientific investigations have highlighted their relevance, particularly in the context of COVID-19-associated coagulopathy and microvascular complications26. Prior studies, including our own, have demonstrated a strong link between inflammatory endotheliopathy and COVID-19 mortality, further validating the role of endothelial damage as a key driver of adverse outcomes in critically ill patients9,25,27. This growing body of evidence strengthens the understanding of DAMPs, nucleosomes, and endotheliopathy as interconnected pathophysiological pathways that contribute to the prothrombotic and inflammatory state observed in severe COVID-195,24. Results from the panel of 21 biomarkers tested on COVID-19 inpatients included in the DisCoVeRy trial establish that the stronger association between a biomarker and clinical outcomes was sST2 blood levels. DNA levels have been found significantly associated to outcomes. However, sST2 appears to be more specifically associated with outcomes, inflammatory and endothelial dysregulation seen in COVID-19. In our study, sST2 demonstrated stronger predictive performance for the two primary outcomes compared to DNA. This difference may be attributed to sST2’s role as a soluble receptor involved in the IL-33/ST2 axis, a pathway critically implicated in inflammation and fibrosis. Indeed, patients with elevated sST2 levels at baseline, regardless of the disease severity at baseline, were less likely to have recovered from initial clinical deterioration by day 15 (assessed by the NEWS-2 score), and less likely to have been discharged from hospital before day 29.
ST2, the soluble form of IL-33 receptor is expressed across various cell types, such as macrophages, neutrophils, lymphocytes, endothelial cells, cardiomyocytes, osteoclasts, osteoblasts, and adipocytes28. Recent years have witnessed extensive deliberation over the role of sST2 as a diagnostic and prognostic biomarker across several clinical conditions, such as acute and chronic heart failure (HF), myocardial fibrosis29 and diabetic and critical limb ischemia30. The lungs are a significant source of sST2 in heart failure and appear to be actively involved in the pathological response associated with ST229. Additionally, the role of sST2 has also been proposed in pulmonary diseases such as acute respiratory distress syndrome31 or idiopathic pulmonary fibrosis 32,33 by contributing to inflammation and fibrotic processes34,35. Regarding COVID-19, sST2/IL-33 axis has been proposed as a relevant predictive factor for severity, intensive care unit direct admission and in-hospital mortality in COVID-19 patients but also immunization level after SARS-CoV-2 infection36,37,38,39,40. However, the various associations between sST2 and preexisting comorbidities could act as a confounding factor in our analysis of the relationship between ST2 and clinical outcomes. To address this, our models incorporate the ISARIC 4C score, which accounts for the number of preexisting comorbidities. This adjustment strengthens the validity of our findings and supports the potential of ST2 as a robust biomarker in COVID-19. Finally, a correlation has been observed between elevated soluble ST2 levels and high scores of Endothelial Activation and Stress Index (EASIX) in COVID-19 cases41. This work indeed proposed a link between endothelial activation and ST2 regulation in COVID-19.
In the large DisCoVery clinical trial, sST2 was a stronger predictor of severity and time to hospital discharge than endotheliopathy biomarkers. Nonetheless, unraveling the precise mechanism and cell origin in this context proves to be unattainable. Given that endotheliopathy4,6 is a focal point in the pathophysiology of COVID-19, and considering the identified associations between sST2 and angiogenic or endotheliopathy markers in the current study, it is crucial to acknowledge the connection between vasculopathy and sST2/IL-33 axis. ST2 has been described to be crucial for revascularization after hind limb ischemia in mice. Indeed, laser doppler imaging showed that hindlimb blood flow remained severely impaired in ST2−/− mice associated with reduced neovascularization in the gastrocnemius muscle and impaired neovascularization in a Matrigel implant model42. In addition, the expression of the angiogenic gene VEGF-A was more regulated in ST2−/− mice than in WT mice42. This result is interesting regarding COVID-19 and its long-term complications. Indeed, we recently described that VEGF-A was increased during follow-up post COVID-19 and its increase was related to impairment in the diffusing capacity of the lung for carbon monoxide and radiological sequelae11. The link between ST2 and endotheliopathy and vasculopathy is of interest. The reason is that intussusceptive angiogenesis and lobular micro-ischemia might be associated with distinctive forms of fibrotic interstitial lung disease that contribute to long COVID-1910, therefore linking ST2 and endotheliopathy/vasculopathy.
