Abstract
Patients with lung metastasis of colorectal cancer typically have a poor prognosis. Therefore, establishing an effective screening and diagnosis model is paramount. Our study seeks to construct and verify a predictive model utilizing machine learning (ML) that can evaluate the risk of lung metastasis with newly diagnosed colorectal cancer (CRC) using Shapley Additive exPlanations (SHAP). Using the Surveillance, Epidemiology, and End Results database, 39,674 were extracted for model development, all of whom had been pathologically diagnosed with CRC. The data spans from 2010 to 2015. Our study has constructed seven ML algorithms based on the data mentioned above, including Random Forest (RF), Decision Tree, Support Vector Machine, Naive Bayes, K-Nearest Neighbor, eXtreme Gradient Boosting, and Gradient Boosting Machine. We selected the best algorithm and visualized it using SHAP. We conducted a validation of the model utilizing data from a Chinese hospital to assess its practicality. Based on this, we have constructed an open web calculator. 39,674 patient data were included in our study, among whom 1369 (3.5%) presented with distant lung metastasis. The Random Forest (RF) algorithm demonstrated the highest predictive capability within the internal test set (AUC of 0.980, AUPR of 0.941). Furthermore, the random forest algorithm also exhibited excellent performance in external validation sets. Meanwhile, we have also established a web calculator (http://121.43.117.60:8003/). The RF algorithm has demonstrated excellent predictive performance. It can assist clinicians in devising more personalized treatment plans.
Similar content being viewed by others
Introduction
Colorectal cancer(CRC) is renowned worldwide for its high prevalence in the population1,2, and the incidence rate of CRC is closely related to global cancer mortality3. About 50% of postoperative deaths among CRC are related to distant metastasis (DM)4. About 75% -90% of patients diagnosed with CRC are considered to be unable to undergo surgical treatment5. For colorectal cancer patients, the Lung is one of the common sites of metastasis6. Studies have indicated that patients with CRC who present with lung metastases constitute approximately 10% to 15% of the total CRC patient population7. Early detection of lung metastasis is crucial in the clinical management of metastasis patients. Generally speaking, Compared with other metastases, such as liver metastasis and peritoneal metastasis, lung metastasis has a relatively better prognosis8,9,10. Studies have shown that early diagnosis of patients with colorectal cancer combined with lung metastasis, coupled with appropriate treatment, can lead to a 5-year survival rate of more than 50% for some colorectal cancer patients11. Therefore, it is imperative to establish an efficient and feasible prediction model. This approach will assist clinicians in making early diagnoses and implementing timely treatments for lung metastasis, ultimately enhancing patient prognosis.
In prior studies, researchers have developed predictive models for lung metastasis in patients with CRC11,12. These predictive models for lung metastasis of colorectal cancer lack external validation data to assess their feasibility. Furthermore, the performance of these models requires additional enhancement.
Machine learning (ML) is characterized by sophisticated algorithmic models that primarily focus on investigating the mechanisms and learning processes inherent in computer research data13. With the advancement of technology, medical data has become increasingly vast and complex. This evolution has created numerous opportunities for machine learning to address clinical challenges effectively. Machine learning techniques have been employed to tackle various clinical issues and have demonstrated superior predictive performance compared to traditional algorithms14. Our study seeks to develop an explainable ML algorithm that can predict CRC lung metastasis.
Methods
Study population
Our data set is sourced from SEER in the United States. Using SEER*stat version 8.4.13 software. The study selected CRC patients from 2010 to 2015. The patient data screening process is shown in Fig. 1. Only colorectal cancer as a primary cancer was considered, excluding incomplete data, unknown pathological diagnosis information, and ambiguous histology. We have collected the following data: age at diagnosis, gender(male or female), race, years of diagnosis, marital status, T stage, N stage, and histological types (8140/3, 8210/3, 8261/3, 8263/3, 8480/3, 8490/3). Additional variables encompassed grade (degree of tumor differentiation), primary tumor site (including colon and rectum), primary tumor size, and CEA levels. The tumor staging utilized in this study was based on version 0204. The ICD-O-3 manual serves as a reference for filtering histological type codes. At the same time, the site codes we selected include (C18.0 through C18.9; C20.9). The AJCC 7th edition TNM staging system was adopted for this study. Given that the SEER database comprises publicly available data, we do not require patients to sign informed consent during this study; likewise, ethical approval was unnecessary.
