Abstract
Leonurus japonicus Houtt. (L. japonicus), as an important plant resource with both ornamental and medicinal value, has now spread worldwide and is widely studied. Currently, its chromosomal genome and chloroplast genome (cpDNA) have been reported, but the mitochondrial genome (mtDNA) has not yet been explored. In this study, we extracted DNA from fresh leaves of L. japonicus and performed sequencing and assembly of its mtDNA using both second-generation and third-generation sequencing technologies. The complete mtDNA of L. japonicus is 382,905 bp in length, with a GC content of 45.13%. This genome includes 15 tRNA genes, 32 protein-coding genes (PCGs), and 4 rRNA genes. In this mtDNA genome, we predicted a total of 480 RNA editing sites among the 32 PCGs. Subsequently, we conducted analyses on repetitive sequences, organelle genome sequence migration, and Relative Synonymous Codon Usage (RSCU). There are 28 homologous sequence fragments between the mtDNA and cpDNA of L. japonicus, which are related to the migration of 10 mtDNA genes. The RSCU analysis predicted 28 high-frequency codons, most of which prefer to end with A/U. Selection pressure analysis indicated that the Ka/Ks ratio for the majority of PCGs is less than 1, suggesting they are highly conserved during evolutionary processes. Phylogenetic results from 24 species indicate that the genera Leonurus and Scutellaria within the Lamiaceae family have the closest relationships. In summary, we have successfully assembled the complete mtDNA of L. japonicus by integrating second-generation and third-generation sequencing data for the first time. Subsequent multi-faceted analyses have allowed us to gain deeper insights into the numerous features of this genome, providing important reference data for the molecular genetics, dynamic evolution, and species identification of this plant. This work promotes the conservation and development of this important resource of medicinal and edible plants.
Similar content being viewed by others
Introduction
Leonurus japonicus Houtt, a member of the Lamiaceae family, is an annual or biennial herbaceous plant native to various parts of Asia but now widely distributed around the world1. L. japonicus is a traditional Chinese medicinal herb first recorded in China’s earliest pharmacopoeia, Shennong Baicao Jing. Its aerial parts are recognized in traditional Chinese medicine for their effects in promoting blood circulation, regulating menstruation, removing blood stasis, reducing swelling, and treating female-related diseases2. Currently, over 280 compounds have been extracted from L. japonicus, including flavonoids, alkaloids, diterpenes, phenylpropanoids, and phenolic acids, which possess significant potential for various applications3. In recent years, it has been discovered that L. japonicus possesses important pharmacological activities such as antibacterial, anti-inflammatory, anti-apoptotic, antioxidant, immune regulation, neuroprotection, and protection of the uterus and cardiovascular system4. These effects are all derived from the secondary metabolites contained in L. japonicus. The flowers and stems of L. japonicus possess an aesthetic appeal, showcasing a fresh and pure natural simplicity. Additionally, L. japonicus is favored by the people in southern China as an edible vegetable. Therefore, it is considered a resource with significant ornamental value and dual uses in medicine and food. Conducting in-depth research on this plant is beneficial for exploring its potential applications. The extensive research on L. japonicus has already provided information on its morphology, genetic diversity, metabolomics, transcriptomics, and genomics5,6,7,8,9,10,11. However, due to the lack of the mitochondrial genome of L. japonicus, elucidating the biological functions of key genes associated with mitochondrial traits remains highly challenging.
It is well known that mitochondria are important energy-producing organelles within cells, and they primarily release the ATP required for life through oxidative phosphorylation12. With the deeper study of mitochondria it was discovered that they are also the processors of the cell. Together with the nucleus and other organelles, they create an information processing system and are involved in all phases of the cell cycle13,14,15,16,17. Recent reports suggest that all mitochondria originated from a common ancestral organelle, which was formed through the symbiotic relationship between an α-proteobacterium and a host cell related to Asgard archaea18. Furthermore, the mtDNA exist independently of the nuclear genome within the cell. During evolution, mtDNA and the nuclear genome have continuously exchanged genetic material and the former has been controlled by the latter, which in turn has led to mitochondria becoming semi-autonomous organelles19. The number of genes in mtDNA is relatively small, with fewer than 40 genes. At the same time, it also possesses several characteristics, including a relatively fast evolutionary rate, relative conservation, and maternal inheritance20. This also gives it an extremely considerable advantage in the study of genetic mechanisms. In addition, mitochondria are also important in plant stress tolerance. Under abiotic stress conditions, mitochondria in plant cells undergo a variety of metabolic reactions to enhance the plant’s ability to adapt to the stress21. Genome-level studies of plant mitochondria can provide important references for the field of molecular genetics. This is significant for species classification, adaptation to adversity, and systematic evolution.
As of today, although the organelle genomes of many species have been extensively studied, the number of reports on cpDNA and mtDNA is not balanced. The NCBI database has nearly 13,000 published cpDNAs, while the number of published plant mtDNAs is less than 10% of that of cpDNAs22. Due to the higher complexity of plant mtDNA and their frequent interference from nuclear genomes, their assembly is more challenging. This may be due to the higher complexity of plant mitochondrial genomes, which are often influenced by the nuclear genome, making their assembly more challenging. Although the chloroplast genome and nuclear genome of Leonurus japonicus have been reported, its mitochondrial genome has not been reported to date. This severely limits our molecular biology research on L. japonicus. In this study, we performed sequencing, assembly, and annotation of the mitochondrial genome of L. japonicus, and conducted multiple analyses, including genomic features, codon usage, RNA editing, and repetitive sequences based on its mitochondrial genome. These results provide important reference data for the molecular genetics research of L. japonicus.
