Abstract
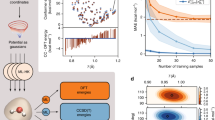
Machine learning plays an important role in quantum chemistry, providing fast-to-evaluate predictive models for various properties of molecules; however, most existing machine learning models for molecular electronic properties use density functional theory (DFT) databases as ground truth in training, and their prediction accuracy cannot surpass that of DFT. In this work we developed a unified machine learning method for electronic structures of organic molecules using the gold-standard CCSD(T) calculations as training data. Tested on hydrocarbon molecules, our model outperforms DFT with several widely used hybrid and double-hybrid functionals in terms of both computational cost and prediction accuracy of various quantum chemical properties. We apply the model to aromatic compounds and semiconducting polymers, evaluating both ground- and excited-state properties. The results demonstrate the model’s accuracy and generalization capability to complex systems that cannot be calculated using CCSD(T)-level methods due to scaling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
99,00 € per year
only 8,25 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Raw computational data files and the training and testing datasets are available with this manuscript through FigShare at https://doi.org/10.6084/m9.figshare.25762212 (ref. 52). Source Data are provided with this paper.
Code availability
The source code to generate the training dataset, train the MEHnet model, and apply the trained MEHnet model to hydrocarbon molecules has been deposited into a publicly available GitHub repository at https://github.com/htang113/Multi-task-electronic (ref. 53), and is also available in the Supplementary Software. The repository contains two branches: the branch v.1.6 is for all results of hydrocarbon molecules in this paper, and the branch v.2.0 is for the benchmark on the QM9 dataset.
Change history
22 January 2025
A Correction to this paper has been published: https://doi.org/10.1038/s43588-025-00767-z
References
Carter, E. A. Challenges in modeling materials properties without experimental input. Science 321, 800–803 (2008).
Kulik, H. J. et al. Roadmap on machine learning in electronic structure. Electron. Struct. 4, 023004 (2022).
Dral, P. O. Quantum Chemistry in the Age of Machine Learning (Elsevier, 2022).
Batzner, S. et al. E(3)-equivariant graph neural networks for data-efficient and accurate interatomic potentials. Nat. Commun. 13, 2453 (2022).
Zhang, L., Han, J., Wang, H., Car, R. & Weinan, E. Deep potential molecular dynamics: a scalable model with the accuracy of quantum mechanics. Phys. Rev. Lett. 120, 143001 (2018).
Takamoto, S., Izumi, S. & Li, J. Teanet: universal neural network interatomic potential inspired by iterative electronic relaxations. Comput. Mater. Sci. 207, 111280 (2022).
Takamoto, S., Okanohara, D., Li, Q. & Li, J. Towards universal neural network interatomic potential. J. Materiomics 9, 447–454 (2023).
Merchant, A. et al. Scaling deep learning for materials discovery. Nature 624, 80–85 (2023).
Chen, C. & Ong, S. P. A universal graph deep learning interatomic potential for the periodic table. Nat. Comput. Sci. 2, 718–728 (2022).
Takamoto, S. et al. Towards universal neural network potential for material discovery applicable to arbitrary combination of 45 elements. Nat. Commun. 13, 2991 (2022).
Zheng, P., Zubatyuk, R., Wu, W., Isayev, O. & Dral, P. O. Artificial intelligence-enhanced quantum chemical method with broad applicability. Nat. Commun. 12, 7022 (2021).
Kirkpatrick, J. et al. Pushing the frontiers of density functionals by solving the fractional electron problem. Science 374, 1385–1389 (2021).
Bogojeski, M., Vogt-Maranto, L., Tuckerman, M. E., Müller, K.-R. & Burke, K. Quantum chemical accuracy from density functional approximations via machine learning. Nat. Commun. 11, 5223 (2020).
Helgaker, T., Jorgensen, P. & Olsen, J. Molecular Electronic-Structure Theory (John Wiley & Sons, 2013).
Schütt, K. T., Gastegger, M., Tkatchenko, A., Müller, K.-R. & Maurer, R. J. Unifying machine learning and quantum chemistry with a deep neural network for molecular wavefunctions. Nat. Commun. 10, 5024 (2019).
