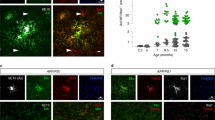
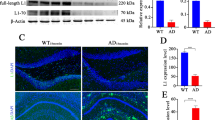
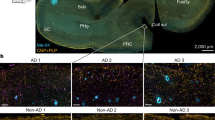
Abstract
Genome-wide association studies have identified many gene polymorphisms associated with an increased risk of developing late-onset Alzheimer’s disease (LOAD). Many of these LOAD risk-associated alleles alter disease pathogenesis by influencing innate immune responses and lipid metabolism of microglia (MG). Here we show that boosting the expression of angiotensin-converting enzyme (ACE), a genome-wide association study LOAD risk-associated gene product, specifically in MG, reduces amyloid-β (Aβ) plaque load, preserves vulnerable neurons and excitatory synapses, and significantly reduces learning and memory abnormalities in the 5xFAD amyloid mouse model of AD. ACE-expressing MG surround plaques more frequently and they have increased Aβ phagocytosis, endolysosomal trafficking and spleen tyrosine kinase activation downstream of the major Aβ receptors, triggering receptor expressed on myeloid cells 2 (Trem2) and C-type lectin ___domain family 7 member A (Clec7a). These findings establish a role for ACE in enhancing microglial immune function and they identify a potential use for ACE-expressing MG as a cell-based therapy to augment endogenous microglial responses to Aβ in AD.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The snRNA-seq and GeoMx data have been deposited with the Gene Expression Omnibus under accession nos. GSE288410 and GSE288280, respectively. The RNA-seq data used the mouse genome sequence GRCm38 (mm10) for reference (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001635.20/). Source data are provided with this paper. Any other data reported in this paper are available from the corresponding author upon reasonable request.
Code availability
No custom code was used for any analysis in these studies.
References
2023 Alzheimer’s disease facts and figures. Alzheimers Dement. 19, 1598–1695 (2023).
Frisoni, G. B. et al. The probabilistic model of Alzheimer disease: the amyloid hypothesis revised. Nat. Rev. Neurosci. 23, 53–66 (2022).
Clayton, K. A., Van Enoo, A. A. & Ikezu, T. Alzheimer’s disease: the role of microglia in brain homeostasis and proteopathy. Front. Neurosci. 11, 680 (2017).
Xu, Y., Jin, M.-Z., Yang, Z.-Y. & Jin, W.-L. Microglia in neurodegenerative diseases. Neural Regen. Res. 16, 270–280 (2021).
Chávez-Gutiérrez, L. & Szaruga, M. Mechanisms of neurodegeneration—insights from familial Alzheimer's disease. Semin. Cell Dev. Biol. 105, 75–85 (2020).
Wightman, D. P. et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282 (2021).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022).
Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019).
Miao, J. et al. Microglia in Alzheimer’s disease: pathogenesis, mechanisms, and therapeutic potentials. Front. Aging Neurosci. 15, 1201982 (2023).
Wingo, A. P. et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nat. Genet. 53, 143–146 (2021).
Kehoe, P. G. et al. Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer disease. Nat. Genet. 21, 71–72 (1999).
Lehmann, D. J. et al. Large meta-analysis establishes the ACE insertion-deletion polymorphism as a marker of Alzheimer’s disease. Am. J. Epidemiol. 162, 305–317 (2005).
Marioni, R. E. et al. GWAS on family history of Alzheimer’s disease. Transl. Psychiatry 8, 99 (2018).
Meng, Y. et al. Association of polymorphisms in the angiotensin-converting enzyme gene with Alzheimer disease in an Israeli Arab community. Am. J. Hum. Genet. 78, 871–877 (2006).
Esther, C. R. et al. The critical role of tissue angiotensin-converting enzyme as revealed by gene targeting in mice. J. Clin. Invest. 99, 2375–2385 (1997).