We selected patient data from Beijing Electric Power Hospital of Capital Medical University for external verification, and included patients who did not receive neoadjuvant radiotherapy before surgery to test the predictive performance of the optimal model. Our research was conducted retrospectively, and there was no violation of any aspects related to patient privacy; therefore, this study was granted an ethical exemption.
SPSS software (version 26.0) was utilized to perform statistical analyses. We performed Spearman correlation tests on all variables included in this study and presented the results as a heatmap to better illustrate the correlation between the data. We chose to express the categorical variables selected in this study as counts and percentages. We used the chi-square test, Fisher’s exact test, and Mann–Whitney U test to compare variables between the two groups. A multivariable logistic regression (LR) was developed, incorporating variables that demonstrated statistical significance in the univariable LR into the multivariable LR. In this study, we chose p < 0.05 as the criterion for statistical significance, and we calculated the two-sided p-value.
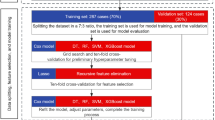
We apply Python 3.9.12 software to construct a machine learning algorithm. Incorporate the variables selected into the ML to establish a model for CRC lung metastasis. The sampled data is completely randomly divided into a training set and a test set in an 8:2 ratio. We used seven standard ML algorithms to establish a model for CRC lung metastasis. By comparing the predictive performance of the models (including accuracy, precision, recall rate, F1 score, and AUC), we selected the optimal model and evaluated its performance using fivefold cross-validation. Random Forest (RF) is an ML algorithm that can develop predictive models based on sample data. Previous studies have employed the RF algorithm to forecast renal disease, demonstrating high levels of accuracy15. Decision Tree (DT) represents one of the more successful algorithms within the realm of machine learning16. There are three key steps for us to establish a model depending on the DT algorithm: variable selection, node splitting, and tree pruning17. Support Vector Machine (SVM) is primarily employed to address classification problems18. Naive Bayes (NB) represents a straightforward variant of Bayesian networks that demonstrates strong predictive performance in addressing classification challenges19. K-Nearest Neighbor (KNN) is a straightforward ML that functions as a non-parametric classifier20. The eXtreme Gradient Boosting (XGBoost), a representative of ensemble learning, is widely popular due to its excellent predictive performance21. Gradient Boosting Machine (GBM) employs a multitude of smaller models, which are subsequently combined to generate the final ag. GBM utilizes a diverse array of smaller models, which are then integrated to produce the final aggregated prediction22. This study uses AUC and AUPR to evaluate the predictive performance of the model.
Sampling methods such as oversampling and undersampling mainly solve the problem of imbalanced categories in the original data. We use the Synthetic Minority Oversampling Technique (SMOTE) to oversample the raw data and the RandomUnderSampler to undersample the raw data, which is widely used to improve prediction models.
We use Shapley Additive exPlanation (SHAP) to visualize and explain the importance of the established optimal model variables. Shap is an ML technique that describes the output of a model by explaining the impact of features on the outcome. It evaluates the effects of variables on outcomes by calculating their Shapley values.
The SEER database indicates that fewer colorectal cancer patients exhibit lung metastasis. We address imbalance data by employing under-sampling and over-sampling on the original data. Subsequently, a correlation matrix is used to evaluate the relationship between sampled data variables. The relationships between variables become more pronounced after sampling. As illustrated in Fig. 2.
Results
This study included 39,674 CRC patients, among which 1,369 (3.5%) had lung metastasis. We collected data from 207 patients in a hospital in China to validate the feasibility of the model from 2010 to 2015. The baseline data for this study are detailed in Table 1. Nine risk factors were included in the multivariate LR model, including age, histological type, grade, primary tumor site, T stage, N stage, tumor size, CEA levels, and tumor deposition. Further details are presented in Table 2. According to the LR model prediction results: AUC = 0.854; 95% CI (0.844–0.863); p < 0.001.