Shao, X., Paetow, L., Tuckerman, M. E. & Pavanello, M. Machine learning electronic structure methods based on the one-electron reduced density matrix. Nat. Commun. 14, 6281 (2023).
Feng, C., Xi, J., Zhang, Y., Jiang, B. & Zhou, Y. Accurate and interpretable dipole interaction model-based machine learning for molecular polarizability. J. Chem. Theory Comput. 19, 1207–1217 (2023).
Fan, G., McSloy, A., Aradi, B., Yam, C.-Y. & Frauenheim, T. Obtaining electronic properties of molecules through combining density functional tight binding with machine learning. J. Phys. Chem. Lett. 13, 10132–10139 (2022).
Cignoni, E. et al. Electronic excited states from physically constrained machine learning. ACS Central Sci. 10, 637–648 (2023).
Dral, P. O. & Barbatti, M. Molecular excited states through a machine learning lens. Nat. Rev. Chem. 5, 388–405 (2021).
Li, H. et al. Deep-learning density functional theory hamiltonian for efficient ab initio electronic-structure calculation. Nat. Comput. Sci. 2, 367–377 (2022).
Gong, X. et al. General framework for E(3)-equivariant neural network representation of density functional theory hamiltonian. Nat. Commun. 14, 2848 (2023).
Unke, O. et al. SE(3)-equivariant prediction of molecular wavefunctions and electronic densities. Advances in Neural Information Processing Systems 34, 14434–14447 (2021).
Mardirossian, N. & Head-Gordon, M. Thirty years of density functional theory in computational chemistry: an overview and extensive assessment of 200 density functionals. Mol. Phys. 115, 2315–2372 (2017).
Bartlett, R. J. & Musiał, M. Coupled-cluster theory in quantum chemistry. Rev. Modern Phys. 79, 291 (2007).
Geiger, M. & Smidt, T. e3nn: Euclidean neural networks. Preprint at https://arxiv.org/abs/2207.09453 (2022).
Tirado-Rives, J. & Jorgensen, W. L. Performance of B3LYP density functional methods for a large set of organic molecules. J. Chem. Theory Comput. 4, 297–306 (2008).
Mulliken, R. S. Electronic population analysis on LCAO–MO molecular wave functions. I. J. Chem. Phys. 23, 1833–1840 (1955).
Mayer, I. Bond order and valence indices: a personal account. J. Comput. Chem. 28, 204–221 (2007).
Caruana, R. Multitask learning. Mach. Learn. 28, 41–75 (1997).
Kozuch, S. & Martin, J. M. Spin-component-scaled double hybrids: an extensive search for the best fifth-rung functionals blending DFT and perturbation theory. J. Comput. Chem. 34, 2327–2344 (2013).
Karton, A. How reliable is DFT in predicting relative energies of polycyclic aromatic hydrocarbon isomers? Comparison of functionals from different rungs of jacob’s ladder. J. Comput. Chem. 38, 370–382 (2017).
Goerigk, L. & Grimme, S. A thorough benchmark of density functional methods for general main group thermochemistry, kinetics, and noncovalent interactions. Phys. Chem. Chem. Phys. 13, 6670–6688 (2011).
Grimme, S., Antony, J., Ehrlich, S. & Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H–Pu. J. Chem. Phys. 132, 154104 (2010).
Becke, A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 (1993).
Goerigk, L. & Grimme, S. Efficient and accurate double-hybrid-meta-GGA density functionals evaluation with the extended GMTKN30 database for general main group thermochemistry, kinetics, and noncovalent interactions. J. Chem. Theory Comput. 7, 291–309 (2011).
Jablonnski, M. & Palusiak, M. Basis set and method dependence in atoms in molecules calculations. J. Phys. Chem. A 114, 2240–2244 (2010).
Slayden, S. W. & Liebman, J. F. The energetics of aromatic hydrocarbons: an experimental thermochemical perspective. Chem. Rev. 101, 1541–1566 (2001).
Heeger, A. J. Nobel lecture: Semiconducting and metallic polymers: the fourth generation of polymeric materials. Rev. Modern Phys. 73, 681 (2001).
Grem, G., Leditzky, G., Ullrich, B. & Leising, G. Realization of a blue-light-emitting device using poly (p-phenylene). Adv. Mater. 4, 36–37 (1992).