Murray, M. D. et al. Antihypertensive medication and dementia risk in older adult African Americans with hypertension: a prospective cohort study. J. Gen. Intern. Med. 33, 455–462 (2018).
Hughes, D. et al. Association of blood pressure lowering with incident dementia or cognitive impairment: a systematic review and meta-analysis. JAMA 323, 1934–1944 (2020).
Ding, J. et al. Antihypertensive medications and risk for incident dementia and Alzheimer’s disease: a meta-analysis of individual participant data from prospective cohort studies. Lancet Neurol. 19, 61–70 (2020).
Li, N.-C. et al. Use of angiotensin receptor blockers and risk of dementia in a predominantly male population: prospective cohort analysis. BMJ 340, b5465 (2010).
van Dalen, J. W. et al. Association of angiotensin II–stimulating antihypertensive use and dementia risk. Neurology 96, e67–e80 (2021).
Barthold, D., Joyce, G., Wharton, W., Kehoe, P. & Zissimopoulos, J. The association of multiple anti-hypertensive medication classes with Alzheimer’s disease incidence across sex, race, and ethnicity. PLoS ONE 13, e0206705 (2018).
Rao, A. et al. Diverse biological functions of the renin-angiotensin system. Med. Res. Rev. 44, 587–605 (2024).
Sinka, L., Biasch, K., Khazaal, I., Péault, B. & Tavian, M. Angiotensin-converting enzyme (CD143) specifies emerging lympho-hematopoietic progenitors in the human embryo. Blood 119, 3712–3723 (2012).
Bernstein, K. E. et al. Angiotensin-converting enzyme in innate and adaptive immunity. Nat. Rev. Nephrol. 14, 325–336 (2018).
Zou, K. et al. Angiotensin-converting enzyme converts amyloid β-protein 1–42 (Aβ1–42) to Aβ1–40, and its inhibition enhances brain Aβ deposition. J. Neurosci. 27, 8628–8635 (2007).
Shen, X. Z. et al. Mice with enhanced macrophage angiotensin-converting enzyme are resistant to melanoma. Am. J. Pathol. 170, 2122–2134 (2007).
Khan, Z. et al. Angiotensin-converting enzyme enhances the oxidative response and bactericidal activity of neutrophils. Blood 130, 328–339 (2017).
Okwan-Duodu, D. et al. Overexpression of myeloid angiotensin-converting enzyme (ACE) reduces atherosclerosis. Biochem. Biophys. Res. Commun. 520, 573–579 (2019).
Bernstein, K. E. et al. Angiotensin-converting enzyme overexpression in myelocytes enhances the immune response. Biol. Chem. 395, 1173–1178 (2014).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
McKinsey, G. L. et al. A new genetic strategy for targeting microglia in development and disease. eLife 9, e54590 (2020).
Parkhurst, C. N. et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Ohno, M. Failures to reconsolidate memory in a mouse model of Alzheimer’s disease. Neurobiol. Learn. Mem. 92, 455–459 (2009).
Ferro, A., Auguste, Y. S. S. & Cheadle, L. Microglia, cytokines, and neural activity: unexpected interactions in brain development and function. Front. Immunol. 12, 703527 (2021).
Stence, N., Waite, M. & Dailey, M. E. Dynamics of microglial activation: a confocal time-lapse analysis in hippocampal slices. Glia 33, 256–266 (2001).
Navarro, V. et al. Microglia in Alzheimer’s disease: activated, dysfunctional or degenerative. Front. Aging Neurosci. 10, 140 (2018).
Roy, E. R. et al. Concerted type I interferon signaling in microglia and neural cells promotes memory impairment associated with amyloid β plaques. Immunity 55, 879–894 (2022).
Eimer, W. A. & Vassar, R. Neuron loss in the 5XFAD mouse model of Alzheimer’s disease correlates with intraneuronal Aβ42 accumulation and Caspase-3 activation. Mol. Neurodegener. 8, 2 (2013).