Seven kinds of ML algorithms were established and contrasted depending on metrics such as accuracy, precision, recall, F1 score, and AUC value. The model trained using oversampling techniques outperformed the one trained with undersampling methods; for detailed information regarding the seven machine learning models constructed through oversampling, please refer to Table 3. We present the performance results in Fig. 3 by employing both oversampling and undersampling approaches in building these 7 ML algorithms. Notably, all models achieved an AUC greater than 0.800 with models built with oversampled data. RF algorithm is superior to other algorithms. In the test set, RF demonstrated excellent predictive performance, with an AUC of 0.980 and an AUPR of 0.941; it showed excellent performance during fivefold cross-validation, with an average accuracy of 0.936.
When testing data from a Chinese hospital, RF also demonstrated impressive predictive ability: accuracy of 0.961, an AUC of 0.927, and an AUPR of 0.657, as illustrated in Fig. 4. We compared the performance of the RF algorithm and LR using AUC values, and the results showed that the RF algorithm (AUC = 0.980) outperformed the LR model (AUC = 0.854).
Interpreting RF using SHAP values. In Fig. 5A, clinical features are ranked according to their average absolute SHAP values to illustrate their relative significance. Figure 5B offers a comprehensive visualization of how various factors influence the RF.
The SHapley Additive exPlanations of the RF model. (A) SHAP feature importance quantified through the average absolute Shapley values. This plot illustrates the significance of each feature in the development of the predictive model. (B) Representation of the influence exerted by each feature on the final model output, assessed via SHAP values distribution. A data point within each row denotes every individual patient. The color indicates whether the continuous feature is at a high level (displayed in red) or a low level (depicted in blue) for that specific observation. When it comes to categorical features, the color blue signifies “no”, while the color red corresponds to “yes”.
We selected two patients diagnosed with CRC—one exhibiting lung metastasis and the other without—based on our constructed model (Fig. 6A). For patients with lung metastasis, significant risk factors included tumor deposits, tumor size ≥ 5 cm, CEA positive status, and age ≥ 60 years. Conversely, in patients without lung metastasis, protective factors encompassed CEA negative status, Grade I tumors, absence of tumor deposits, colon involvement (T3), and belonging to other ethnicities (Fig. 6B).
SHAP force plot for interpreting individual’s prediction outcomes. This plot offers a visual illustration of the RF model’s predictions, wherein the blue and red bars signify risk factors and protective factors, respectively. The length of the bars corresponds to the extent of feature importance. (A) Poor outcome; (B) favorable outcome.
As illustrated in Fig. 5, tumor deposits emerge as the most significant predictive factors for lung metastasis with CRC.
Our study constructs an online network calculator utilizing the RF algorithm. (http://121.43.117.60:8003/).
Discussion
CRC is a prevalent malignancy. Approximately 25% of patients are diagnosed with distant metastasis, which remains the primary cause of mortality among this patient population23. Generally speaking, the process of colorectal cancer metastasis primarily involves tumor cells entering the liver via the portal vein system. Subsequently, these cells can disseminate to other organs, including the lungs24. However, the metastatic pathway of lung metastases may circumvent the portal system by utilizing venous drainage, thereby entering the systemic circulation25. It is essential to investigate the risk factors associated with lung metastasis in CRC patients separately, as the patterns of lung metastasis may differ, and early detection and timely intervention are crucial. Studies have indicated that patients who identify lung metastasis at an early stage and undergo surgical intervention experience a 30% higher five-year survival rate compared to those who do not receive surgical treatment26. At present, the existing detection methods for lung metastasis, such as PET-CT and biopsy, exhibit certain limitations; PET-CT is not suitable for early screening due to its prohibitive cost and potential risk of radiation damage. Likewise, while biopsy can confirm lung metastasis, it also carries the risks of tumor dissemination and false negatives27. With artificial intelligence already widely applied in the medical field, it serves as a crucial tool for precision medicine, assisting in the selection of optimal diagnostic and treatment strategies1; the machine learning model, established on clinical and pathological data, not only circumvents the risks associated with high examination costs, limited scalability, and potential bodily harm but also exhibits strong predictive performance (with an accuracy of 0.961), assisting clinicians in identifying high-risk patients and formulating tailored treatment plans. We utilized an explainable ML algorithm that integrates clinical and pathological features to develop predictive models for lung cancer metastasis. Additionally, we performed a comparative analysis of these models.
