Abstract
Background
Cardiovascular diseases (CVDs) rank amongst the leading causes of long-term disability and mortality. Predicting CVD risk and identifying associated genes are crucial for prevention, early intervention, and drug discovery. The recent availability of UK Biobank Proteomics data enables investigation of blood proteins and their association with a variety of diseases. We sought to predict 10 year CVD risk using this data modality and known CVD risk factors.
Methods
We focused on the UK Biobank participants that were included in the UK Biobank Pharma Proteomics Project. After applying exclusions, 50,057 participants were included, aged 40–69 years at recruitment. We employed the Explainable Boosting Machine (EBM), an interpretable machine learning model, to predict the 10 year risk of primary coronary artery disease, ischemic stroke or myocardial infarction. The model had access to 2978 features (2923 proteins and 55 risk factors). Model performance was evaluated using 10-fold cross-validation.
Results
The EBM model using proteomics outperforms equation-based risk scores such as PREVENT, with a receiver operating characteristic curve (AUROC) of 0.767 and an area under the precision-recall curve (AUPRC) of 0.241; adding clinical features improves these figures to 0.785 and 0.284, respectively. Our models demonstrate consistent performance across sexes and ethnicities and provide insights into individualized disease risk predictions and underlying disease biology.
Conclusions
In conclusion, we present a more accurate and explanatory framework for proteomics data analysis, supporting future approaches that prioritize individualized disease risk prediction, and identification of target genes for drug development.
Plain language summary
Cardiovascular diseases (CVDs) are a major cause of long-term disability and death. However, current prediction models are limited in their approach. We aimed to predict individual risk of CVD using blood protein markers, or biomarkers, that can serve as indicators of CVDs using an artificial intelligence model. Our findings show that this model was accurate and performed better than traditional risk prediction methods. The model provided personalized insights into disease risk and helped identify genes that could be targeted for prevention or development of new treatments. This research could improve early detection and prevention of CVD, leading to better health outcomes for many people in the future.
Similar content being viewed by others
Introduction
Cardiovascular disease (CVD), primarily coronary heart disease and stroke, collectively ranks among the leading causes of death worldwide. The primary pathology underlying these diseases is atherosclerosis, which is characterized by the buildup of plaque in major arteries. CVD impacts people in both developed and developing countries, encompassing individuals from diverse ethnic backgrounds, and has an increasing impact on women and younger individuals1. Current treatment approaches include lifestyle adjustments, regular medical monitoring, and pharmaceutical interventions targeting major risk factors such as elevated cholesterol (statins) and blood pressure2,3. Although the incidence rate of CVD has decreased slightly, the global burden remains substantial, with 55.45 million CVD incidences and 18.56 million deaths in year 20194. There is a pressing need to identify individuals who may be at increased risk of developing disease to enhance screening strategies for prevention, improve existing treatments, and enable precision medicine approaches to improve patient outcomes. Traditional clinical predictors, such as blood pressure, body mass index (BMI), cholesterol levels, and medical and family history, have been employed to estimate individual disease risk, but their accuracy remains limited5,6. Therefore, there is a need to develop more precise predictive methods incorporating state-of-the-art omics technologies such as proteomics.
The UK Biobank recruited over 500,000 participants to gather comprehensive baseline data and long-term follow-up of health outcomes7. To promote an in-depth understanding of disease biology and accelerate drug development, proteomics data were collected from over 54,000 participants8. The circulating concentration of 2923 plasma proteins was quantified using the Proximity Extension Assay with the Olink Explore Platform. These proteomics data can be utilized for drug target and biomarker discovery, to improve disease understanding, and to inform patient stratification as well as disease prediction. Here, we focus on building machine learning models for two primary objectives: predicting disease risk and identifying genes associated with it.
Previous studies have used proteomics to predict cardiovascular events9, type II diabetes10, and chronic kidney disease11. Incorporating additional information, such as clinical12, lipidomics13, and metabolomics data14,15, has been demonstrated to further enhance the predictive capabilities of disease risk. For instance, Schuermans et al. recently applied LASSO regression to predict common cardiac diseases in the initial proteomics data released by the UK Biobank, identifying 820 potential protein–disease associations16. However, such linear models may not fully capture non-linear relationships between predictors and outcomes. This modeling limitation makes LASSO inadequate at providing in-depth insights into feature importance and individualized risk predictions.
Explainable Boosting Machines (EBMs) are interpretable and non-linear machine learning models that belong to the family of Generalized Additive Models (GAMs)17. These models involve a response variable that depends linearly on shape functions, which are unknown smooth functions of predictor variables. EBMs use bagged ensembles of boosted depth-restricted trees to represent these shape functions18. In other words, EBMs use a combination of simple, single-feature models to accurately represent how each of the features (e.g., age or plasma level of a protein) predict an outcome (disease risk) by using a technique called boosting. In boosting models, simple models are created sequentially, with each new model trying to correct the errors made by the previous ones. This process helps to create a robust final model that can provide accurate and reliable predictions. When the final model is built, the per-feature models get combined into the shape functions, which provide valuable insights into the risk profile for subranges of features’ values. EBM models offer both local and global explanations. Local explanations predict the features (e.g., molecular or clinical factors) that are important for each participant. For instance, the local explanation for a participant’s prediction might show that a particular protein is an important predictor of disease risk based on its expression level in that participant. On the other hand, global explanations focus on the importance of features across a spectrum of values. For instance, the global explanation for a protein would show how risk changes across the different expression levels as observed in the whole cohort. For comparison, we also considered predictive models using other gradient-boosting approaches in addition to EBM that are popular in the literature. Gradient-boosted trees are well-established strong baselines for tabular data19. Moreover, they are robust to uninformative features and outperform other methods on skewed data distributions. Coupled with SHapley Additive exPlanation (SHAP) values20, these classifiers provide a way to explain the global and local feature importance as well.