Results
Features of the L. japonicus mtDNA
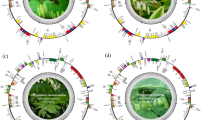
The assembly of the L. japonicus mtDNA reveals (Fig. 1) that its total length is 382,905 bp, with a base composition of 27.31% A, 27.56% T, 22.39% C, and 22.74% G, resulting in a GC content of 45.13%. As shown in Table 1, a total of 51 genes were identified in the complete mtDNA, including 15 tRNA genes, 32 protein-coding genes (PCGs), and 4 rRNA genes. The 32 PCGs are classified into 10 categories: NADH dehydrogenase (9), ATP synthase (5), Small ribosomal subunit proteins (5), Cytochrome c biogenesis (4), Cytochrome c oxidase (3), Large ribosomal subunit proteins (2), Ubiquinol cytochrome c reductase (1), Succinate dehydrogenase (1), Transport membrane protein (1), and Maturases (1). During the annotation of this mtDNA, we found that trnI-GAT, cox1, ccmFc, cox2, rps3, and rps10 each contain one intron; nad7 and nad4 each contain three introns; and nad5, nad2, and nad1 each contain four introns. Additionally, there are two copies of rrn18 and five copies of trnM-CAT in the genome.
Leonurus japonicus mt genome map. The outer and inner sides of the largest circle in the diagram represent genes that are transcribed in the reverse and forward directions, respectively. The dark portions of the inner circle indicate GC content. Genes are represented in different colored blocks based on their functions.
Prediction of RNA editing sites
RNA editing primarily occurs in various organelles of plants and is closely related to their functions. RNA editing is an important molecular mechanism in mtDNA. As shown in Fig. 2, we predicted a total of 480 RNA editing sites among the 32 PCGs in the Motherwort mtDNA. The majority of these RNA editing sites (84.79%) are of the cytosine-uracil (C-U) type. Of these, 184 (38.33%) RNA editing sites are located at the first position of the codon, while 296 (61.67%) are at the second position. The gene ccmB has the most RNA editing sites, with 36, followed by ccmFn with 35. Genes ccmC, nad4, and nad6 each have more than 30 RNA editing sites. Conversely, atp9 has the fewest RNA editing sites, with only one. Additionally, we speculate that RNA editing may lead to premature termination of the rps10 gene. RNA editing results in 7.80% of amino acids changing from hydrophobic to hydrophilic, 49.27% changing from hydrophilic to hydrophobic, and 42.69% of hydrophilic amino acids remaining unchanged (Table 2).
Repeat sequence analysis
In the L. japonicus mtDNA, there are numerous dispersed, tandem, and simple sequence repeats (DSR, TSR, and SSR), which are widely distributed throughout various regions of the genome (Fig. 3A). A total of 49 DSRs were identified in this mtDNA (Fig. 3B), including 17 forward repeats, 21 palindromic repeats, and 11 reverse repeats. The majority of these DSRs (67.35%) are between 30 and 70 bp in length. These DSRs may be closely related to the function of the mtDNA. A total of 10 TSRs were predicted in this mtDNA (Table 3), all with a match rate higher than 90% and lengths ranging from 6 to 41 bp. Additionally, 74 SSRs were identified in this mtDNA (Fig. 3C,D), with di-nucleotide repeats being the most common (27%), followed by mono-nucleotide repeats (23%), tetra-nucleotide repeats (23%), and tri-nucleotide repeats (21.6%). The least common were penta-nucleotide and hexa-nucleotide repeats, each accounting for only 2.7% of the total. Among mono-nucleotide SSRs, adenine (A) repeats and thymine (T) repeats are the most prevalent, each constituting 47.06% of the total. In di-nucleotide SSRs, AG and CT repeats are the most common, each making up 30% of the total. These SSRs can serve as molecular markers, providing important genetic information for plant classification and identification.
Repeat sequence analysis of the L. japonicus mt genome. (A) The distribution of repeat sequences. The outer circle displays different genes. The blue inner arc indicates repeat sequences that are greater than or equal to 30 bp in length. (B) Histograms of lengths and classes of different DSRs. Different colors represent different types of DSRs. The x-axis and y-axis indicate the length and quantity of DSRs, respectively. (C) Proportion of different types of SSRs. Different colors represent different types of SSRs. (D) Statistical histograms of various SSRs. The x-axis displays different SSRs, while the y-axis shows the length of each SSR.
Homologous sequence analysis between organellar genomes
Mitochondria and chloroplasts are semi-autonomous organelles that are endosymbiotic within the cytoplasm, and they share many similarities, including a number of identical genes. Previous studies have shown that during plant evolution, frequent DNA sequence transfers occur between the mitochondria, chloroplasts, and the nuclear genome23,24,25,26. In this study, we found that the mtDNA size of L. japonicus (382,905 bp) is approximately 2.5 times larger than that of the cpDNA (151,654 bp). Due to the relatively fewer number of genes in the mtDNA, the distribution of genes in homologous regions is more dispersed, whereas the distribution of chloroplast genes is more compact (Fig. 4). As shown in Supplementary Table 1, we detected 28 segments in the L. japonicus mtDNA that may be involved in DNA migration between the two organelles, with a total length of 21,113 bp (5.51%). These homologous segments range in length from 30 to 5541 bp. The mtDNA of L. japonicus contains 5 genes that are fully matched with the cpDNA (trnI-GAT, trnW-CCA, trnD-GTC, trnH-GTG, and trnM-CAT) and 5 genes with partial matches (nad2, nad1, rrn18, nad5, and rrn26). The cpDNA of L. japonicus contains 7 genes that are fully matched with the mtDNA (rrn23S, trnA-TGC, trnD-GTC, trnH-GTG, trnI-CAT, trnI-GAT, and trnP-TGG) and 12 genes with partial matches (psaA, rbcL, rpoB, rpoC1, rpoC2, rps3, rrn16S, rrn23S, rrn4.5S, trnM-CAT, trnW-CCA, and ycf2).