Among the various approaches evaluated, the machine learning model demonstrated commendable performance. In our previous research, explainable ML algorithms have not been used to make corresponding predictions. The AUC of 7 algorithms predominantly exceeds 0.800. Consequently, we believe that the models we have built based on ML have good predictive performance. The RF algorithm is the best-performing model. Currently, research on CRC patients primarily emphasizes prognostic factors28,29,30. In prior studies, researchers have developed models while investigating the associated risk factors12,31. Some studies have predicted lung metastasis and assessed overall survival rates among these individuals32. Previous studies on lung metastasis of colorectal cancer were primarily based on traditional logistic regression, and the AUC of these models was often less than 0.812,33; the predictive performance of these models still requires improvement. Simultaneously, the adoption of interpretable machine learning algorithms for our models has significantly enhanced their explanation in practical applications, thereby facilitating users’ understanding of the model’s operational rules.
The LR revealed that age, histological type, grade, primary tumor site, T stage, N stage, tumor size, carcinoembryonic antigen (CEA) levels, and tumor deposits were all independent predictors of lung metastasis in CRC patients. The RF feature selection, consistent with those from the multivariable logistic regression analysis, indicated that tumor deposits are a key predictor. CEA levels and the T stage follow this. Notably, the T stage is a critical criterion for assessing tumor progression. Tumor deposits are observed in 20%–25% of patients with colon cancer33. At present, there is a limited body of research on tumor deposits, with the majority concentrating on their impact on prognosis34. CEA is one of the essential predictive factors in this study; the level of CEA is closely related to the distant metastasis of patients35. Studies have demonstrated that the T stage is directly associated with distant metastasis in various regions of the tumor32. In this study, T staging emerged as the most significant predictor within the RF model.
Furthermore, multivariable logistic analysis demonstrated a positive correlation between T staging and the risk of lung metastasis. This association may be attributed to the direct invasion of cancer cells into the systemic circulation via the venous system35. The primary tumor site emerged as a significant predictor in this study. It was observed that the rectum exhibited a higher propensity for lung metastasis compared to the colon (including the sigmoid), which aligns with previous findings12,31. However, some studies suggest that there is minimal difference in the impact of lung metastasis between these factors32. Consistent with previous research12, the N stage serves as a significant predictor of lung metastasis in CRC. Stage N is associated with the risk of lung metastasis in CRC patients. This may be attributed to the lymphatic system being the primary pathway for metastasis36, and the Lung is among the organs that contain the highest abundance of lymph27. Furthermore, it has been indicated that the lymphatic drainage from positive regional lymph nodes can directly reach the lungs, thereby facilitating the occurrence of lung metastasis37.
ML is a scientific discipline that employs computational techniques to learn from data. While statistics focuses on elucidating the relationships among data, computer science prioritizes the development of more efficient algorithms for computation. Machine learning thus represents the convergence of these two fields38. Whereas traditional statistics primarily emphasizes the testing of causal hypotheses, machine learning places greater emphasis on the predictive performance of models39. As a compilation of diverse algorithms, machine learning must take into account a range of factors, including data availability, data type, and the specific aspects that require prediction when addressing practical problems to identify the most suitable algorithm40. Predictive models developed through machine learning can enable healthcare professionals to deliver more personalized and precise diagnostic and therapeutic strategies for patients. As databases continue to expand and algorithms undergo optimization, machine learning algorithms are poised to assume an even more significant role within the medical ___domain.