Bhembre, N., Bonthron, C. & Opazo, P. Synaptic compensatory plasticity in Alzheimer’s disease. J. Neurosci. 43, 6833–6840 (2023).
Hong, S. et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716 (2016).
Roy, E. R. et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J. Clin. Invest. 130, 1912–1930 (2020).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017).
Koutsodendris, N. et al. Neuronal APOE4 removal protects against tau-mediated gliosis, neurodegeneration and myelin deficits. Nat. Aging 3, 275–296 (2023).
Chen, X. & Holtzman, D. M. Emerging roles of innate and adaptive immunity in Alzheimer’s disease. Immunity 55, 2236–2254 (2022).
Palmieri, M. et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20, 3852–3866 (2011).
Settembre, C. & Medina, D. L. Chapter 3—TFEB and the CLEAR network. Methods Cell Biol. 126, 45–62 (2015).
Wang, S. et al. TREM2 drives microglia response to amyloid-β via SYK-dependent and -independent pathways. Cell 185, 4153–4169.e19 (2022).
Torres-Hernandez, A. et al. Targeting SYK signaling in myeloid cells protects against liver fibrosis and hepatocarcinogenesis. Oncogene 38, 4512–4526 (2019).
Ennerfelt, H. et al. SYK coordinates neuroprotective microglial responses in neurodegenerative disease. Cell 185, 4135–4152.e22 (2022).
Cao, D.-Y. et al. ACE overexpression in myeloid cells increases oxidative metabolism and cellular ATP. J. Biol. Chem. 295, 1369–1384 (2020).
Schafer, D. P. & Stillman, J. M. Microglia are SYK of Aβ and cell debris. Cell 185, 4043–4045 (2022).
Schweig, J. E. et al. Alzheimer’s disease pathological lesions activate the spleen tyrosine kinase. Acta Neuropathol. Commun. 5, 69 (2017).
Makhoul, S., Dorschel, S., Gambaryan, S., Walter, U. & Jurk, K. Feedback regulation of Syk by protein kinase C in human platelets. Int. J. Mol. Sci. 21, 176 (2019).
Tsang, E. et al. Molecular mechanism of the Syk activation switch. J. Biol. Chem. 283, 32650–32659 (2008).
Liu, S. et al. A clinical dose of angiotensin-converting enzyme (ACE) inhibitor and heterozygous ACE deletion exacerbate Alzheimer’s disease pathology in mice. J. Biol. Chem. 294, 9760–9770 (2019).
Ryan, D. K. et al. Genetic evidence for protective effects of angiotensin-converting enzyme against Alzheimer disease but not other neurodegenerative diseases in European populations. Neurol. Genet. 8, e200014 (2022).
Okwan-Duodu, D. et al. Angiotensin-converting enzyme overexpression in mouse myelomonocytic cells augments resistance to Listeria and methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 285, 39051–39060 (2010).
Bernstein, K. E. et al. Angiotensin-converting enzyme overexpression in myelomonocytes prevents Alzheimer’s-like cognitive decline. J. Clin. Invest. 124, 1000–1012 (2014).
Bernstein, K. E. et al. Classical and nonclassical effects of angiotensin-converting enzyme: how increased ACE enhances myeloid immune function. J. Biol. Chem. 300, 107388 (2024).
Sudwarts, A. & Thinakaran, G. Alzheimer’s genes in microglia: a risk worth investigating. Mol. Neurodegener. 18, 90 (2023).
Liu, C.-C. et al. Cell-autonomous effects of APOE4 in restricting microglial response in brain homeostasis and Alzheimer’s disease. Nat. Immunol. 24, 1854–1866 (2023).
Zhao, Y. et al. TREM2 is a receptor for β-amyloid that mediates microglial function. Neuron 97, 1023–1031.e7 (2018).
Atagi, Y. et al. Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2). J. Biol. Chem. 290, 26043–26050 (2015).
Song, W. M. et al. Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J. Exp. Med. 215, 745–760 (2018).
Zhou, Y. et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020).