In this study, we showcase not only the high predictive power of our machine learning models for CVD risk but also their explainability. Specifically, we apply the EBM, an explainable machine learning framework, for predicting CVD risk. Our analysis reveals insights into three aspects of explainability: (1) features with high predictive power; (2) participant-specific risk factors; and (3) distinct risks associated with varying feature values. We further substantiate our findings through statistical approaches and reviewing existing literature.
Methods
UK Biobank dataset
The UK Biobank is a large-scale prospective study designed to investigate the impact of biological and environmental factors on human health. It enrolled ~500,000 participants aged 40–69 years old between 2006 and 2010 in England, Scotland, and Wales. In this study, we focused on the 54,181 participants that were included in the UK Biobank Pharma Proteomics Project (UKB-PPP) and had maintained their consent up until April 20, 2023. The UKB-PPP participants were representative of the entire UK Biobank cohort8. The North West Multi-Centre Research Ethics Committee approved the study (06/MRE08/65). All participants provided written informed consent. We obtained access to the UK Biobank by submitting applications numbered 53639 and 65851, which were approved by the UK Biobank access committee.
Each sample was characterized by a rich set of 2941 protein features derived from two major releases: 1472 features from the explore release and 1469 features from the expansion release. Since some proteins were measured multiple times, the number of unique proteins measured was 2923. In addition to proteomic features, 55 clinical data fields were extracted to provide a comprehensive overview of each participant’s health and medical history.
Defining cardiovascular disease
Our objective is to predict which participants will experience a primary CVD event within 10 years of recruitment. First, we excluded 4119 participants with pre-existing CVD. Pre-existing CVD events were defined as coronary artery disease, ischemic stroke, and myocardial infarction using ICD 9/10 diagnostic codes from secondary care hospital episode statistics (Supplementary Table 1), along with self-reported CVD events such as heart attack, angina, and stroke (validated by a trained nurse). The cutoff for determining pre-existing CVD was set as the date of the participant’s initial assessment center visit: any event occurring before this visit was classified as pre-existing CVD. This filtering step resulted in a dataset with 50,057 participants with proteomics measurements.
Subsequently, we defined our outcome of interest using a combination of hospital inpatient data and death register records (ONS). We defined CVD as any of the following diagnoses: coronary artery disease, ischemic stroke, and myocardial infarction (Supplementary Table 1). The study end date varied for participants, based on individual administrative censoring dates. Importantly, we included deaths as relevant events not only when CVD was the primary cause of death but also when it was a contributory (secondary) cause. Participants not enrolled for the entire study duration (e.g., end date <10 years or participants dying of complications unrelated to CVD) were excluded from our analyses, which yielded a final set of 46,009 participants. Out of these, 3287 were considered cases, 60% of which were men (Supplementary Table 2).
To account for potential delays in ICD code recording, we conducted sensitivity analyses by excluding participants who experienced a CVD event within 3 and 6 months after their initial assessment center visit, respectively. Excluding these participants reduced the datasets by 47 (for the 3-month window) and 94 (for the 6-month window), resulting in adjusted cohorts of 45,962 and 45,915 participants, respectively. Since these exclusions had minimal impact on model performance, we proceeded with all analyses using the main cohort of 46,009 participants (Supplementary Table 3).
Train and test data split
We divided all 46,009 participants into 10 groups. Specifically, we sorted male and female participants by date of diagnosis (if the participant is a case) or by date of recruitment (if the participant is a control). Participants were then iteratively assigned to each of the 10 groups. Groups were used for imputation, feature selection, and model training: we iteratively conducted each procedure on 9 of the 10 groups (“train set”) and only used the remaining group to measure performance (“test set”). This rendered 10 different splits of the data by taking different train and test sets, which allowed us to estimate the variance of our models’ performance across slightly different datasets. We ensured that the groups were roughly equally sized and matched by age, ethnicity and assessment center, although these variables that were not used to define the groups (Supplementary Fig. 1).
Preprocessing of plasma proteomics measurements
The plasma levels of 2923 proteins were measured at the baseline visit using the Olink platform, which employs Proximity Extension Assay technology8. This technology uses pairs of antibodies attached to unique oligonucleotides, binding specifically to target proteins and allowing for precise and sensitive plasma protein quantification. The protein levels, reported as Normalized Protein eXpression (NPX) values, were directly used as input of the machine learning models. In the case of the proteins that were measured multiple times, all the measurements that passed QC were used for model training. Five participants with invalid data entries were excluded, particularly those lacking available normalized protein expression data.
Encoding clinical fields
We selected 55 clinical fields that were potentially useful predictors of CVD complications (Supplementary Data 1). We excluded fields: (1) describing complications of CVD; (2) recorded after the initial assessment; (3) with identical values across all samples; or (4) with values missing in >99% of the participants. Subsequently, we encoded the selected fields according to their value type21. Continuous fields were left unchanged. Categorical fields with single values, where a single answer is selected from a coded list, were also used as they were, except for two fields with relatively large sets of values — Assessment center (field 54) and Ethnic background (field 21,000) — which were mapped to smaller value sets. Fields with multiple values, where sets of answers are selected from a coded list, such as Qualifications (field 6138) and Illnesses of father (field 20,107), underwent a one-hot encoding, resulting in one binary feature for each possible value. After encoding, the 55 fields were transformed into 173 features.