We developed seven predictive models to predict lung metastasis in colorectal cancer. The performance of these seven algorithms was assessed based on accuracy, precision, recall, F1 score, and AUC. Among them, RF demonstrated superior predictability with an AUC of 0.980, surpassing that of the logistic regression, which had an AUC of 0.854. Consequently, RF emerged as the most effective algorithm.
Our study has several limitations: (1) The validation cohort consisted of single-center data with a limited number of patients, all of whom were Asian. (2) We anticipate that the accuracy of the model can be further enhanced by incorporating more risk factors associated with metastasis in future research. (3) Additionally, the SEER database does not provide details on specific treatment plans; further analysis is required to assess their impact on patient prognosis. Furthermore, regional differentiation was not included in the decision-making process within this model.
Our study developed and validated a model utilizing explainable ML algorithms, which incorporates clinical features to quantify the primary factors contributing to lung metastasis. Among these factors, tumor deposits, CEA levels, and T stage emerged as the three most significant predictors of lung metastasis in CRC patients. In comparison to logistic regression models, the random forest algorithm demonstrated superior predictive capability; thus, it offers the potential for personalized treatment strategies. We have built a web calculator (http://121.43.117.60:8003/).
Data availability
The datasets generated for this study are available on request to the first author or the corresponding author.
Abbreviations
- ML:
-
Machine learning
- CRC:
-
Colorectal cancer
- SHAP:
-
Shapley additive exPlanations
- DM:
-
Distant metastasis
- LR:
-
Logistic regression
- RF:
-
Random forest
- DT:
-
Decision tree
- SVM:
-
Support vector machine
- NB:
-
Naive Bayes
- KNN:
-
K-nearest neighbor
- XGBoost:
-
EXtreme gradient boosting
- GBM:
-
Gradient boosting machine
References
Mao, Y. et al. Machine learning algorithms are comparable to conventional regression models in predicting distant metastasis of follicular thyroid carcinoma. Clin. Endocrinol. (Oxf.) 98(1), 98–109. https://doi.org/10.1111/cen.14693 (2023).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424. https://doi.org/10.3322/caac.21492 (2018).
Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 70(4), 313. https://doi.org/10.3322/caac.21609 (2020).
Li, T. et al. Predictive models based on machine learning for bone metastasis in patients with diagnosed colorectal cancer. Front. Public Health 10, 984750. https://doi.org/10.3389/fpubh.2022.984750 (2022).
Yoon, K. D. & Suman, P. Invited commentary: Survival outcome of palliative primary tumor resection for colorectal cancer patients with synchronous liver and/or lung metastases: A retrospective cohort study in the SEER database by propensity score matching analysis. Int. J. Surg. 82, 85–86. https://doi.org/10.1016/j.ijsu.2020.08.008 (2020).
Kopetz, S. et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J. Clin. Oncol. 27(22), 3677–3683. https://doi.org/10.1200/jco.2008.20.5278 (2009).
Limmer, S. & Unger, L. Optimal management of pulmonary metastases from colorectal cancer. Expert Rev. Anticancer Ther. 11(10), 1567–1575. https://doi.org/10.1586/era.11.123 (2011).
Zhang, G. Q. et al. Aggressive multimodal treatment and metastatic colorectal cancer survival. J. Am. Coll. Surg. 230(4), 689–698. https://doi.org/10.1016/j.jamcollsurg.2019.12.024 (2020).
Luo, D. et al. Prognostic value of distant metastasis sites and surgery in stage IV colorectal cancer: A population-based study. Int. J. Colorectal Dis. 33(9), 1241–1249. https://doi.org/10.1007/s00384-018-3091-x (2018).
Yi, C. et al. Is Primary tumor excision and specific metastases sites resection associated with improved survival in stage IV colorectal cancer? Results from SEER database analysis. Am. Surg. 86(5), 499–507. https://doi.org/10.1177/0003134820919729 (2020).
Huang, Y. et al. Pulmonary metastasis in newly diagnosed colon-rectal cancer: A population-based nomogram study. Int. J. Colorectal Dis. 34(5), 867–878. https://doi.org/10.1007/s00384-019-03270-w (2019).