Kisselev, A. F., van der Linden, W. A. & Overkleeft, H. S. Proteasome inhibitors: an expanding army attacking a unique target. Chem. Biol. 19, 99–115 (2012).
Donato, R. et al. Functions of S100 proteins. Curr. Mol. Med. 13, 24–57 (2013).
Liew, F. Y., Girard, J.-P. & Turnquist, H. R. Interleukin-33 in health and disease. Nat. Rev. Immunol. 16, 676–689 (2016).
Yamada, K. M. & Sixt, M. Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 20, 738–752 (2019).
Maeda, M. et al. ARHGAP18, a GTPase-activating protein for RhoA, controls cell shape, spreading, and motility. Mol. Biol. Cell 22, 3840–3852 (2011).
Vizovišek, M., Fonović, M. & Turk, B. Cysteine cathepsins in extracellular matrix remodeling: extracellular matrix degradation and beyond. Matrix Biol. 75-76, 141–159 (2019).
Bergeron, S. et al. The serine protease inhibitor serpinE2 is a novel target of ERK signaling involved in human colorectal tumorigenesis. Mol. Cancer 9, 271 (2010).
Reyes, R., Cardeñes, B., Machado-Pineda, Y. & Cabañas, C. Tetraspanin CD9: a key regulator of cell adhesion in the immune system. Front. Immunol. 9, 863 (2018).
Baloh, R. H. et al. Transplantation of human neural progenitor cells secreting GDNF into the spinal cord of patients with ALS: a phase 1/2a trial. Nat. Med. 28, 1813–1822 (2022).
Ifediora, N., Canoll, P. & Hargus, G. Human stem cell transplantation models of Alzheimer’s disease. Front. Aging Neurosci. 16, 1354164 (2024).
Chadarevian, J. P. et al. Therapeutic potential of human microglia transplantation in a chimeric model of CSF1R-related leukoencephalopathy. Neuron 112, 2686–2707.e8 (2024).
Goodwin, L. O. et al. Large-scale discovery of mouse transgenic integration sites reveals frequent structural variation and insertional mutagenesis. Genome Res. 29, 494–505 (2019).
Herre, J. et al. Dectin-1 uses novel mechanisms for yeast phagocytosis in macrophages. Blood 104, 4038–4045 (2004).
Krasemann, S. et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581 (2017).
Acknowledgements
This study was supported by NIH grant nos. RF1-AG074365 and R01-AG074365 to W.G.T., NIH grant no. R01-AI164519 to K.E.B. and a Cedars-Sinai Goldrich Alzheimer’s Center grant to W.G.T. and K.E.B. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank L. Karman for technical assistance and A. Moser for reviewing the paper.
Author information
Authors and Affiliations
Contributions
W.G.T. conceived the project and designed the studies. A.R.G., S.W., H.R.B., A.K.M.G.M, L.L., A.M., J.I. and J.C. performed the experiments. W.G.T., A.R.G., S.W., H.R.B., A.K.M.G.M, K.E.B., L.L. and R.A. analyzed the data. W.G.T. and A.R.G. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 ACE expression in microglia from wildtype, 5xFAD and ACE10/10 mice.
Representative immunofluorescence images from 6-month-old brain cut in the coronal plane at -2mm Bregma from wildtype (WT), homozygous 5xFAD (5xFADTg/Tg) and homozygous ACE10/10 mice using antibodies to Iba1 to label microglia (green) and ACE (magenta). Representative images depicted were derived from analysis of n = 3 mice of each genotype and 2 brain sections per mouse with consistent results. (scale, 100 µm).