Understanding the sequence of pathophysiology regarding respiratory sequelae could help researchers to appreciate and identify new therapeutic approaches for SARS-CoV-2 infection. First, several strategies have been proposed to block the ST2/IL33 axis in various clinical conditions. Anti-ST2-conjugated nanoparticles have been shown to control lung inflammation by reducing the ability of group 2 innate lymphoid cells to produce IL-5 and IL-13, thereby reducing CD4+T cells43. Targeting the IL33-ST2 axis with monoclonal antibodies has been proposed as an alternative treatment for cancer44 and organ fibrosis45. Exploring additional treatment approaches beyond antiviral medications and steroids is essential for individuals with severe COVID-19, especially in the case of immunocompromised patients, who continue to experience elevated morbidity and mortality rates. Blocking ST2 could be a potential approach and should to be tested in appropriate animal models and preclinical studies. Second, fibrosis may be linked to micro-ischemia and angiogenesis remodeling processes that are observed in COVID-1910. If so, blocking angiogenesis-induced micro-ischemia and fibrotic processes with anti-angiogenic drugs could help to stop the progression of respiratory sequelae.
Thus, despite being a non-specific inflammatory biomarker46,47, sST2 exhibited strong predictive value for hospitalization duration. The unique aspect of sST2 lies in its involvement in both systemic inflammation and endotheliopathy, two hallmarks of severe COVID-195. Unlike other biomarkers, sST2 reflects a broader spectrum of pathophysiological changes, including endothelial activation, fibrosis, and vascular remodeling. These processes are critical in determining clinical deterioration and recovery timelines in COVID-19. Moreover, our statistical analysis showed that sST2 had a higher discriminative performance compared to other biomarkers, as evidenced by its superior AUC values. This strengthens the case for sST2 as a key prognostic biomarker in COVID-19. However, this study has certain limitations. Although the multivariate models were adjusted for covariables that were strongly associated with the outcomes, we cannot exclude the possibility of residual confusion arising from unmeasured variables. In addition, given the exploratory nature of this study, the models were fit on the entire dataset; we did not employ training and validation sets. Although post-hoc analyses of an external cohort of hospitalized COVID-19 patients (SARCODO) confirmed the strong association between sST2 and time to hospital discharge before day 29, further studies are needed to obtain external validation of our findings. Finally, this study has been realized during the first wave of pandemic and we need further studies to ascertain whether this holds true for all variants of SARS-CoV-2.
All in all, this randomized controlled trial unequivocally establishes a robust association between circulating sST2 levels and clinical severity upon hospitalization. Remarkably, sST2 emerges as the best biomarker for predicting the time to hospital discharge. Our study, pioneering in its evaluation of an extensive spectrum of biomarkers within a large cohort, not only positions sST2 as a pivotal biomarker but also suggests its potential as an active participant in the pathophysiology of COVID-19. The prospect of blocking sST2 emerges as a groundbreaking therapeutic avenue, offering a new and promising approach to treating individuals afflicted with severe SARS-CoV-2 infection.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
Horita, N. & Fukumoto, T. Global case fatality rate from COVID-19 has decreased by 96.8% during 2.5 years of the pandemic. J. Med. Virol. 95, e28231. https://doi.org/10.1002/jmv.28231 (2023).
Diehl, J.-L. et al. Respiratory mechanics and gas exchanges in the early course of COVID-19 ARDS: A hypothesis-generating study. Ann. Intensive Care 10, 95. https://doi.org/10.1186/s13613-020-00716-1 (2020).
Ackermann, M. et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. N. Engl. J. Med. 383, 120–128. https://doi.org/10.1056/NEJMoa2015432 (2020).
Heinrich, F. et al. Using autopsies to dissect COVID-19 pathogenesis. Nat. Microbiol. 8, 1986–1994. https://doi.org/10.1038/s41564-023-01488-7 (2023).
Smadja, D. M. et al. COVID-19 is a systemic vascular hemopathy: Insight for mechanistic and clinical aspects. Angiogenesis 24, 755–788. https://doi.org/10.1007/s10456-021-09805-6 (2021).