Imputation of clinical features
The 173 clinical features exhibited various degrees of missingness (Supplementary Data 1): some had no missing values, like genetic sex, age at assessment date, or age at recruitment; while others had missingness rates ranging from 0.18% (e.g., smoking status, alcohol intake frequency) to over 90% (e.g., number of cigarettes). Missingness rates were correlated within feature groups. For example, missingness for white blood cell count is identical to missingness for red blood cell count and hemoglobin concentration, likely indicating that the test used to assess these measures was not performed or invalid.
Crucially, none of the clinical features have values missing completely at random22. For instance, whether a value is missing for a given participant in the private healthcare feature (missingness rate of 65.65%) is highly dependent on values of features such as assessment center and deprivation index for that participant. In such cases, imputation with the mean or the median is not advised. Even state-of-the-art imputation methods such as MissForest23 may cause unexpected problems24. Hence, we only imputed features with a low missingness rate (<1.5%), reducing the potential impact of incorrect imputations. For features that had a missingness rate above 1.5%, we replaced missing entries with a special value indicating “unknown.”
For features with a missingness rate below 1.5%, a Random Forest Regressor was utilized to model each feature with missing values as a function of other clinical features that exhibited low missingness. The imputation process was carried out through multiple iterative rounds, adhering to an iterated round-robin approach. Specifically, at each step within an iteration, one feature column with missing values was designated as the output variable, while the remaining feature columns were treated as input variables. It is worth noting that the imputer was trained exclusively on the training dataset to prevent data leakage and ensure the generalizability of the imputed values. This procedure was systematically applied to each feature requiring imputation. The iterative nature of the process allowed for refinement of the imputed values, with the algorithm proceeding for a maximum of 10 rounds or until a predefined stopping criterion was met. The stopping criterion in our study was set to trigger when the difference in imputed values between consecutive rounds became negligible, ensuring convergence to reliable estimates. For each split, the imputer, once trained on the training dataset, was then used to fill in missing values in both the training and the test datasets of that split. This ensured that the same imputation model was used across both datasets within each split, maintaining consistency in data handling and preserving the integrity of the imputation process.
EBM models
In this manuscript, we focus on Explainable Boosting Machines (EBMs), an interpretable and highly performant model18. In EBM models, the response variable depends linearly on unknown smooth functions of predictor variables (shape functions) with a link function, e.g., identity function for regression and the logistic function for classification. EBMs use bagged ensembles of boosted depth-restricted trees to represent the shape functions. We used the implementation in Python’s interpret package25. To minimize overfitting and utilize the most predictive proteins to train our final models, we performed a feature selection step. We trained 11 EBM models on the train set (Train and test data split) and all the proteins in the panel: one model on the outcome; three additional models that predict the phenotype at three different time horizons (3, 5, and 15 years); one model on a subset of samples in which cases are over-represented (all cases, only 20% of the controls); one model for each sex; one model for each age split (split at the median age of 58); and one model for each BMI split (split at the median BMI of 26.6). We then calculated feature importance and selected the top 70 features with the highest feature importance for each model. The union of these features was then used to train the models. This yielded between 288 and 330 proteins across the 10 data splits (Train and test data split). The selected proteins were consistent across the splits: 248 were selected in at least half of the splits, and 138 appeared in all the splits.
We used machine learning to predict the outcome from all the clinical features and the selected proteins from the feature selection step. For hyperparameter tuning, we first split the training set into train (80%) and validation (20%) sets. We trained models with different hyperparameter configurations on the train portion of the training set and chose the optimal configuration based on the performance of these models on the validation set. The test set was not used in the optimization to avoid data leakage and overfitting. We used the Bayesian optimization algorithm26 implemented in the hyperopt python package27 to propose the series of tries leading to finding the optimal configuration. This algorithm allows us to find the optimal or near-optimal configuration in significantly fewer tries than an exhaustive grid search and with higher probability than a random search of the configuration space. We averaged hyperparameter settings from the top M tries. We used M = 10% of all tries. We repeated this procedure on each fold independently. In EBM models, feature importance values are calculated as mean absolute values of the logit function. To calculate the number, we looked up the logit contribution for each feature for each training sample, took the absolute value of the logit, and then averaged those absolute values across all samples for each feature.
To calculate feature importance for groups of features (e.g., those participating in different pathways), we added the contributions of each feature in the group for each individual, took the absolute value of the sum, and then averaged over all individuals.
To investigate temporal changes in proteomic profiles associated with CVD risk, we trained separate EBM models for different prediction horizons (3, 5, and 10 years). For each time horizon, we used the outcome variable as the occurrence of a CVD event within that specific period. We trained year-specific EBM models following the procedures outlined in our general EBM training, including the feature selection and hyperparameter tuning steps.
Clinical scores
We compared our models for CVD risk prediction with four established clinical scores: PCE28, PREVENT29, QRISK330, and SCORE231. The Pooled Cohort Equations (PCE) provide sex- and race-specific estimations for 10-year atherosclerotic CVD risk, considering variables such as age, total cholesterol, high-density lipoprotein cholesterol, systolic blood pressure, diabetes mellitus, and current smoking status. PREVENT is a family of sex-specific and race-independent models to predict the risk of CVD events using risk factors like age, systolic blood pressure, medications and HbA1c. Specifically we used the PREVENT Enhanced for HbA1c equation to compute 10-year cardiovascular risk. QRISK3, a Cox proportional hazards model, predicts the 10-year risk of CVD in both men and women. The model considers 14 clinical factors, like age, ethnicity, and systolic blood pressure, along with eight additional risk factors, such as chronic kidney disease and migraine. We reimplemented the QRISK3 computation used in ref. 32. Last, the Systematic COronary Risk Evaluation (SCORE2) is a risk prediction model that estimates the 10-year CVD risk across different sexes, age groups, and regions, using factors like age, sex, smoking status, history of diabetes mellitus, systolic blood pressure, and total and HDL cholesterol. We provide the distribution of these scores in Supplementary Fig. 2.