Extended Data Fig. 2 Generating and characterizing knock-in mice with ACE expression restricted to microglia.
a, Rosa-hACE-flox mice were generated by targeting the Gt(Rosa) safe harbor locus in mouse embryonic stem cells. The targeting construct contains a loxP-site flanked transcriptional stop sequence upstream of a bicistronic TurboGFP and c-terminal flag-tagged human ACE (hACE-3xF) expression unit that is expressed after loxP site recombination in the genome. Embryonic stem cell clones were screened by PCR (3′ screen) to identify positive clones that generated a 4,355 bp PCR amplicon. Secondary screening was used to detect 5’ homologous recombination by Southern blotting on MfeI restricted genomic DNA to identify a 10.3 kb fragment indicative of heterozygous homologous recombination and germline heterozygous ACE-flox (Rosa+/hACE-flox) mice were generated. b, ACE-flox mice were mated to Cx3Cr1+/CreERT2 mice to generate RACE- (Cx3Cr1+/CreERT2; Rosa+/+) and RACE+ (Cx3Cr1+/CreERT2; Rosa+/hACE-flox) mice. c, RACE- and RACE+ mice were injected with Tamoxifen (TMX; 125 mg/kg, IP) for four consecutive days to activate Cre-recombinase in R- and R+ myeloid-derived cells. d, Myeloid cells isolated from brain, spleen and blood were gated on DAPI, CD11b and Cx3Cr1 and then sub-gated on GFP to detect transgene expression, 7, 21 and 180 days after TMX injection. e, Transgene expression persisted in myeloid cells in the brain and became undetectable in the blood and spleen by 3 weeks after TMX injection (data are plotted as mean ± SEM).
Extended Data Fig. 3 ACE expression in the brain of R+ mice.
Representative immunofluorescence images from 9-month-old R+ mouse brain cut in the coronal plane at -2mm Bregma and two representative brain regions shown (Parietal Association Cortex - PtA and hippocampus CA1, stratum radiatum – CA1:SR). Labeling for ACE expression in microglia (green) does not co-localize with a, neurons (NeuN; magenta), b, astrocytes (GFAP; magenta) or c, oligodendroglia cells (Sox10; magenta). Experiments depicted in representative images were derived from analysis of n = 2; R+ mice and 2 sections per mouse with consistent results in all areas of the brain. (scale, 50 µm).
Extended Data Fig. 4 Amyloid plaque burden in 5xFAD mice with and without expression of ACE in microglia.
Representative images of 9-month-old a, R−; AD+ and b, R+; AD+ brain cut in the coronal plane at -2mm Bregma and stained with Thioflavin S (Thio. S), showing representative regions of the brain where plaque burden is quantified; hippocampus CA1, stratum oriens (CA1:SO), Parietal Association (PtA) cortex and Amygdala (Amg) (scale, 500 µm). c, Quantification of the percent of Thio. S coverage in R+; AD+ normalized to coverage in R−; AD+ brain in the CA1:SO, PtA and Amg. Representative immunofluorescence images of 9-month-old d, R−; AD+ and e, R+; AD+ brain cut in the coronal plane at -2mm Bregma labeled with 6E10 antibody to label amyloid precursor protein and showing representative regions of the CA1:SO, PtA and Amg that are quantified (scale, 500 µm). f, Quantification of the percent of 6E10 coverage in R+; AD+ and R−; AD+ brains in the CA1:SO, PtA and Amg. For quantification in c, f, n = 3-4 males and 4-5 females from each genotype, sexes are combined since no significant differences were detected between males and females, plotted as mean ± SEM normalized to the mean of R−; AD+, two-way analysis of variation (ANOVA) and Sidak’s post-hoc test for multiple comparisons, * = p < 0.05, ** = p < 0.01, and *** = p < 0.001. (male values = square symbol and female values = round symbol).
Extended Data Fig. 5 Cytokine levels in the hippocampus of wild type and 5xFAD mice with and without ACE expression in microglia.