Otto, P., Piris, M., Martinez, A. & Ladik, J. Dynamic (hyper) polarizability calculations for polymers with linear and cyclic π-conjugated elementary cells. Synth. Metals 141, 277–280 (2004).
Champagne, B. et al. Assessment of conventional density functional schemes for computing the polarizabilities and hyperpolarizabilities of conjugated oligomers: an ab initio investigation of polyacetylene chains. J. Chem. Phys. 109, 10489–10498 (1998).
Ramakrishnan, R., Dral, P. O., Rupp, M. & Von Lilienfeld, O. A. Quantum chemistry structures and properties of 134 kilo molecules. Sci. Data 1, 140022 (2014).
Szalay, P. G., Muller, T., Gidofalvi, G., Lischka, H. & Shepard, R. Multiconfiguration self-consistent field and multireference configuration interaction methods and applications. Chem. Rev. 112, 108–181 (2012).
Dunning Jr, T. H. Gaussian basis sets for use in correlated molecular calculations. i. The atoms boron through neon and hydrogen. J. Chem. Phys. 90, 1007–1023 (1989).
Neese, F., Wennmohs, F., Becker, U. & Riplinger, C. The ORCA quantum chemistry program package. J. Chem. Phys. 152, 224108 (2020).
Perdew, J. P. Density-functional approximation for the correlation energy of the inhomogeneous electron gas. Phys. Rev. B 33, 8822 (1986).
Löwdin, P.-O. On the non-orthogonality problem connected with the use of atomic wave functions in the theory of molecules and crystals. J. Chem. Phys. 18, 365–375 (1950).
Kesharwani, M. K., Brauer, B. & Martin, J. M. Frequency and zero-point vibrational energy scale factors for double-hybrid density functionals (and other selected methods): can anharmonic force fields be avoided? J. Phys. Chem. A 119, 1701–1714 (2015).
Kim, S. et al. PubChem 2023 update. Nucl. Acids Res. 51, D1373–D1380 (2022).
Krylov, A. I. Equation-of-motion coupled-cluster methods for open-shell and electronically excited species: the hitchhiker’s guide to Fock space. Annu. Rev. Phys. Chem. 59, 433–462 (2008).
Tang, H. et al. Training and testing datasets in ‘Multi-task learning for molecular electronic structure approaching coupled-cluster accuracy’. (FigShare, 2024); https://doi.org/10.6084/m9.figshare.25762212
htang113/Multi-task-electronic (GitHub, 2024); https://github.com/htang113/Multi-task-electronic
Linstrom, P. & W.G. Mallard, E. NIST Chemistry WebBook, NIST Standard Reference Database Number 69 (National Institute of Standards and Technology, 2024).
Wilson Jr, E. B. The normal modes and frequencies of vibration of the regular plane hexagon model of the benzene molecule. Phys. Rev. 45, 706 (1934).
Acknowledgements
This work was supported by Honda Research Institute (HRI-USA). H.T. acknowledges support from the Mathworks Engineering Fellowship. The calculations in this work were performed in part on the Matlantis high-speed universal atomistic simulator, the Texas Advanced Computing Center (TACC), the MIT SuperCloud, and the National Energy Research Scientific Computing (NERSC).
Author information
Authors and Affiliations
Contributions
All authors contributed to the discussions of theory and results and to writing the manuscript. J.L. designed and guided the project and formulated the research goals. H.T., H.X., B.X. and W.H. designed and developed the computational method and code package, generated the quantum chemistry dataset, implemented the machine learningtraining and applications, and did data analysis and visualization. A.H. initiated the theme and formulated the research goals. Y.W., F.L. and P.S. provided important comments for the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Debashree Ghosh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6, Tables 1 and 2, and Supplementary Sections 1–5.
Supplementary Data
Relaxed structure models and example input files.
Supplementary Code
Python code package of MEHnet.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tang, H., Xiao, B., He, W. et al. Approaching coupled-cluster accuracy for molecular electronic structures with multi-task learning. Nat Comput Sci 5, 144–154 (2025). https://doi.org/10.1038/s43588-024-00747-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-024-00747-9