Relative synonymous codon usage (RSCU) analysis
The RSCU value is commonly used to assess the frequency of codon usage under random conditions, with an RSCU of 1 indicating that the codon usage rate aligns with the theoretical frequency under random conditions. The RSCU values of L. japonicus mtDNA are shown in Fig. 5. In this mtDNA, 28 codons were identified with RSCU > 1, indicating that these codons are preferentially used. The average RSCU value of the GCU codon encoding Ala is the highest at 1.63, suggesting it is used most frequently. Moreover, over 90% (28) of the high-frequency codons end with A/U, which further indicates a preference for A/U at the third position of these codons.
Ka/Ks analysis
The synonymous substitution rate (Ks) and non-synonymous substitution rate (Ka) are often useful for analyzing the dynamic evolution and phylogenetic relationships of protein-coding sequences between closely related species27. The ratio of non-synonymous to synonymous substitutions (Ka/Ks) can predict the type of selective pressure experienced during the genetic evolution of species. When Ka/Ks > 1, it indicates positive selection; when Ka/Ks = 1, it suggests neutral selection; and when Ka/Ks < 1, it implies negative selection. Comparing the Ka/Ks ratios of L. japonicus with those of five other Lamiaceae mtDNAs (Fig. 6), it is observed that the Ka/Ks values for the genes ccmB, mttB, and rps10 all exceed 1, suggesting they are likely under positive selection. In contrast, the majority of genes (90%) have Ka/Ks values below 1, indicating they are under negative selection. This suggests that most PCGs in the L. japonicus mtDNA are highly conserved during genetic evolution.
Phylogenetic analysis
The high conservation of PCGs in mtDNA is advantageous for constructing phylogenetic trees and can aid in classifying plant genetic evolution. To determine the phylogenetic position of L. japonicus, a phylogenetic tree was constructed using 21 shared PCGs from the mtDNAs of 24 species, including 9 Lamiaceae plants, 3 Cucurbitaceae plants, 2 Poaceae plants, 2 Solanaceae plants, 1 Caricaceae plant, 1 Bataceae plant, 1 Apiaceae plant, 1 Asparagaceae plant, 1 Araceae plant, 1 Arecaceae plant, 1 Ginkgoaceae plant (outgroup), and 1 Marchantiaceae plant (outgroup). The results showed that the phylogenetic tree is consistent with traditional taxonomic relationships and strongly supports the separation of different plant families (Fig. 7). The phylogenetic tree divides all plants into three major groups: Lamiaceae, Solanaceae, and Apiaceae are grouped together; Cucurbitaceae, Caricaceae, and Bataceae are grouped together; and Poaceae, Asparagaceae, Araceae, and Arecaceae are grouped together. The phylogenetic analysis indicates that Lamiaceae and Solanaceae are the closest related families, suggesting that these two families are closely related. In addition, the genera Leonurus and Scutellaria in the Lamiaceae family have the closest relationship, indicating that these two genera are sister groups.
Discussion
Both chloroplasts and mitochondria are crucial organelles that play essential roles in the growth, development, and life activities of plant cells28,29. Both are semi-autonomous organelles in plants, and their genomes exhibit complex structures, including circular, branched forms, linear, and mixed structures with multiple rings22,30,31. This suggests that they may have some mysterious potential values that need to be further revealed. In this study, we report for the first time the mitochondrial DNA (mtDNA) of the important medicinal and edible plant L. japonicus. Similar to Perilla frutescens, the mtDNA of L. japonicus exhibits a typical circular structure (Fig. 1)32. This conserved topological feature may be associated with its relatively low frequency of homologous recombination, thereby maintaining the stability of the genomic structure. In previous studies, the mitochondrial genomes of some Lamiaceae species have been shown to possess complex multimeric structures, as observed in Prunella vulgaris, Salvia officinalis, and Scutellaria tsinyunensis33,34,35. The mtDNA of phylogenetically closely related species exhibits remarkable conformational diversity, a characteristic that fully demonstrates the high degree of adaptability and structural complexity displayed by Lamiaceae mtDNA during evolutionary processes34. The full length of this mtDNA is 382,905 bp, which is similar to the mtDNA size of S. tsinyunensis (354,073 bp). GC content serves as a crucial factor in studies of biological evolution and genomic adaptation36. The GC content of L. japonicus mtDNA is 45.13%, which is similar to that of other Lamiaceae plants, such as Vitex rotundifolia (45.54%)37, S. tsinyunensis (45.26%)35, and P. frutescens (45.23%)32. It appears that the GC content of mtDNA in Lamiaceae plants has been relatively conserved throughout long genetic evolution. Angiosperm mtDNA contains 24 core protein-coding genes, which are primarily associated with functional categories including ATPase subunits, cytochrome c biogenesis, and cytochrome c oxidase subunits35. A total of 51 genes were identified in L. japonicus mtDNA, including 15 tRNA genes, 32 protein-coding genes (PCGs), and 4 rRNA genes. The types and numbers of important core protein-coding genes in the mtDNA of L. japonicus are generally consistent with those of most Lamiaceae plants, but its tRNA genes show partial deletions38. This significant feature may be due to the combined action of various molecular mechanisms during the long-term evolution of L. japonicus mtDNA, including homologous recombination, horizontal gene transfer, and retrograde signaling from the nuclear genome39,40,41. These mechanisms have collectively promoted more frequent gene exchange between the mtDNA, nuclear genome, and cpDNA. The sequence transfers between chloroplasts and mitochondria within plant cells may also be a dynamically evolving endosymbiotic process42. By comparison we identified 28 sequence fragments that may be involved in the migration of DNA between the genomes of the two organelles. This sequence migration may be the primary reason for the differences in gene numbers among the mtDNAs of different plants.