Polygenic risk scores
We additionally compared our models to the polygenic risk scores (PRSs). We used four such genetics-based predictors for cardiovascular disease, coronary artery disease, ischemic stroke and hypertension, provided by the UK Biobank33. We provide the distribution of these PRSs in Supplementary Fig. 2.
Gradient-boosting decision trees
We established a machine learning-based baseline based on gradient-boosting decision trees. Such models are a common choice for high-dimensional tabular data due to their speed, flexibility in handling non-linear relationships, and high classification performance. Specifically, we used the LightGBM framework34. To extract interpretations from them, we used SHAP (SHapley Additive exPlanations) values, a game-theoretic approach used to explain the output of machine learning models20. They provide insights into how each feature contributes to the prediction of a specific instance, allowing for a detailed understanding of the model’s decision-making process. SHAP values provide local interpretations, i.e., the contribution of each feature toward the individual prediction. To compute global interpretation, we took the absolute value of the feature contribution and averaged over all the training points in a split to get the global mean SHAP for each split.
We implemented a feature selection process to improve model performance and interpretability for LightGBM models. Initially, we trained our LightGBM models using the complete set of proteomics and clinical features based on the training data for each split. Subsequently, we computed the SHAP values to evaluate the global importance of each feature. The top 10% of features demonstrating the highest global importance, as determined by their SHAP values, were retained and used to train the the LightGBM model for each split. Notably, when testing the EBM-selected features with the LightGBM model, a slight decrease in predictive performance was observed compared to using the features selected through the abovementioned SHAP-value approach.
To search the hyperparameter space for the best-performing model, we used the Azure Machine Learning’s AutoML framework. It uses a combination of Bayesian optimization and collaborative filtering to search for the optimal feature transformations and hyperparameter choices. For each of the 10 splits, we utilized the train portion of the data to tune hyperparameters. Four-fold cross-validation was performed on the train portion, and the best hyperparameter configuration was the one having highest mean dev AUROC. The process was repeated for each split, leading to 10 separate best models, one for each split.
We also used SHAP values to understand the feature importance of the final models. There may be slight variations in feature importance and their relative ranking across splits. To ensure further robustness to global feature importance, we finally took the mean across splits and reported those top features.
LASSO model
To compare the performance of the EBM model with a linear approach, we trained a LASSO (Least Absolute Shrinkage and Selection Operator) model using the LogisticRegression function from the Python scikit-learn library with an L1 penalty. This model was trained using both proteomics and clinical data, and the regularization strength (C parameter) was optimized using the Bayesian optimization algorithm implemented in the hyperopt package. We applied the same hyperparameter tuning strategy as described for the other models. The optimized LASSO model was then evaluated using the same train-test data splits as the non-linear models.
Statistics and reproducibility
We split the data into 10 parts (Train and test data split) and used nine for data mining and the remaining one for performance evaluation. We repeated this procedure 10 times, obtaining 10 unbiased measurements of performance. We evaluated the metrics above on the whole cohort and across multiple stratifications, including sex, age, and self-reported ethnicity.
We used two metrics of model performance: the area under the receiver operating characteristic curve (AUROC) and the area under the precision-recall curve (AUPRC). The AUROC measures the ability of a model to discriminate between positive and negative classes. A value of 1 indicates perfect prediction capability, while 0.5 indicates the model is not better than a random guess. The AUPRC measures the ability of the model to balance precision and recall, which is crucial in situations with highly imbalanced classes like the current setting. A perfect classifier has an AUPRC of 1, while the AUPRC of a random classifier would have a score around the proportion of positive classes.
Additionally, we used two additional metrics to assess the performance benefits of a new model: the net reclassification improvement (NRI) and integrated discrimination improvement (IDI). The NRI quantifies the net change in the number of individuals correctly reclassified as high-risk or low-risk when transitioning from Model 1 to Model 2. A positive NRI value indicates that Model 2 correctly reclassifies more individuals than Model 1, while a negative NRI value indicates that Model 2 incorrectly reclassifies more individuals than Model 1. The IDI, on the other hand, measures the overall improvement in the predictive model’s ability to distinguish between high-risk and low-risk individuals when transitioning from Model 1 to Model 2. A positive IDI value indicates that Model 2 is better at distinguishing between high-risk and low-risk individuals than Model 1, while a negative IDI value indicates that Model 2 is worse at distinguishing between high-risk and low-risk individuals than Model 1.
Given the stochastic nature of the CVD event, there might not be fundamental differences between a participant developed CVD at year 9.5 compared to those at year 11. Consequently, participants who developed CVD between 10 and 15 years would not serve as suitable negative cases for evaluation, as they could be more similar to positive cases than to negative cases where participants never develop CVD diseases. Therefore, we excluded those participants from model evaluation. Similarly, we excluded participants who developed CVD between 5–10 years when evaluating 5 year CVD risk; we exclude participants who developed CVD between 3 and 5 years when evaluating 3 year CVD risk.
Computing pathway importances
We grouped proteins into pathways involved in atherosclerosis from the Kyoto Encyclopedia of Genes and Genomes (KEGG). This set included 13 pathways: the KEGG pathway map05417 and its related pathways. To compute a pathway feature importance, we followed the procedure described in the EBM feature importance section.
Quantifying feature importance differences by sex/age/BMI
To test if the observed differences in feature importance across models trained on data split by sex, age, and BMI were due to limited sample size and stochastic noise, we generated a baseline experiment where we randomly split the population into two halves and trained a model using each half of the data.