Li, Y. et al. Predictive and prognostic factors of synchronous colorectal lung-limited metastasis. Gastroenterol. Res. Pract. 2020, 6131485. https://doi.org/10.1155/2020/6131485 (2020).
Liu, W. et al. Prediction of lung metastases in thyroid cancer using machine learning based on SEER database. Cancer Med. 11(12), 2503–2515. https://doi.org/10.1002/cam4.4617 (2022).
Frizzell, J. D. et al. Prediction of 30-day all-cause readmissions in patients hospitalized for heart failure: Comparison of machine learning and other statistical approaches. JAMA Cardiol. 2(2), 204–209. https://doi.org/10.1001/jamacardio.2016.3956 (2017).
Xing, F. et al. A new random forest algorithm-based prediction model of postoperative mortality in geriatric patients with hip fractures. Front. Med. (Lausanne) 9, 829977. https://doi.org/10.3389/fmed.2022.829977 (2022).
Che, D., Liu, Q., Rasheed, K. & Tao, X. Decision tree and ensemble learning algorithms with their applications in bioinformatics. Adv. Exp. Med. Biol. 696, 191–199. https://doi.org/10.1007/978-1-4419-7046-6_19 (2011).
Chern, C. C., Chen, Y. J. & Hsiao, B. Decision tree-based classifier in providing telehealth service. BMC Med. Inform. Decis. Mak. 19(1), 104. https://doi.org/10.1186/s12911-019-0825-9 (2019).
Lee, Y. W., Choi, J. W. & Shin, E. H. Machine learning model for predicting malaria using clinical information. Comput. Biol. Med. 129, 104151. https://doi.org/10.1016/j.compbiomed.2020.104151 (2021).
Lee, S. M., Park, J. H. & Park, H. J. Implications of systematic review for breast cancer prediction. Cancer Nurs. 31(5), E40–E46. https://doi.org/10.1097/01.NCC.0000305765.34851.e9 (2008).
Pal, M., Parija, S., Panda, G., Dhama, K. & Mohapatra, R. K. Risk prediction of cardiovascular disease using machine learning classifiers. Open Med. (Wars) 17(1), 1100–1113. https://doi.org/10.1515/med-2022-0508 (2022).
Ma, B. et al. Diagnostic classification of cancers using extreme gradient boosting algorithm and multi-omics data. Comput. Biol. Med. 121, 103761. https://doi.org/10.1016/j.compbiomed.2020.103761 (2020).
Lynch, C. M. et al. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int. J. Med. Inform. 108, 1–8. https://doi.org/10.1016/j.ijmedinf.2017.09.013 (2017).
Roth, E. S. et al. Does colon cancer ever metastasize to bone first? A temporal analysis of colorectal cancer progression. BMC Cancer 9, 274. https://doi.org/10.1186/1471-2407-9-274 (2009).
Mitry, E. et al. Epidemiology, management and prognosis of colorectal cancer with lung metastases: A 30-year population-based study. Gut 59(10), 1383–1388. https://doi.org/10.1136/gut.2010.211557 (2010).
Robinson, J. R., Newcomb, P. A., Hardikar, S., Cohen, S. A. & Phipps, A. I. Stage IV colorectal cancer primary site and patterns of distant metastasis. Cancer Epidemiol. 48, 92–95. https://doi.org/10.1016/j.canep.2017.04.003 (2017).
Heinemann, V. et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 15(10), 1065–1075. https://doi.org/10.1016/s1470-2045(14)70330-4 (2014).
Qiu, B., Shen, Z., Yang, D. & Wang, Q. Applying machine learning techniques to predict the risk of lung metastases from rectal cancer: a real-world retrospective study. Front. Oncol. 13, 1183702. https://doi.org/10.3389/fonc.2023.1183072 (2023).
Deng, S. et al. Development and validation of a prognostic scoring system for patients with colorectal cancer hepato-pulmonary metastasis: a retrospective study. BMC Cancer 22(1), 643. https://doi.org/10.1186/s12885-022-09738-3 (2022).