After transcription is complete, RNA editing occurs, a process that modifies the RNA sequence. This can result in differences in protein function and expression compared to what is defined in the genome43. This process plays a crucial regulatory role in the life cycle of plants. In this study, we identified a total of 480 RNA editing sites in the PCGs of mtDNA, with the vast majority (84.79%) classified as C-U type. The preferential selection of G/C codons during RNA editing may be attributed to their relatively high binding free energy, a molecular characteristic that contributes to the maintenance of translational accuracy44. Interestingly, RNA editing events in the mitochondrial DNA (mtDNA) of Leonurus japonicus lead to the premature introduction of a stop codon in the rps10 gene and generate start codons in three genes: cox2, nad4L, and rps10. The premature termination of translation or abnormal generation of start codons could potentially result from either the organism’s adaptive response to environmental conditions or the occurrence of nonsense mutations45,46. These editing events are typically associated with the production of highly conserved homologous proteins, a mechanism that may contribute to the optimization of gene expression efficiency within mitochondria47. Additionally, the frequency of editing sites in different PCGs varies significantly among species. In Abelmoschus esculentus, Siberian larch, and Fritillaria ussuriensis, the genes rpl2, nad5, and nad4 have the most editing sites, respectively48,49,50. In the mtDNA of L. japonicus, the gene ccmB has the highest number of editing sites (Fig. 2). Additionally, we predicted that certain genes related to cytochrome c biogenesis and NADH dehydrogenase (ccmB, ccmC, ccmFn, nad2, nad4, and nad5) in L. japonicus mtDNA have a relatively higher number of editing sites. This indicates that the frequency of editing sites for genes associated with cytochrome c biogenesis and NADH dehydrogenase is higher in the mtDNA of L. japonicus. Based on previous studies, we have observed that RNA editing events in the mitochondrial mtDNA of many Lamiaceae species exhibit a strong bias towards genes associated with cytochrome c biogenesis and NADH dehydrogenase33,37,38,51. This editing preference likely reflects the Lamiaceae family’s critical dependence on energy metabolism and redox homeostasis, which is closely linked to their biological characteristics such as high essential oil content, rapid growth rates, and environmental adaptability52,53,54. This editing strategy serves a dual purpose: it acts as a compensatory mechanism for the high mutation rate in mitochondrial genomes, while simultaneously optimizing metabolic efficiency and enhancing stress resistance43,44,55,56. Hydrophilic amino acids interact with water molecules, guiding the proper folding of protein structures30. When a large number of hydrophilic amino acids are replaced by hydrophobic ones, forming a more stable hydrophobic core, the overall stability of the protein structure increases. In L. japonicus mtDNA, most amino acids are converted from hydrophilic to hydrophobic to enhance protein stability. This early termination could be an adaptive response to environmental conditions or the result of a nonsense mutation45,46. In the genomes of species, codon usage bias often arises due to the combined effects of dynamic changes in the external environment and internal factors. Based on RSCU analysis, we identified 28 highly frequent codons in the mtDNA of L. japonicus, the majority of which tend to end with A/U. This phenomenon is also observed in the mtDNAs of various other plants19,57,58. Additionally, the RSCU results can provide a reference for exploring the genetic mapping of L. japonicus during the evolutionary process. Ka/Ks analysis is commonly used to determine whether protein-coding genes (PCGs) are influenced by selective pressure during genetic evolution. In this study, we found that the majority of PCGs in the mtDNA of L. japonicus are affected by purifying selection. However, genes under positive selection, such as ccmB, mttB, and rps10, have already been identified, indicating that they may play a significant role in the evolution of the species.
Large amounts of repeat sequences are present in the mtDNAs of angiosperms, and they play an important role in genetic molecular markers, genomic structural variation, and genetic stability studies across species59,60. DSR refers to the identical or similar DNA sequences that are scattered at different locations within the genome. In mtDNA, these sequences are often regarded as segmental duplications involved in the DNA double-strand break repair process61. DSR includes four types: Forward repeat, Reverse repeat, Complement repeat, and Palindromic repeat. According to a report by Li et al. in the mtDNA of nine Lamiales plants, DSRs mainly exist in the forms of Forward repeat and Reverse repeat35. In contrast, L. japonicus mtDNA also contains a notable number of DSRs in the form of Complement repeats. This phenomenon may suggest that specific evolutionary pressures or changes in genomic repair mechanisms have led to the accumulation of these Complement repeats. SSRs (simple sequence repeats) possess high polymorphism, ease of detection, extensive genomic coverage, relative abundance, and codominant inheritance, making them uniquely valuable for genetic marker development, cultivar breeding, and population genetics research62. In 1997, Szibor et al. discovered dinucleotide polymorphisms in the D-loop region of human mtDNA, which can be used to distinguish between different human populations63. As early as 1999, Soranzo et al. were the first to discover the polymorphism of SSRs in the mtDNA of 15 species of conifers64. Khera et al. also developed SSR markers in the mtDNA of Cajanus cajan, and they noted in their report that these SSR markers can distinguish genotypes based on mtDNA type in phylogenetic studies65. In this study, we identified 74 SSRs distributed across the mtDNA. We identified 74 SSRs distributed throughout the mtDNA of L. japonicus. In contrast to the predominance of tetranucleotide repeats reported in most Lamiaceae species, dinucleotide repeats constitute the most abundant SSR type in L. japonicus mtDNA, accounting for 27% of the total35,51. This distinctive pattern may reflect unique selective pressures acting on the mitochondrial genome during its evolutionary history or could be associated with its specific ecological adaptations. These identified SSRs will aid in the identification and conservation of important species resources.