Linear correlations between feature importance were calculated for the feature importance of the top 100 features. For non-overlapping features, we assigned a dummy feature importance as that of the 100th feature in order of importance. To evaluate the linear correlation between the experimental group (data split by sex, age or BMI) against the random split, we performed the Fisher’s Z-transformation on both correlation coefficients and calculated the standard error for the differences in the two Z-scores. Then, we calculated the P-value to assess if the correlation coefficient between the two splits deviates significantly from that of a random split. For instance, if there are significant differences in feature importance across sexes, we would anticipate that the splits between sexes would exhibit a significantly lower linear correlation of feature importance compared to that of the random split.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Model accuracy of cardiovascular disease risk prediction
Using data from the UK Biobank participants with proteomics measurements, we aimed to predict the risk of a primary cardiovascular disease (CVD) event occurring within 10 years from the initial assessment center visit, in which blood samples were collected. Results for 3- and 5 year risk predictions are provided in the Supplementary material.
After applying quality control exclusions (see Methods), our analytic cohort included 46,009 participants without pre-existing CVD at baseline. The plasma levels of 2923 proteins were quantified in these participants and used alongside clinical features to train the models (Fig. 1a). During the 10 year follow-up, 3287 participants (7.1%) experienced a primary CVD event, with 60.8% of cases being men (Supplementary Table 2). These events consisted of 1870 coronary artery disease events (56.9%), 938 myocardial infarctions (28.5%), and 479 ischemic strokes (14.6%).
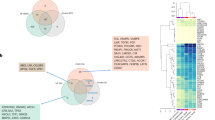
a Schematic overview of the study design. This study focuses on using the UK Biobank proteomics and clinical data to predict target disease risk for cardiovascular disease and identify marker proteins using advanced machine learning models. Detailed insights were revealed by the risk profiles for proteins and individual risk factors for patients. b, c EBM models outperform the other models at predicting the 10-year risk of CVD. b Area under the receiver operating characteristic curve (AUROC) and c area under the precision-recall curve (AUPRC) are displayed for the polygenic risk scores (PRSs), the clinical scores, and the machine learning approaches trained on different sets of variables. The PRSs are for cardiovascular disease (CVD), coronary artery disease (CAD), hypertension (HT), and ischemic stroke (ISS). The machine learning approaches are EBM and LightGBM. d Mean AUROC across the two genetic sexes and the self-reported ethnicities with at least 5 cases. The bars represent the 95% confidence interval. The dashed line represents the AUROC over the whole cohort, and the shaded area is its 95% confidence interval. e The predicted risk aligns well with the observed risk in the evaluation set. In (b–d) 10 individual datapoints are shown, grouped into 30 equally sized bins across the data range. These datapoints are available in Supplementary Data 2.
For model training and evaluation, we implemented a 10-fold cross-validation strategy by dividing the cohort into 10 approximately equal-sized groups (~4601 participants each), matched by sex, age, ethnicity, assessment center, and time until CVD diagnosis (Supplementary Fig. 1). In each experiment, the union of nine groups served as the training set, and the one remaining group was used as the test set.
Our proteomics-only model (“EBM Proteomics”) achieved an area under the receiver operating characteristic curve (AUROC) of 0.767 and an area under the precision-recall curve (AUPRC) of 0.241. Its performance surpassed four equation-based CVD risk prediction models (PCE, PREVENT, QRISK3, and SCORE2), which rely on clinical features, and four relevant polygenic risk scores (PRSs) (Fig. 1b, c, Tables 1 and 2). Our results were consistent for events in shorter timeframes (Supplementary Fig. 3). We measured the model’s improvement using the net reclassification improvement (NRI) and the integrated discrimination improvement (IDI) metrics. The EBM Proteomics model displayed significantly better performance than the best-performing traditional score (PREVENT) for both metrics (Supplementary Table 4).
To further enhance predictive accuracy, we trained EBM models on both proteomics and clinical features (“EBM Proteomics & Clinical,” Fig. 1b, c), which increased the AUROC to 0.785 (Table 1) and the AUPRC to 0.284 (Table 2). The EBM Proteomics & Clinical model, trained on both proteomics and clinical data, outperformed both the EBM Proteomics model (AUROC increase = 2.3%, AUPRC increase = 17.9%; ΔAUROC = 0.018, ΔAUPRC = 0.043), which was trained solely on proteomics features, and the EBM Clinical model (AUROC increase = 2.0%, AUPRC increase = 9.3%; ΔAUROC = 0.015, ΔAUPRC = 0.024), which focuses solely on clinical features. The EBM model also outperformed the LightGBM model, a state-of-the-art machine learning model for tabular data (AUROC increase = 1.1%, AUPRC increase = 16.9%; ΔAUROC = 0.009, ΔAUPRC = 0.041). At shorter time horizons, EBM models remained competitive compared with the LightGBM models (Supplementary Fig. 3). Last, we compared the EBM model to LASSO, a linear model, and found that EBM also exhibited a higher predictive performance (AUROC increase = 1.5%, AUPRC increase = 16.1%; ΔAUROC = 0.012, ΔAUPRC = 0.039), demonstrating the benefit of capturing non-linear relationships (Fig. 1b, c, Tables 1 and 2).
Model fairness and calibration
The EBM Proteomics & Clinical model shows consistent performance across genetic sexes and self-reported ethnicities (Fig. 1d). Crucially, it remains competitive across ethnicities, being the best-performing model in most of them (Supplementary Fig. 4). We evaluated whether our model remains predictive among the participants taking statins, commonly used to treat elevated LDL cholesterol (Supplementary Fig. 5). The AUROC is 0.786 for people not taking statins and 0.685 for people taking statins. This performance gap might be explained by the relatively small number of statin users who also had a primary CVD event: among 2967 statins users, only 318 (10.7%) had an event by year 10. Moreover, the performance across subpopulations with different income, blood pressure, and LDL levels was largely consistent, all exceeding the current state-of-the-art clinical model (Supplementary Fig. 5).