Smadja, D. M. et al. Circulating Von Willebrand factor: A consistent biomarker predicting in-hospital mortality across different waves of the COVID-19 pandemic. Angiogenesis https://doi.org/10.1007/s10456-023-09901-9 (2023).
Chocron, R. et al. D-dimer at hospital admission for COVID-19 are associated with in-hospital mortality, independent of venous thromboembolism: Insights from a French multicenter cohort study. Arch. Cardiovasc. Dis. 114, 381–393. https://doi.org/10.1016/j.acvd.2021.02.003 (2021).
McCracken, I. R. et al. Lack of evidence of angiotensin-converting enzyme 2 expression and replicative infection by SARS-CoV-2 in human endothelial cells. Circulation 143, 865–868. https://doi.org/10.1161/CIRCULATIONAHA.120.052824 (2021).
Philippe, A. et al. Circulating Von Willebrand factor and high molecular weight multimers as markers of endothelial injury predict COVID-19 in-hospital mortality. Angiogenesis 24, 505–517. https://doi.org/10.1007/s10456-020-09762-6 (2021).
Ackermann, M. et al. The fatal trajectory of pulmonary COVID-19 is driven by lobular ischemia and fibrotic remodelling. EBioMedicine 85, 104296. https://doi.org/10.1016/j.ebiom.2022.104296 (2022).
Philippe, A. et al. VEGF-A plasma levels are associated with impaired DLCO and radiological sequelae in long COVID patients. Angiogenesis https://doi.org/10.1007/s10456-023-09890-9 (2023).
Ader, F. et al. An open-label randomized controlled trial of the effect of lopinavir/ritonavir, lopinavir/ritonavir plus IFN-β-1a and hydroxychloroquine in hospitalized patients with COVID-19. Clin. Microbiol. Infect. 27, 1826–1837. https://doi.org/10.1016/j.cmi.2021.05.020 (2021).
Ader, F. et al. Final results of the DisCoVeRy trial of remdesivir for patients admitted to hospital with COVID-19. Lancet Infect. Dis. 22, 764–765. https://doi.org/10.1016/S1473-3099(22)00295-X (2022).
Gidari, A., De Socio, G. V., Sabbatini, S. & Francisci, D. Predictive value of national early warning score 2 (NEWS2) for intensive care unit admission in patients with SARS-CoV-2 infection. Infect. Dis. 52, 698–704. https://doi.org/10.1080/23744235.2020.1784457 (2020).
Kostakis, I. et al. The performance of the National Early Warning Score and National Early Warning Score 2 in hospitalised patients infected by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Resuscitation 159, 150–157. https://doi.org/10.1016/j.resuscitation.2020.10.039 (2021).
Williams, B. Evaluation of the utility of NEWS2 during the COVID-19 pandemic. Clin. Med. (Lond.) 22, 539–543. https://doi.org/10.7861/clinmed.2022-news-covid (2022).
Royal College of Physicians. National Early Warning Score (NEWS): Standardising the Assessment of Acute-Illness Severity in the NHS; Report of a Working Party (2012)
Royal College of Physicians. National Early Warning Score (NEWS) 2: Standardising the Assessment of Acute-Illness Severity in the NHS; Updated Report of a Working Party (2017).
Knight, S. R. et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: Development and validation of the 4C mortality score. BMJ 370, m3339. https://doi.org/10.1136/bmj.m3339 (2020).
Gupta, R. K. et al. Development and validation of the ISARIC 4C deterioration model for adults hospitalised with COVID-19: A prospective cohort study. Lancet Respir. Med. 9, 349–359. https://doi.org/10.1016/S2213-2600(20)30559-2 (2021).
Smadja, D. M. et al. Placental growth factor level in plasma predicts COVID-19 severity and in-hospital mortality. J. Thromb. Haemost. 19, 1823–1830. https://doi.org/10.1111/jth.15339 (2021).
Garcia, G. et al. Impaired balance between neutrophil extracellular trap formation and degradation by DNases in COVID-19 disease. J. Transl. Med. 22, 246. https://doi.org/10.1186/s12967-024-05044-7 (2024).