The mtDNAs are conserved and slow to mutate during species evolution, and they often play an important role in phylogenetic studies19. To investigate the systematic evolutionary relationships of L. japonicus, we collected high-quality mtDNAs from various plants within the Lamiaceae family and other families. Phylogenetic analysis based on their shared conserved PCGs showed that the Lamiaceae family is more closely related to the Solanaceae family among the numerous families studied. Additionally, within the Lamiaceae family, Leonurus and Scutellaria are likely to be sister genera. Compared to cpDNA, plant mtDNA is generally larger and more structurally variable, which means that, in certain cases, mtDNA may provide richer phylogenetic information than cpDNA66. Currently, research on plant mtDNA is receiving increasing attention, and it is believed that it will play an important role in future systematic evolutionary studies. The mtDNA of L. japonicus provides important reference data for exploring the evolutionary history of Lamiaceae species and further contributes valuable information for molecular genetic studies of L. japonicus.
Conclusions
In this study, we have for the first time assembled the complete mtDNA of L. japonicus using both second-generation and third-generation sequencing data. This assembly strategy has resulted in a more complete and reliable genome assembly. Through genomic characterization of the complete mtDNA and its annotation results, we found that the mtDNA of L. japonicus is 382,905 bp in length, including 15 tRNA genes, 32 PCGs, and 4 rRNA genes. By combining RNA editing site prediction results with previous studies, we observed a higher frequency of editing sites in genes related to cytochrome c biogenesis and NADH dehydrogenase. The Ka/Ks results also indicate that the majority of PCGs in mtDNA are highly conserved. Additionally, phylogenetic analysis based on shared PCGs from 24 mtDNAs revealed the evolutionary positioning of L. japonicus. Our study provides important reference data for the molecular genetics, dynamic evolution, and species identification of L. japonicus, thereby advancing the conservation and development of this important medicinal and edible plant resource.
Materials & methods
Plant collection, DNA extraction, and sequencing
L. japonicus plants cultivated for 5 months at the Baiyun Experimental Base of Guangdong Academy of Agricultural Sciences (N23°07ʹ29ʺ, E113°10ʹ31ʺ) were selected as experimental materials. In June 2024, fresh leaves of L. japonicus were collected using liquid nitrogen for quick freezing. The collected samples were stored in a deep freezer (− 80 °C). The plant material used in this study was formally identified by Professor Keming Liu, a retired professor of plant taxonomy from Hunan Normal University. Specimens were preserved in the Key Laboratory Building of the College of Forestry and Horticulture at Hubei Minzu University (N30°17ʹ51ʺ, E109°29ʹ54ʺ, person: Qun Hu, email: [email protected]) with the voucher number 20240627009. Total DNA was extracted using a plant genome DNA extraction kit (Tsingke Biotech, Beijing, China). The quantity and quality of extracted DNA were evaluated using a NanoDrop One spectrophotometer and agarose gel electrophoresis. Qualified samples were then sent to Guangzhou Yuda Biotechnology Company Limited (Guangzhou, China) for Illumina and SMRT sequencing to obtain high-accuracy reads.
Assembly and annotation of mtDNA
We used SMRT Analysis v2.3.0 software to assess the quality of sequencing data generated by the PacBio RS II system, identifying and filtering out low-quality reads. We extracted the mtDNA sequence of L. japonicus from the filtered reads data. The obtained sequences were aligned to plastid genome data in the NCBI database, and sequences with alignment rates greater than 90% were filtered out to maximize the quality and accuracy of the mitochondrial genome assembly. Subsequently, we first used Canu v2.2 software to correct the third-generation data, and then used Bowtie 2-v2.5.2 software to align the second-generation data to the corrected sequences67,68. Finally, we used Unicycler v0.5.0 software to assemble the final sequences from the second-generation and third-generation data (with default parameters), resulting in the complete circular mtDNA of L. japonicus69.
We used BLAST to align rRNAs and coding proteins in the assembled mtDNA with published plant mt sequences, which were manually calibrated using closely related species as the primary reference. Annotation of tRNAs and ORFs using tRNAscan-SE (https://lowelab.ucsc.edu/tRNAscan-SE/) and Open Reading Frame Finder (https://www.ncbi.nlm.nih.gov/orffinder/)70,71. Finally, we used OGDRAW software (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) to draw the mtDNA map of L. japonicus72.
Prediction of RNA editing sites
To predict the RNA editing sites of L. japonicus mtDNA, we imported the sequence files of the PCGs into the PREPACT online website (http://www.prepact.de/prepact-main.php), then chose to use the complete mtDNA of Arabidopsis thaliana as a template for predicting the editing sites73. Finally, we compiled the predicted results from the comparison and plotted a histogram.
Repetitive sequence analysis
Repeated sequences include SSRs, TSRs, and DSRs. We used MISA software to predict SSRs in the L. japonicus mtDNA (parameters: 10 5 4 3 3 3)74. We predicted TSRs in the L. japonicus mtDNA using Tandem Repeats Finder software (TRF-v4.09.1, parameters: 2 7 7 80 10 50 2000, − f − d − m)75. Subsequently, we predicted DSRs in the mtDNA using the REPuter website (https://bibiserv.cebitec.uni-bielefeld.de/reputer)76. We used the Advanced Circos program in TBtools software to visualize the repeated sequences77.