The EBM Proteomics & Clinical model showed similar performance at predicting specific acute complications like ischemic stroke and myocardial infarction, as well as diagnosis of coronary artery disease (CAD), with AUROC scores ranging from 0.785 for CAD to 0.802 for stroke (Supplementary Fig. 6, Supplementary Tables 5 and 6). For each subtype, the performance was comparable between sexes (Supplementary Tables 5 and 6). The largest gap occurred for infarction, with an AUROC for women of 0.787 and for men of 0.767; however, this difference was not statistically significant (P = 0.123, Wilcoxon test two-sided test).
We also assessed the calibration of the EBM Proteomics & Clinical model by examining whether the predicted risk probability aligns with the actual disease risk, ensuring that the model produces reliable predictions with clinical relevance, i.e., the model score corresponds to the probability of a primary CVD event. Overall, our model is well calibrated (Fig. 1e).
Local and global explanations of feature importance
EBM models are highly interpretable, providing both local and global measures of feature importance. Local explanations quantify how different features contribute to an individual participant’s predicted CVD risk. EBM can produce risk factors specific to each participant (Fig. 2a–c), which can vary considerably. For example, participants with similarly high predicted risks may have distinct sets of predictive proteomic features (Fig. 2a, b), which are distinct from a low-risk individual’s (Fig. 2c).
a–c Local explanations quantify the contribution of different features to an individual participant’s predicted CVD risk. The ribbon of each panel represents the label and the probability of the label, as provided by the EBM Proteomics model. For instance, “Case, P(Case) = 0.564” means that the participant was indeed a case, and the model assessed (without knowledge of the outcome) that they had a 56.4% probability of being a case. The model quantifies the contribution of each protein to CVD risk for every participant. Red bars represent increased risks based on the plasma protein expression level, while blue bars denote reduced disease risks according to the expression level. Note participants in (a, b) show similar risk but different contributing proteins, while the participant in (c) shows low disease risk. The intercept, with a value of −3.04, is not shown. d Global explanations aggregate the contribution of features across the cohort. The graph displays the top 10 contributing proteins in the EBM model. Known CVD markers were highlighted in red.
Global explanations, on the other hand, assess the overall feature importance of each proteomic feature in the population. Notably, 5 out of the 10 most important predictors in the EBM Proteomics model are established biomarkers of cardiovascular health (Fig. 2d): NT-proBNP35, NPPB35, PLA2G7 (Lp-PLA2)36, MMP1237 and GDF1538,39,40. When comparing these 10 proteins to those identified by models predicting on a shorter time frame (3- and 5 years), the overlap was considerable (Supplementary Fig. 7). The top features revealed by EBM models were also largely consistent with those highlighted by the LightGBM model using SHAP values (Supplementary Fig. 8). Similarly, when examining the top 50 proteins identified by the Proteomics models predicting different time frames, we found a 66% overlap between the 3 year and 5 year models; a 54% overlap between the 5 year and 10 year models; and a 38% overlap between the 3 year and 10 year models. Additionally, EBM models enable exploration of risk contribution of each feature across its entire value range (Fig. 3).
a FASLG and (b) GDF15 have similar feature importance in the EBM Proteomics & Clinical model, but the risk profiles differ drastically. The two clinical features (c) Cystatin C (field 30720) and d None of the medications* (field 6153, value −7) in the EBM Clinical model with similar risk profiles show drastically different risk profiles and feature distributions. c A small number of participants with high Cystatin C levels show greatly increased CVD risk. d Participants who selected “none of the above” when reporting medication use* showed reduced risk of CVD. This is a weak effect affecting a large number of individuals. Risk profiles for (e) HDL (field 30780) and f MSR1 show high consistency. The top panels of each subfigure display the risk score on the logarithmic scale (y-axis) for different feature values (x-axis), with gray-shaded regions representing the standard deviation for estimations. The dotted line corresponds to the population mean risk for the given feature. Positive values indicate increased risk; negative values indicate decreased risk. The bottom panels show histograms of the feature values across all participants. *Medications for cholesterol, blood pressure, diabetes, or exogenous hormones.
Studying importance of clinical risk factors in the EBM Proteomics & Clinical model reveals their contribution to CVD risk in the context of plasma proteomics. Among the 20 most important features (Supplementary Table 7), we find clinical variables related to medications, age, sex, family history of heart disease, and LDL levels. Notably, when features are correlated, the importance might get split across them. For instance, NT-proBNP shows reduced importance after incorporating clinical features (Supplementary Fig. 9).
Pathway analysis and feature importance comparisons
To better understand the contributions from various molecular pathways, we grouped proteomic features into Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways involved in atherosclerosis and calculated the aggregated contribution of each pathway (Fig. 4a). PI3K-Akt signaling, reflecting apoptosis and cell-cycle contributions to atherosclerosis, and cholesterol metabolism were the two pathways with the highest contributions.
a The aggregated contribution from top pathways predictive of CVD risk. b Models built based on a split by gender, age and BMI showed lower linear correlations of feature importance between the subgroup models compared with random splits shown as a dotted line (gender P = 2e-9, age P = 3e-4 and BMI P 0.10, two-sided Fisher’s Z-test). An asterisk denotes a significant difference, using a significance threshold of 0.05.