Prével, R. et al. Plasma markers of neutrophil extracellular trap are linked to survival but not to pulmonary embolism in COVID-19-related ARDS patients. Front Immunol. 13, 851497. https://doi.org/10.3389/fimmu.2022.851497 (2022).
Bonaventura, A. et al. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID-19. Nat. Rev. Immunol. 21, 319–329. https://doi.org/10.1038/s41577-021-00536-9 (2021).
Smadja, D. M. et al. Angiopoietin-2 as a marker of endothelial activation is a good predictor factor for intensive care unit admission of COVID-19 patients. Angiogenesis 23, 611–620. https://doi.org/10.1007/s10456-020-09730-0 (2020).
Libby, P. Endothelial inflammation in COVID-19. Science 386, 972–973. https://doi.org/10.1126/science.add2962 (2024).
Khider, L. et al. Curative anticoagulation prevents endothelial lesion in COVID-19 patients. J. Thromb. Haemost. 18, 2391–2399. https://doi.org/10.1111/jth.14968 (2020).
Pascual-Figal, D. A. & Januzzi, J. L. The biology of ST2: The international ST2 consensus panel. Am. J. Cardiol. 115, 3B-7B. https://doi.org/10.1016/j.amjcard.2015.01.034 (2015).
Pascual-Figal, D. A. et al. Pulmonary production of soluble ST2 in heart failure. Circ. Heart Fail. 11, e005488. https://doi.org/10.1161/CIRCHEARTFAILURE.118.005488 (2018).
Caporali, A. et al. Soluble ST2 is regulated by p75 neurotrophin receptor and predicts mortality in diabetic patients with critical limb ischemia. Arterioscler Thromb. Vasc. Biol. 32, e149-160. https://doi.org/10.1161/ATVBAHA.112.300497 (2012).
Bajwa, E. K. et al. Prognostic and diagnostic value of plasma soluble suppression of tumorigenicity-2 concentrations in acute respiratory distress syndrome. Crit. Care Med. 41, 2521–2531. https://doi.org/10.1097/CCM.0b013e3182978f91 (2013).
Tajima, S., Oshikawa, K., Tominaga, S. & Sugiyama, Y. The increase in serum soluble ST2 protein upon acute exacerbation of idiopathic pulmonary fibrosis. Chest 124, 1206–1214. https://doi.org/10.1378/chest.124.4.1206 (2003).
Xu, J. et al. IL-33/ST2 pathway in a bleomycin-induced pulmonary fibrosis model. Mol. Med. Rep. 14, 1704–1708. https://doi.org/10.3892/mmr.2016.5446 (2016).
Li, D. et al. IL-33 promotes ST2-dependent lung fibrosis by the induction of alternatively activated macrophages and innate lymphoid cells in mice. J. Allergy Clin. Immunol. 134, 1422-1432.e11. https://doi.org/10.1016/j.jaci.2014.05.011 (2014).
Tajima, S. et al. ST2 gene induced by type 2 helper T cell (Th2) and proinflammatory cytokine stimuli may modulate lung injury and fibrosis. Exp. Lung Res. 33, 81–97. https://doi.org/10.1080/01902140701198583 (2007).
Cabrera-Garcia, D. et al. Plasma biomarkers associated with survival and thrombosis in hospitalized COVID-19 patients. Int. J. Hematol. 116, 937–946. https://doi.org/10.1007/s12185-022-03437-2 (2022).
Zeng, Z. et al. Serum-soluble ST2 as a novel biomarker reflecting inflammatory status and illness severity in patients with COVID-19. Biomark. Med. 14, 1619–1629. https://doi.org/10.2217/bmm-2020-0410 (2020).
Omland, T. et al. Soluble ST2 concentrations associate with in-hospital mortality and need for mechanical ventilation in unselected patients with COVID-19. Open Heart 8, e001884. https://doi.org/10.1136/openhrt-2021-001884 (2021).
Motloch, L. J. et al. Cardiovascular biomarkers for prediction of in-hospital and 1-year post-discharge mortality in patients with COVID-19 pneumonia. Front Med. (Lausanne) 9, 906665. https://doi.org/10.3389/fmed.2022.906665 (2022).
Stanczak, M. A. et al. IL-33 expression in response to SARS-CoV-2 correlates with seropositivity in COVID-19 convalescent individuals. Nat. Commun. 12, 2133. https://doi.org/10.1038/s41467-021-22449-w (2021).