DNA migration between mitochondria and chloroplasts
Download the complete cpDNA sequence of L. japonicus from the NCBI database (https://www.ncbi.nlm.nih.gov/nuccore/OQ417592.1). We used BLAST software to compare homologous sequences between the cpDNA and mtDNA of L. japonicus, using length greater than 30 bp, e-value less than 1e−5, and alignment rate greater than 70% as screening criteria78. Subsequently, we used the Advanced Circos program in TBtools software to create plots of the homologous sequences77.
Codon usage patterns of mtDNA
To understand the codon usage pattern of the mtDNA of L. japonicus, we first used a Perl script to filter out all CDS sequences from its mtDNA and import them into a text file. Then, we uploaded this text file to an online cloud platform (http://112.86.217.82:9919/#/) to calculate the RSCU values. Finally, we compiled the results and plotted a stacked chart and a histogram.
Ka/Ks analysis
We imported the mtDNAs of five Lamiaceae plants (Ajuga reptans KF709392.1, Pogostemon heyneanus MK728874.1, S. tsinyunensis MW553042.1, Scutellaria barbata MZ127834.1, and Scutellaria franchetiana MZ127835.1) along with the mtDNA of L. japonicus into the Genepioneer cloud platform (http://112.86.217.82:9919/#/) to calculate the Ka/Ks values. Subsequently, a box plot was created using Origin 2021 software based on the conserved proteins.
Prediction of the systematic evolutionary relationships
We selected Ginkgo biloba and Marchantia paleacea as outgroups and performed systematic evolutionary analysis using species from the Lamiaceae family and other plant families included in the NCBI database. The mtDNAs of 24 plant species were imported into PhyloSuite v1.2.2 software, where duplicate gene copies and non-shared genes were removed79. Subsequently, we performed multiple sequence alignments of the filtered shared genes using MAFFT v7.313 software80. The concatenated multiple sequence alignment results were used to construct the phylogenetic tree. The optimal substitution model (GTR + F + I + G4) was selected based on the Bayesian Information Criterion (BIC). Maximum Likelihood (ML) analysis with 1000 bootstrap replicates was performed using IQ-TREE v1.6.8 to assess nodal support81. Finally, we imported the constructed phylogenetic tree into the ITOL online website (https://itol.embl.de/) for optimization82.
Data availability
The sequence and annotation of L. japonicus mtDNA was submitted to the CNCB (https://ngdc.cncb.ac.cn/genbase/). The accession number in GenBase is C_AA079493.1.
References
Shang, X., Pan, H., Wang, X., He, H. & Li, M. Leonurus japonicus Houtt.: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 152, 14–32 (2014).
Xu, C., Tan, B., Zhang, S. & Li, W. Corrosion inhibition of copper in sulfuric acid by Leonurus japonicus Houtt. extract as a green corrosion inhibitor: Combination of experimental and theoretical research. J. Taiwan Inst. Chem. Eng. 139, 104532 (2022).
Miao, L.-L., Zhou, Q.-M., Peng, C., Liu, Z.-H. & Xiong, L. Leonurus japonicus (Chinese motherwort), an excellent traditional medicine for obstetrical and gynecological diseases: A comprehensive overview. Biomed. Pharmacother. 117, 109060 (2019).
Zhang, R.-H. et al. Phytochemistry and pharmacology of the genus Leonurus: The herb to benefit the mothers and more. Phytochemistry 147, 167–183 (2018).
Yang, D.-J. et al. Comparative genomic analysis between Leonurus japonicus and Leonurus sibiricus. bioRxiv https://doi.org/10.1101/2022.11.27.518111 (2022).
Chen, C. et al. Comprehensive analysis of the effects of climate change on the species distribution and active components of Leonurus japonicus Houtt. Ind. Crops Prod. 218, 119017 (2024).
Wang, X. et al. De novo chromosome-level genome assembly of Chinese motherwort (Leonurus japonicus). Sci. Data 11, 55 (2024).
Garran, T. A. et al. Elucidation of metabolite isomers of Leonurus japonicus and Leonurus cardiaca using discriminating metabolite isomerism strategy based on ultra-high performance liquid chromatography tandem quadrupole time-of-flight mass spectrometry. J. Chromatogr. A 1598, 141–153 (2019).
Wang, Y. et al. Genetic diversity and population divergence of Leonurus japonicus and its distribution dynamic changes from the last interglacial to the present in China. BMC Plant Biol. 23, 276 (2023).
Pitschmann, A., Waschulin, C., Sykora, C., Purevsuren, S. & Glasl, S. Microscopic and phytochemical comparison of the three Leonurus species L. cardiaca, L. japonicus, and L. sibiricus. Planta Med. 83, 1233–1241 (2017).
Li, P. et al. Multiomics analyses of two Leonurus species illuminate leonurine biosynthesis and its evolution. Mol. Plant 17, 158–177 (2024).
Verdin, E., Hirschey, M. D., Finley, L. W. S. & Haigis, M. C. Sirtuin regulation of mitochondria: Energy production, apoptosis, and signaling. Trends Biochem. Sci. 35, 669–675 (2010).
Picard, M. & Shirihai, O. S. Mitochondrial signal transduction. Cell Metab. 34, 1620–1653 (2022).
Kroemer, G. & Reed, J. C. Mitochondrial control of cell death. Nat. Med. 6, 513–519 (2000).
Rehman, J. et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 26, 2175–2186 (2012).
Bonora, M. et al. Tumor necrosis factor-α impairs oligodendroglial differentiation through a mitochondria-dependent process. Cell Death Differ. 21, 1198–1208 (2014).
Cheng, Y. et al. Assembly and comparative analysis of the complete mitochondrial genome of Suaeda glauca. BMC Genom. 22, 167 (2021).
Roger, A. J., Muñoz-Gómez, S. A. & Kamikawa, R. The origin and diversification of mitochondria. Curr. Biol. CB 27, R1177–R1192 (2017).