While the overall model performances are similar across genetic sexes, age groups, and BMI profiles, we observe differences in predictive features in models trained separately on these subgroups. Only 2 and 3 of the top 10 features are shared between models trained separately on data split by age and sex, respectively, while 5 features are shared between models trained on data split by BMI (Supplementary Fig. 10).
To ensure that these differences between subgroups are not due to stochastic noise, we randomly divided the participant population into two equal-sized subgroups and reran the experiment. Seven of the top 10 features are shared across models trained on the two random subgroups, with correlated feature importance for the top 100 features (R = 0.67, Supplementary Fig. 11). In contrast, we observe significant differences in feature importance for models built for different sexes (R = 0.13, P = 2e-9, two-sided Fisher’s Z-test, compared with correlation between the random splits) and age groups (R = 0.38, P = 3e-4, two-sided Fisher’s Z-test), suggesting that the underlying mechanisms might vary among these groups. However, no significant differences were found between models trained on the low and high BMI groups compared to the random split (R = 0.58, P = 0.10, two-sided Fisher’s Z-test; Fig. 4b, Supplementary Fig. 11).
Discussion
In this study, we employed the EBM model to predict 10-year CVD risk using the UK Biobank proteomic data, generating models with high predictive power and explainability. By jointly considering the proteomics and clinical features, our model offered a comprehensive and coherent understanding of disease mechanisms, suggesting candidates for further examination. The EBM Proteomics & Clinical model performance surpassed traditional models for CVD risk prediction like PREVENT (AUROC increase = 6.7%, AUPRC increase = 58.4%; ΔAUROC = 0.049, ΔAUPRC = 0.105) and the competing machine learning model, LightGBM (AUROC increase = 1.1%, AUPRC increase = 16.9%; ΔAUROC = 0.009, ΔAUPRC = 0.041). The improvement in the AUPRC is particularly significant, given that CVD only affected 7.1% of our studied cohort.
The EBM Clinical model and the EBM Proteomics model demonstrate similar performance, with the Clinical model performing slightly better than the Proteomics model (AUROC increase = 0.3%, AUPRC increase = 7.8%; ΔAUROC = 0.003, ΔAUPRC = 0.019). However, the combination of both Proteomics and Clinical features further improves the performance (AUROC increase = 2.0%, AUPRC increase = 9.3%; ΔAUROC = 0.015, ΔAUPRC = 0.024 compared to the Clinical model). This underscores the complementarity of proteomics and clinical features and emphasizes that each offers unique information not provided by the other. Moreover, as shown in Supplementary Tables 8 and 9, a model utilizing only the 10 most predictive plasma proteins performs well, although significantly worse than the EBM Proteomics model (AUROC increase = 4.1%; ΔAUROC = 0.030) and slightly worse than PREVENT (AUROC increase = 4.3%; ΔAUROC = 0.031). This is consistent with previous observations that the top protein predictors for atherosclerotic CVD explain most of the protein risk score9,41, and also highlights the benefit of using the full proteomics data in predicting CVD risk. Notably, the AUPRC of the small proteomics models is also superior to that of PREVENT, suggesting that they could reduce the number of false negatives.
Our approach enables individualized risk prediction and generates enriched risk profiles for predictive features. As shown earlier, two participants exhibiting similar risks of CVD might have different features contributing to that prediction (Fig. 2a, b). By incorporating feature range-associated disease risk, our model can offer a comprehensive and holistic understanding of the prediction for each individual with a high level of interpretability. Uncovering such individualized risk factors will aid in better understanding the underlying etiology and has potential to influence disease management. Furthermore, our approach also shows comparatively good performance at predicting CVD in shorter timeframes. This might be key to identifying patients at high immediate risk. However, the most predictive features differ between models trained to predict events over different timeframes, and this departure is largest between the shortest and the longest timeframe (3 and 10 years). These findings highlight the importance of considering specific time frames when developing predictive models, as the proteomic signatures relevant for short-term risk may differ from those predictive of longer-term risk.
Our model offers detailed insights into the predicted disease risks associated with various circulating levels for each plasma protein, contributing valuable insights into the disease mechanisms. In many machine learning models, feature importance is represented by a single value, which is not ideal for interpretability due to two main shortcomings. Firstly, a single value of feature importance tends to conflate the prevalence of an effect with the effect’s magnitude. That is, a protein with a very high risk restricted to a small number of participants would appear indistinguishable from that of a protein with a slightly elevated risk over many participants. For example, FASLG and GDF15 show similar global feature importance values, but their risk profiles differ drastically (Fig. 3a, b). Similarly, two clinical features with similar feature importance could have different feature distributions and risk profiles in the EBM Clinical model (Fig. 3c, d). Second, many traditional machine learning models evaluate the overall importance of variables but ignore the fact that the risk may not vary linearly or even monotonically with the value of a variable. For instance: while high plasma levels of GDF15 are linked to high CVD risk, our non-linear models show that the risk does not increase linearly; instead, it plateaus at high GDF15 levels (Fig. 3b). When looking at the features’ profile plots, the EBM model recapitulated how elevated LDL levels are predictive of a higher risk of CVD (Fig. 3e). Similarly, the HDL risk profile identifies the middle level as predictive of lower CVD risk while both high and low levels indicate higher risk (Supplementary Fig. 13A). The model also recapitulated risk profiles of plasma proteins involved in cholesterol homeostasis. Macrophage scavenger receptor 1 (MSR1; ranked 18 in the EBM Proteomics model) is responsible for mediating the endocytosis of modified LDLs and the uptake of modified lipoprotein42. In line with previous studies, elevated MSR1 levels were associated with an increased atherosclerosis risk (Fig. 3f)43. Inhibitors of Proprotein convertase subtilisin/kexin type 9 (PCSK9; ranked 19 in the EBM Proteomics model), which inhibits the clearance of plasma LDL by downregulating the LDL-C receptor, have been shown to reduce the risk of cardiovascular events (Supplementary Fig. 13B)44,45.