Luft, T. et al. EASIX for prediction of outcome in hospitalized SARS-CoV-2 infected patients. Front. Immunol. 12, 634416. https://doi.org/10.3389/fimmu.2021.634416 (2021).
Dong, Q. et al. Interleukin-33 protects mice against hindlimb ischemic injury by enhancing endothelial angiogenesis. Int. Immunopharmacol. 109, 108850. https://doi.org/10.1016/j.intimp.2022.108850 (2022).
Wu, Y. et al. Anti-ST2 nanoparticle alleviates lung inflammation by targeting ILC2s-CD4+T response. Int. J. Nanomed. 15, 9745–9758. https://doi.org/10.2147/IJN.S268282 (2020).
Kudo-Saito, C. et al. IL33 is a key driver of treatment resistance of cancer. Cancer Res. 80, 1981–1990. https://doi.org/10.1158/0008-5472.CAN-19-2235 (2020).
Kim, Y. C. et al. ST2 blockade mitigates peritoneal fibrosis induced by TGF-β and high glucose. J. Cell Mol. Med. 23, 6872–6884. https://doi.org/10.1111/jcmm.14571 (2019).
Feng, Y. & He, L. Soluble ST2: A novel biomarker for diagnosis and prognosis of cardiovascular disease. Curr. Med. Sci. 44, 669–679. https://doi.org/10.1007/s11596-024-2907-x (2024).
Arora, H. et al. ST2 levels and neurodegenerative diseases: Is this a significant relation?. Ann. Med. Surg. (Lond.) 86, 2812–2817. https://doi.org/10.1097/MS9.0000000000001939 (2024).
Acknowledgements
We acknowledge all the patients enrolled in the DisCoVeRy and SARCODO trial and all the daily care staffs. We acknowledge the DisCoVeRy Steering Committee (by alphabetical order): Voting members: Florence Ader, Dominique Costagliola, Alpha Diallo, Alexander Egle, Hélène Espérou, Monika Halanova, Maya Hites, Bruno Lina, France Mentré, José-Artur Paiva, Marie-Thérèse Staub, Yazdan Yazdanpanah. Non-voting members: Pascale Augé, Drifa Belhadi, Diana Brainard, Charles Burdet, Mireille Caralp, Sandrine Couffin-Cadiergues, Aline Dechanet, Marta Del Alamo, Christelle Delmas, Jacques Demotes, Marina Dumousseaux, Monica Ensini, Joe Eustace, Richard Greil, Christophe Hezode, Fionnuala Keane, Marie-Paule Kiény, Soizic Le Mestre, Bernd Muehlbauer, Mickael Ohana, Nathan Peiffer Smadja, Ventzislava Petrov-Sanchez, Gilles Peytavin, Julien Poissy, Isabel Püntmann, Cécile Rabian, Jean Reuter, Juliette Saillard, Benjamin Terrier, Vida Terzic. We acknowledge the French DisCoVeRy Trial Management Team (by alphabetical order): Laurent Abel, Florence Ader, Toni Alfaiate, Claire Andrejak, Basma Basli, Drifa Belhadi, Lila Bouadma, Maude Bouscambert, Charles Burdet, Carole Cagnot, Anissa Chair, Dominique Costagliola, Sandrine Couffin-Cadiergues, Eric D’Ortenzio, Aline Dechanet, Christelle Delmas, Alpha Diallo, Axelle Dupont, Hélène Esperou, Claire Fougerou, Alexandre Gaymard, Ambre Gelley, Jérémie Guedj, Benjamin Hamze, Vinca Icard, Samira Laribi, Minh-Patrick Lê, Soizic Le Mestre, Delphine Lebrasseur- Longuet, François-Xavier Lescure, Benjamin Leveau, Bruno Lina, France Mentré, Noémie Mercier, Laëtitia Moinot, Florence Morfin-Sherpa, Marion Noret, Nathan Peiffer-Smadja, Ventzislava Petrov-Sanchez, Gilles Peytavin, Julien Poissy, Oriane Puéchal, Juliette Saillard, Marion Schneider, Caroline Semaille, Marie-Capucine Tellier, Jean-François Timsit, Sarah Tubiana, Priyanka Velou, Linda Wittkop, Yazdan Yazdanpanah. We acknowledge Working Groups of the DisCoVeRy Trial (by alphabetical order): Methodology and Statistics: Drifa Belhadi, Charles Burdet, Dominique Costagliola, Axelle Dupont, France Mentré. Data: Toni Alfaiate, Drifa Belhadi, Charles Burdet, Aline Dechanet, Christelle Delmas, Mouhamadou Diallo, Yakhara Diawara, Claire Fougerou-Leurent, Annabelle Metois, Priyanka Velou. Virology: Maude