Song, Y. et al. Assembly and analysis of the complete mitochondrial genome of Forsythia suspensa (Thunb.) Vahl. BMC Genom. 24, 708 (2023).
Li, K. et al. The phylogenetic relationship of Lamiinae (Coleoptera: Cerambycidae) using mitochondrial genomes. Genes 15, 13 (2023).
Aliyari Rad, S., Dehghanian, Z., Asgari Lajayer, B., Nobaharan, K. & Astatkie, T. Mitochondrial respiration and energy production under some abiotic stresses. J. Plant Growth Regul. 41, 3285–3299 (2022).
Wang, J. et al. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 29, 754–769 (2024).
Blanchard, J. L. & Schmidt, G. W. Pervasive migration of organellar DNA to the nucleus in plants. J. Mol. Evol. 41, 397–406 (1995).
Wynn, E. L. & Christensen, A. C. Repeats of unusual size in plant mitochondrial genomes: Identification incidence and evolution. G3 GenesGenomesGenet. 9, 549–559 (2019).
Bergthorsson, U., Adams, K. L., Thomason, B. & Palmer, J. D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424, 197–201 (2003).
Guo, W. et al. Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol. Biol. Evol. 33, 1448–1460 (2016).
Zhang, Z. et al. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinf. 4, 259–263 (2006).
Friedman, J. R. & Nunnari, J. Mitochondrial form and function. Nature 505, 335–343 (2014).
Jensen, P. E. & Leister, D. Chloroplast evolution, structure and functions. F1000Prime Rep. 6, 40 (2014).
Wang, L. et al. Assembly and comparative analysis of the first complete mitochondrial genome of a traditional Chinese medicine Angelica biserrata (Shan et Yuan) Yuan et Shan. Int. J. Biol. Macromol. 257, 128571 (2024).
Li, X. et al. Comparative analysis of the complete mitochondrial genomes of Apium graveolens and Apium leptophyllum provide insights into evolution and phylogeny relationships. Int. J. Mol. Sci. 24, 14615 (2023).
Wang, R., Luo, Y., Lan, Z. & Qiu, D. Insights into structure, codon usage, repeats, and RNA editing of the complete mitochondrial genome of Perilla frutescens (Lamiaceae). Sci. Rep. 14, 13940 (2024).
Sun, Z. et al. Assembly and analysis of the mitochondrial genome of Prunella vulgaris. Front. Plant Sci. 14, 1237822 (2023).
Yang, H. et al. Mitochondrial genome sequence of Salvia officinalis (Lamiales: Lamiaceae) suggests diverse genome structures in cogeneric species and finds the stop gain of genes through RNA editing events. Int. J. Mol. Sci. 24, 5372 (2023).
Li, J. et al. Assembly of the complete mitochondrial genome of an endemic plant, Scutellaria tsinyunensis, revealed the existence of two conformations generated by a repeat-mediated recombination. Planta 254, 36 (2021).
Šmarda, P. et al. Ecological and evolutionary significance of genomic GC content diversity in monocots. Proc. Natl. Acad. Sci. 111, E4096–E4102 (2014).
Yu, X., Duan, Z., Wang, Y., Zhang, Q. & Li, W. Sequence analysis of the complete mitochondrial genome of a medicinal plant, Vitex rotundifolia Linnaeus f. (Lamiales: Lamiaceae). Genes 13, 839 (2022).
Chetri, B. K. et al. The evolutionary landscape of mitogenomes: Insights from Elsholtzia blanda and comparative analysis of ten related species. Ecol. Genet. Genom. 34, 100328 (2025).
Lonsdale, D. M. et al. The plant mitochondrial genome: Homologous recombination as a mechanism for generating heterogeneity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 319, 149–163 (1988).
Richardson, A. O. & Palmer, J. D. Horizontal gene transfer in plants. J. Exp. Bot. 58, 1–9 (2006).
Butow, R. A. & Avadhani, N. G. Mitochondrial signaling: The retrograde response. Mol. Cell 14, 1–15 (2004).
Hong, Z. et al. A complete mitochondrial genome for fragrant Chinese rosewood (Dalbergia odorifera, Fabaceae) with comparative analyses of genome structure and intergenomic sequence transfers. BMC Genom. 22, 672 (2021).
Takenaka, M., Zehrmann, A., Verbitskiy, D., Härtel, B. & Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 47, 335–352 (2013).
Hao, W. et al. RNA editing and its roles in plant organelles. Front. Genet. 12, 757109 (2021).
Picardi, E. et al. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 38, 4755–4767 (2010).
Schuster, W. & Brennicke, A. RNA editing makes mistakes in plant mitochondria: Editing loses sense in transcripts of a rps19 pseudogene and in creating stop codons in coxl and rps3 mRNAs of Oenothera. Nucleic Acids Res. 19, 6923–6928 (1991).
Hanson, M. R., Sutton, C. & Luis, B. Plant organelle gene expression: Altered by RNA editing. Trends Plant Sci. 1, 57–64 (1996).
Li, J. et al. The complete mitochondrial genome of okra (Abelmoschus esculentus): Using nanopore long reads to investigate gene transfer from chloroplast genomes and rearrangements of mitochondrial DNA molecules. BMC Genom. 23, 481 (2022).
Putintseva, Y. A. et al. Siberian larch (Larix sibirica Ledeb.) mitochondrial genome assembled using both short and long nucleotide sequence reads is currently the largest known mitogenome. BMC Genom. 21, 654 (2020).
Xie, P. et al. Assembly and comparative analysis of the complete mitochondrial genome of Fritillaria ussuriensis Maxim. (Liliales: Liliaceae), an endangered medicinal plant. BMC Genom. 25, 773 (2024).