Cytokine levels for a, TNF-α, b, IL-1β, c, CXCL1, d, IFN-γ, e, IL-6, f, IL-10, g, IL-12p70, h, IL-2 and i, IL-5 measured from 100 µg of total soluble hippocampal protein (THP) from 9-month-old, R−; AD-, R+; AD-, R−; AD+ and R+; AD+ mice. For quantification in a-i, n = 3-5 males and n = 3-5 females, sexes are combined since no significant differences were detected between males and females, plotted as mean ± SEM of analyte mass normalized to unit mass of THP, one-way analysis of variation (ANOVA) and Tukey’s post-hoc test for multiple comparisons, ** = p < 0.01 and **** = p < 0.0001 and ns = non-significant. (male values = square symbol and female values = round symbol).
Extended Data Fig. 6 Aβ1-40, Aβ1-42 and ratio of Aβ1-42/Aβ1-40 protein concentrations in the hippocampus from 5xFAD mice with and without ACE expression in microglia.
Quantification of a, Aβ1-40, b, Aβ1-42 and c, Aβ1-42/Aβ1-40 ratio measured from 10 μg of total hippocampal soluble protein (THP) in 9-month-old R−; AD+ and R+; AD+ mice. For quantification, a–c, n = 8-10 males and n = 10 females, sexes are combined since no significant differences were detected between males and females, plotted as mean ± SEM of analyte mass normalized to unit mass of THP, unpaired T-test, * = p < 0.05 and ns = non-significant. (male values = square symbol and female values = round symbol).
Extended Data Fig. 7 Phagocytosis and endolysomal trafficking in primary microglia with and without ACE expression.
a, Representative Incucyte plots from R- (blue) and R+ (red) microglia treated with oAβ1-42 conjugated to pHrodo, which fluoresces after phagocytosis and loading into the acidic intracellular lysosomal compartment. Data are plotted as integrated intensity of pHrodo fluorescence normalized to the confluency of cells and plotted over 120 h in vitro. Each point represents the mean ± SD of values measured in 8 separate regions of each well. b, Quantification of the amount of oAβ1-42 phagocytosis and lysosome loading relative to R- microglia when it is left in the culture media during the first 30 h of incubation (pulse). c, Quantification of the amount of intracellular oAβ1-42 degraded within the cell between 30 and 120 h after it is removed from the media (chase). d, Representative Incucyte plots identical to a but with R- and R+ microglia are treated with Zymosan conjugated to pHrodo, which binds to microglia CLEC7A receptors. e, Quantification of the amount of Zymosan phagocytosis and lysosome loading relative to R- microglia when it is left in the culture media during the first 30 h of incubation (pulse). f, Quantification of the amount of Zymosan degraded between 30 and 120 h after it is removed from the media (chase). For quantification in b, n = 7 R- and n = 6R+ independent microglia cultures; c, n = 7 R- and n = 6R+ independent microglia cultures; e, n = 13 R- and n = 15R+ independent microglia cultures and f, n = 13 R- and n = 15R+ independent microglia cultures, plotted as mean ± SEM and normalized to the mean of R-, two-tailed unpaired t-test, ** = p < 0.01, *** = p < 0.001 and **** = p < 0.0001.
Extended Data Fig. 8 Experimental design for RNAseq analysis of single nuclei isolated from cortex and hippocampus of wild type and 5xFAD mice with and without ACE expression in microglia.
a, Single nuclei were isolated from cortex and hippocampus of 9-month-old mice, and pooled into samples representing two mice from each genotype and sex (SN1-8). The samples were divided into two groups (Group 1 and Group 2) containing 4 samples that were each labeled with a unique hash-tag antibody. b, The samples were sorted using fluorescence activated nuclear sorting to capture nuclei in the P1 gate that were enriched for microglia by rejecting many neurons (NeuN+), oligodendroglia cells (Sox10+) and astrocytes (Sox9+). c, cDNA libraries were generated from Group 1 and Group 2 samples and the sequences were demultiplexed into samples according to the antibody hash-tag that was used to label the nuclear samples (SN1-SN8). Nuclei that were labeled by more than a single hash-tag antibody (cell colored red) were removed from the datasets within each sample. The TotalSeq hash-tag antibody used and the number of single labeled nuclei per sample are shown.