Bouscambert, Charles Burdet, Alexandre Gaymard, Benjamin Leveau, Bruno Lina, France Mentré, Florence Morfin-Sherpa. Immunology/Biology: Laurent Abel, Florence Ader, Alexandre Belot, Maude Bouscambert, Charles Burdet, Darragh Duffy, Véronique Fremeaux-Bacchi, Xavier Lescure, France Mentré, Nathan Peiffer-Smadja, David M. Smadja, Sophie Susen, Benjamin Terrier, Eric Vivier. Collection of biological human samples: Aline Dechanet, Christelle Delmas, Vinca Icard, Ouifiya Kalif, Benjamin Leveau, Bruno Lina, Florence Morfin-Sherpa, Valentine Piquard, Chaimae Saji, Céline Sakonda, Sarah Tubiana. Clinical Research Unit CHU Bichat-Claude Bernard: Camille Couffignal. Data hosting: the COVID-19 Partners Platform and Hervé Le Nagard. We gratefully acknowledge the RENARCI network: Marion Noret, Albert Sotto and Pierre Tattevin. We thank all the staff members involved in data monitoring. We acknowledge the monitoring management team: Lydie Beniguel, Christine Betard, Chloé Birklé, Maud Brossard, Charlotte Cameli, Alain Caro, Asma Essat, Michèle Génin, Mathilde Ghislain, Mélanie Grubner, Axel Levier, Cécile Moins, Emmanuelle Netzer, Marie-José Ngo Um Tégué, Isabelle Pacaud and Yoann Riault. We acknowledge the Clinical research associates: Belgium: Nathalie Van Sante, Zineb Khalil; France: Malek Ait Djoudi, Lydie Antoine, Christelle Back, Marcellin Bellonet, Assia Benlakhryfa, Nour Boudjoghra, Fabrice Bouhet, Isabelle Calmont, Sabine Camara, Asma Cherifi, Camille Collette, Alexandra De Lemos, Marie Diesel, Elodie Donet, Marine Douillet, Caroline Dubois-Gache, Edith Faillet, Volanantenaina Fanomezantsoa, Stéphanie Flasquin, Shervin Fonooni, Euma Fortes Lopes, Isabelle Gaudin, Blandine Gautier, Quentin Gerome, Marion Ghidi, Pauline Ginoux, Lyna Gouichiche, Marie Granjon, Valérie Guerard, Elina Haerrel, Camille Harpon, Morgane Herbele, Lorrie Lafuente, Dominique Lagarde, Audrey Langlois, Aude Le Breton, Stéphanie Lejeune, Hend Madiot, Bercelin Maniangou, Eric Marquis, Anne-Sophie Martineau, Murielle Mejane, Béatrice Mizejewski, Victoria Mouanga, Brigitte Mugnier, Issraa Osman, Maxence Passageon, Manon Pelkowski, Véronique Pelonde-Erimée, Christine Pintaric, Celina Pruvost, Brigitte Risse, Justine Rousseaux, Christine Schiano, Alexandra Seux, Marielle Simon, Marie-Laure Stupien, Sophie Tallon, Jérémy Tobia, Chaima Traika, Solange Tréhoux, Alice Verdier, Adele Wegang-Nzeufo, Rachida Yatimi; Luxembourg: Gloria Montanes. Portugal: Catarina Madeira. We acknowledge all the SARCODO study participants: We would like to extend our gratitude to the technicians of the Hematology Department at the George Pompidou European Hospital for their assistance with participant inclusion, with special thanks to Julie Brichet and Nadège Ochat for their specific technical organization within the department. We also thank AP-HP for promoting the SARCODO Project. Our appreciation goes to the Unit of Clinical Research URC HEGP CIC-EC1418 (Natacha Nohile, Pauline Jouany, and Dr. Juliette Djadi-Prat) and Helene Cart-Grandjean from AP-HP for their involvement in the project. Grants: The DisCoVeRy trial was sponsored by INSERM and initially launched in France as a WHO Solidarity trial "add-on" study. EU-RESPONSE facilitated its expansion to other European countries. The trial received support from GILEAD, SANOFI, MERCK, and ABBVIE, who provided the study drugs. The SARCODO study was sponsored by ANR (Agence Nationale de la Recherche), Fondation de France (ANR Flash COVID), APHP Mécénat Collecte Crise COVID, and the Mécénat Crédit Agricole Ile de France Programme Jeune Talent.