Wang, J., Liu, X., Zhang, M. & Liu, R. The mitochondrial genome of Lavandula angustifolia Mill. (Lamiaceae) sheds light on its genome structure and gene transfer between organelles. BMC Genom. 25, 929 (2024).
Nieto, G. Biological activities of three essential oils of the Lamiaceae family. Medicines 4, 63 (2017).
Carović-Stanko, K. et al. Medicinal plants of the family Lamiaceae as functional foods-a review. Czech J. Food Sci. 34, 377–390 (2016).
Lichman, B. R., Godden, G. T. & Buell, C. R. Gene and genome duplications in the evolution of chemodiversity: Perspectives from studies of Lamiaceae. Curr. Opin. Plant Biol. 55, 74–83 (2020).
Gott, J. M. & Emeson, R. B. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34, 499–531 (2000).
Cai, J., Shen, L., Kang, H. & Xu, T. RNA modifications in plant adaptation to abiotic stresses. Plant Commun. 6, (2025).
Ke, S.-J. et al. Apostasia mitochondrial genome analysis and monocot mitochondria phylogenomics. Int. J. Mol. Sci. 24, 7837 (2023).
Guo, Y. et al. Assembly and comparative analysis of the complete mitochondrial genome of two species of Calla Lilies (Zantedeschia, Araceae). Int. J. Mol. Sci. 24, 9566 (2023).
Chevigny, N., Schatz-Daas, D., Lotfi, F. & Gualberto, J. M. DNA repair and the stability of the plant mitochondrial genome. Int. J. Mol. Sci. 21, 328 (2020).
Novák, P. et al. Repeat-sequence turnover shifts fundamentally in species with large genomes. Nat. Plants 6, 1325–1329 (2020).
Choi, I.-S. et al. Born in the mitochondrion and raised in the nucleus: Evolution of a novel tandem repeat family in Medicago polymorpha (Fabaceae). Plant J. Cell Mol. Biol. 110, 389–406 (2022).
Powell, W., Machray, G. C. & Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1, 215–222 (1996).
Szibor, R. et al. Mitochondrial D-loop 3′ (CA)n repeat polymorphism: Optimization of analysis and population data. Electrophoresis 18, 2857–2860 (1997).
Soranzo, N., Provan, J. & Powell, W. An example of microsatellite length variation in the mitochondrial genome of conifers. Genome 42, 158–161 (1999).
Khera, P., Saxena, R., Sameerkumar, C. V., Saxena, K. & Varshney, R. K. Mitochondrial SSRs and their utility in distinguishing wild species, CMS lines and maintainer lines in pigeonpea (Cajanus cajan L.). Euphytica 206, 737–746 (2015).
Evans, D. L., Hlongwane, T. T., Joshi, S. V. & Pachón, D. M. R. The sugarcane mitochondrial genome: Assembly, phylogenetics and transcriptomics. PeerJ 7, e7558 (2019).
Koren, S. et al. Canu: Scalable and accurate long-read assembly via adaptive k -mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 13, e1005595 (2017).
Chan, P. P., Lin, B. Y., Mak, A. J. & Lowe, T. M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49, 9077–9096 (2021).
Rombel, I. T., Sykes, K. F., Rayner, S. & Johnston, S. A. ORF-FINDER: A vector for high-throughput gene identification. Gene 282, 33–41 (2002).
Lohse, M., Drechsel, O., Kahlau, S. & Bock, R. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581 (2013).
Lenz, H. & Knoop, V. PREPACT 2.0: Predicting C-to-U and U-to-C RNA editing in organelle genome sequences with multiple references and curated RNA editing annotation. Bioinf. Biol. Insights 7, BBI.S11059 (2013).
Beier, S., Thiel, T., Münch, T., Scholz, U. & Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 33, 2583–2585 (2017).
Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Kurtz, S. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642 (2001).
Chen, C., Wu, Y. & Xia, R. A painless way to customize Circos plot: From data preparation to visualization using TBtools. iMeta 1, e35 (2022).
Ye, J., McGinnis, S. & Madden, T. L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 34, W6–W9 (2006).
Zhang, D. et al. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355 (2020).
Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82 (2024).
Acknowledgements
We gratefully acknowledge Professor Keming Liu, retired plant taxonomy expert from Hunan Normal University, for his valuable assistance in identifying the plant material and providing taxonomic guidance for this research.
Funding
This work was supported by the Li Qing Youth Mentorship Project (0002002094), Guangdong Crop Germplasm Resources investigation project (No.2022-NBH-00-004, 2023-NBH-00-001) and the 2023 Provincial Rural Revitalization Strategy Special Funds Seed Industry Revitalization Project, Operation and Maintenance of Perennial Special Cash Crop Resource Nurseries in Guangdong Province (2023-NBH-00-017).
Author information
Authors and Affiliations
Contributions
W.Z. (Wenhu Zhang) and Z.D. (Zhijun Deng) planned and supervised the project, conceived and designed the experiments, and participated in obtaining funding; Q.H. (Qun Hu) and Q.L. (Qing Li) wrote the manuscript; Y.M. (Yin Mao) and Y.L. (Yongjian Luo) performed the data organization and formal analysis. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The use of plant parts in this study was in accordance with international, national and/or institutional guidelines. The L. japonicus plants used in this experiment were grown in the Baiyun Experimental Base of Guangdong Academy of Agricultural Sciences. Ethical approval or consent was not required for this study, as no endangered or protected species were involved.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hu, Q., Li, Q., Mao, Y. et al. Assembly and analysis of the complete mitochondrial genome of Leonurus japonicus (Lamiaceae). Sci Rep 15, 13372 (2025). https://doi.org/10.1038/s41598-025-97594-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-97594-z