Extended Data Fig. 9 Quality control of snRNAseq data from 9-month-old wild type and 5xFAD cortex and hippocampus with and without ACE expression in microglia.
a, The number of cells per cluster. b, Violin plot of the number of genes expressed per cell in each cluster. Each dot represents the number of genes detected in a single cell. c, Violin plot of the nUMI per cell in each cluster. Each dot represents the nUMI detected in a single cell. d, Violin plot of the percent of mitochondrial genes per cell in each cluster. Each dot represents the percent of mitochondrial genes detected in a single cell. For a-d, the cluster legend correlates the informatically generated cluster number with their cellular identity. e, Violin plot of the number of genes per cell in each sample (SN1-SN8). Each dot represents the number of genes detected in a single cell. f, Violin plot of nUMI per cell in each sample. Each dot represents the nUMI detected in a single cell. (See Methods for detailed quality control measures applied.)
Extended Data Fig. 10 Distinct cell types identified by snRNAseq from 9-month-old wildtype and 5xFAD cortex and hippocampus with and without expression of ACE in microglia.
a, UMAP principal component analysis of 10 clusters and the relative percent of nuclei within each cluster from SN1-SN8 (cluster number shown in parentheses). b, Cell lineage restricted gene expression defines cluster cellular identity. c, C1qa, Hexb and P2ry12, genes expressed relatively specifically in microglia, map to clusters annotated as microglia (all cell nuclei shown in grey).
Supplementary information
Supplementary Information
Supplementary key resources, methods, data figures and data source information, data tables and references.
Supplementary Table 1
Volcano plot data showing association between DAM-A R+; AD+ and R− AD− gene expression in Fig. 6c.
Supplementary Table 2
Ingenuity Pathway Analysis (IPA) showing all pathways highlighted from genes differentially expressed in from R+; AD+ relative to R− AD+ DMA-A as abbreviated in Fig. 6d.
Supplementary Table 3
Average gene expression in CLEAR signaling, phagosome maturation and production of ROS pathways highlighted by IPA analysis in R−; AD+ and R+; AD+ DAM-A and shown in Fig. 6e.
Supplementary Table 4
Average gene expression in mTOR signaling, PI3K/Akt signaling and oxidative phosphorylation pathways highlighted by IPA analysis in R−; AD+ and R+; AD+ DAM-A and shown in Fig. 6f.
Supplementary Table 5
Average gene expression of DAM-A genes highlighted in IPA function Movement of Cells in R− ; AD+ and R+; AD+ as shown in Supplementary Data Fig. 4.
Supplementary Table 6
Gene expression for volcano plot of GeoMx gene expression of PAM in R+; AD+ versus R−; AD+ shown in Fig. 7c.
Supplementary Table 7
Complete list of Reactome pathways identified from GeoMx expression analysis of R+; AD+ versus R−; AD+ PAM. Highlighted pathways are color coated the same as in Fig. 7d.
Supplementary Table 8
Average gene expression in C-lectin receptor signaling and Clec7a signaling pathways from Reactome analysis in R+; AD+ versus R−; AD+ PAM and shown in Fig. 7e.
Supplementary Table 9
Average gene expression in NF-κB signaling and IL-1 family signaling pathways from Reactome analysis in R+; AD+ versus R−; AD+ PAM and shown in Fig. 7f.
Supplementary Table 10
Average gene expression in translation signaling and rRNA synthesis signaling pathways from Reactome analysis in R+; AD+ versus R−; AD+ PAM and shown in Fig. 7g.
Supplementary Table 11
Volcano plot data of GeoMx gene expression of PAM from R−; AD+ PAM versus nPAM shown in Supplementary Data Fig. 5a.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gomez, A.R., Byun, H.R., Wu, S. et al. Boosting angiotensin-converting enzyme (ACE) in microglia protects against Alzheimer’s disease in 5xFAD mice. Nat Aging (2025). https://doi.org/10.1038/s43587-025-00879-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43587-025-00879-1