Author information
Authors and Affiliations
Consortia
Contributions
All the undersigning authors have substantially contributed to the paper. All other authors included patients, reviewed all patients’ characteristics, interpreted data, drafted and revised the manuscript, and approved the final version. All authors declare that the submitted work is original and has not been published before (neither in English nor in any other language) and that the work is not under consideration for publication elsewhere. David M. Smadja: PI of SARCODO trial, designed the present study, performed and/or analyzed the data and wrote the manuscript Clément R. Massonnaud: designed the present study, performed and/or analyzed the data and wrote the manuscript Aurélien Philippe: performed and/or analyzed the data Mickael Rosa: performed and/or analyzed the data Sophie Luneau: performed and/or analyzed the data Antoine Rauch: performed and/or analyzed the data Nathan Peiffer-Smadja: PI of Discovery trial, inclusion of patients Amandine Gagneux-Brunon: inclusion of patients Julien Poissy: inclusion of patients Maxime Gruest: performed and/or analyzed the data Alexandre Ung: performed and/or analyzed the data Valérie Pourcher inclusion of patients François Raffi inclusion of patients Lionel Piroth inclusion of patients Kévin Bouiller inclusion of patients Hélène Esperou work on Discovery data base Christelle Delmas work on Discovery data base Drifa Belhadi inclusion of patients Alpha Diallo inclusion of patients Juliette Saillard inclusion of patients Aline Dechanet inclusion of patients Noémie Mercier inclusion of patients Axelle Dupont inclusion of patients François-Xavier Lescure inclusion of patients François Goehringer inclusion of patients Stéphane Jaureguiberry inclusion of patients François Danion inclusion of patients Violaine Tolsma inclusion of patients André Cabie inclusion of patients Johan Courjon inclusion of patients Sylvie Leroy inclusion of patients Joy Mootien inclusion of patients Bruno Mourvillier inclusion of patients Sébastien Gallien inclusion of patients Jean-Philippe Lanoix inclusion of patients Elisabeth Botelho-nevers inclusion of patients Florent Wallet inclusion of patients Jean-Christophe Richard inclusion of patients Jean Reuter inclusion of patients Alexandre Gaymard inclusion of patients Richard Greil inclusion of patients Guillaume Martin-Blondel inclusion of patients Claire Andrejak inclusion of patients Yazdan Yazdanpanah: PI of Discovery trial, designed the present study, performed and/or analyzed the data and wrote the manuscript Charles Burdet: PI of Discovery trial, designed the present study, performed and/or analyzed the data and wrote the manuscript Jean-Luc Diehl: PI of SARCODO trial, designed the present study, performed and/or analyzed the data and wrote the manuscript Maya Hites: PI of Discovery trial, designed the present study, performed and/or analyzed the data and wrote the manuscript Florence Ader: PI of Discovery trial, designed the present study, performed and/or analyzed the data and wrote the manuscript Sophie Susen: designed the present study, performed and/or analyzed the data and wrote the manuscript France Mentré: PI of Discovery trial, designed the present study, performed and/or analyzed the data and wrote the manuscript Annabelle Dupont: designed the present study, performed and/or analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Smadja, D.M., Massonnaud, C.R., Philippe, A. et al. sST2 is a key outcome biomarker in COVID-19: insights from discovery randomized trial. Sci Rep 15, 14348 (2025). https://doi.org/10.1038/s41598-025-95122-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-95122-7