Huang, B. et al. Smaller tumor size is associated with poor survival in stage II colon cancer: An analysis of 7,719 patients in the SEER database. Int. J. Surg. 33(Pt A), 157–163. https://doi.org/10.1016/j.ijsu.2016.07.073 (2016).
Ding, X. et al. Risk and prognostic nomograms for colorectal neuroendocrine neoplasm with liver metastasis: A population-based study. Int. J. Colorectal Dis. 36(9), 1915–1927. https://doi.org/10.1007/s00384-021-03920-y (2021).
Nordholm-Carstensen, A., Krarup, P. M., Jorgensen, L. N., Wille-Jørgensen, P. A. & Harling, H. Occurrence and survival of synchronous pulmonary metastases in colorectal cancer: A nationwide cohort study. Eur. J. Cancer 50(2), 447–456. https://doi.org/10.1016/j.ejca.2013.10.009 (2014).
Mo, S. et al. Nomograms for predicting specific distant metastatic sites and overall survival of colorectal cancer patients: A large population-based real-world study. Clin. Transl. Med. 10(1), 169–181. https://doi.org/10.1002/ctm2.20 (2020).
Nagtegaal, I. D. et al. Tumor deposits in colorectal cancer: Improving the value of modern staging-a systematic review and meta-analysis. J. Clin. Oncol. 35(10), 1119–1127. https://doi.org/10.1200/jco.2016.68.9091 (2017).
Guo, Z. et al. Machine learning for predicting liver and/or lung metastasis in colorectal cancer: A retrospective study based on the SEER database. Eur. J. Surg. Oncol. 50(7), 108362. https://doi.org/10.1016/j.ejso.2024.108362 (2024).
Hughes, E. S. & Cuthbertson, A. M. Recurrence after curative excision of carcinoma of the large bowel. JAMA 182, 1303–1306. https://doi.org/10.1001/jama.1962.03050520001001 (1962).
Dumont, F. et al. Significance of lymph node involvement in local recurrence of colorectal cancer. J. Surg. Oncol. 120(4), 722–728. https://doi.org/10.1002/jso.25631 (2019).
Kato, Y. et al. Lymph node metastasis is strongly associated with lung metastasis as the first recurrence site in colorectal cancer. Surgery 170(3), 696–702. https://doi.org/10.1016/j.surg.2021.03.017 (2021).
Deo, R. C. Machine learning in medicine. Circulation 132(20), 1920–1930. https://doi.org/10.1161/circulationaha.115.001593 (2015).
Handelman, G. S. et al. eDoctor: Machine learning and the future of medicine. J. Intern. Med. 284(6), 603–619. https://doi.org/10.1111/joim.12822 (2018).
Weiss, J., Kuusisto, F., Boyd, K., Liu, J. & Page, D. Machine learning for treatment assignment: Improving individualized risk attribution. AMIA Annu. Symp. Proc. 2015, 1306–1315 (2015).
Funding
This study was supported by Beijing Municipal Science & Technology Commission (No. Z171100000417056) and the Key Support Project of Guo Zhong Health Care of China General Technology Group (GZKJ-KJXX-QTHT-20240429).
Author information
Authors and Affiliations
Contributions
Zhentian Guo: Conceptualization, methodology, software, Investigation, Writing—original draft. Zongming Zhang: Conceptualization, Funding acquisition, Investigation, Formal Analysis, Supervision, Writing—review and editing. Limin Liu: Data curation, Investigation. Yue Zhao: Data curation, Investigation. Zhuo Liu: Data curation, Investigation. Chong Zhang: Data curation, Investigation. Hui Qi: Data curation, Investigation. Jinqiu Feng: Data curation, Investigation, Writing—review and editing. Peijie Yao: Data curation, Investigation.
Corresponding author
Ethics declarations
Competing of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, Z., Zhang, Z., Liu, L. et al. Explainable machine learning for predicting lung metastasis of colorectal cancer. Sci Rep 15, 13611 (2025). https://doi.org/10.1038/s41598-025-98188-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-98188-5