Many of the proteins featured in this study have previously been reported as biomarkers for CVD. Matrix metalloproteinase-12 (MMP12) plasma levels have previously been shown to predict CVD46, as well as being directly involved in atherosclerosis pathogenesis47, including from genetic studies37. N-terminal pro b-type natriuretic peptide (NT-proBNP; Supplementary Fig. 12A) and natriuretic peptide B (NPPB; Supplementary Fig. 12B) exhibit similar risk profiles, as they are both derived from different fragments of the pro-B-type natriuretic peptide precursor. Each participates in the natriuretic peptide system, a vital pathway for regulating blood pressure and fluid balance, and is used in diagnosing cardiac dysfunction35. Phospholipase A2 group VII (PLA2G7), also known as lipoprotein-associated phospholipase A2 (Lp-PLA2), is an inflammatory enzyme expressed in atherosclerotic plaques and is a well-established marker of coronary heart disease36. Growth/differentiation factor 15 (GDF15) is another prognostic marker for cardiovascular disease, and its elevated expression was shown to be associated with higher risks, which is in alignment with our findings38. Furthermore, we also demonstrated the predictive ability of proteins that were not previously used as CVD markers. For example, the hepatitis A virus cellular receptor 1 (HAVCR1) was previously used as a biomarker for kidney injury48.
We used a composite outcome, comprising coronary artery disease, ischemic stroke, and myocardial infarction. Composite outcomes are common in the field9,29,30. The endpoints share cardiovascular risk factors and allowed us to work with a larger number of events, thereby improving the predictive power of our model. The high performance of the EBM model on each of the outcomes (Supplementary Fig. 6) supports this rationale.
However, our approach has some limitations. First, we leveraged the UK Biobank for our prediction model development, and future work would be to replicate our study in other cohorts and could boost the generalizability of our conclusions to other populations. For instance, most participants in the UK Biobank cohort are of European origin, and tend to be healthier and wealthier than the general UK population49. Although the performance of the EBM Proteomics & Clinical model is consistent across ancestries (Supplementary Fig. 4), the performance across different income levels is not (Supplementary Fig. 5); further work is needed to confirm these trends and address the disparities. There remains a huge need to increase diversity in medical research, as only then will we be able to improve medical outcomes for all. Second, while certain features may be highly predictive, they do not necessarily imply a causal relationship. As such, further validation of our findings through experimental approaches and independent cohorts is essential to confirm causal relationships. Mendelian randomization and reviewing existing literature can be employed to further support or refute the causal relationships. Last, the proteomics panel contains a non-random subset of 2923 proteins. A more extensive panel might lead to better models and the discovery of novel disease–protein associations.
Our findings underscore the importance and feasibility of individualized risk prediction and information-rich analysis of feature importance. The high accuracy of our EBM Proteomics & Clinical model supports its potential for future clinical application in disease risk prediction using standalone proteomics blood tests. Further work is needed to translate our approach to the clinical practice, for instance by reducing the number of protein markers or focusing on those measurable in routine blood tests. However, our experiments using only a subset of the plasma proteins show that this would compromise model performance; sensitivity analyses could help strike the best trade-off. Additionally, our explanatory modeling framework can be readily adapted for predicting risk of other diseases and phenotypes, providing valuable insights to facilitate drug development and individualized risk assessments.
Data availability
The data used for training the models was obtained from the UK Biobank. Adhering to privacy protection policy, the raw data cannot be made publicly available, but can be accessed through a research application through the UK Biobank’s Access Management System (http://amsportal.ukbiobank.ac.uk/). The source data for Tables 1 and 2; panels b, c and d in Fig. 1; Supplementary Figs. 3, 4 and 6; and Supplementary Tables 5 and 6 is in Supplementary Data 2.
Code availability
The code for the equation-based CVD risk models and for model training and evaluation of the EBM and LightGBM models is available at https://github.com/novonordisk-research/ML-Proteomics-CVD-Prediction. A snapshot of the repository has been archived in Zenodo50 (DOI: 10.5281/zenodo.14591534). We used Python 3, with the following packages: azure-ai-ml, azureml-automl-runtime, azureml-fsspec, hyperopt, interpret, lightgbm, mltable, numpy, pandas, scikit-learn, shap.
References
Libby, P. The changing landscape of atherosclerosis. Nature 592, 524–533 (2021).
Charo, I. F. & Taub, R. Anti-inflammatory therapeutics for the treatment of atherosclerosis. Nat. Rev. Drug Discov. 10, 365–376 (2011).
Chen, W. et al. Macrophage-targeted nanomedicine for the diagnosis and treatment of atherosclerosis. Nat. Rev. Cardiol. 19, 228–249 (2022).
Li, Y., Cao, G.-Y., Jing, W.-Z., Liu, J. & Liu, M. Global trends and regional differences in incidence and mortality of cardiovascular disease, 1990−2019: findings from 2019 global burden of disease study. Eur. J. Prevent. Cardiol. 30, 276–286 (2022).
Singh, S. S., Pilkerton, C. S., Shrader, C. D. Jr & Frisbee, S. J. Subclinical atherosclerosis, cardiovascular health, and disease risk: is there a case for the Cardiovascular Health Index in the primary prevention population? BMC Public Health 18, 429 (2018).
Huang, B. et al. Prediction of subclinical atherosclerosis in low Framingham risk score individuals by using the metabolic syndrome criteria and insulin sensitivity index. Front Nutr. 9, 979208 (2022).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Sun, B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622, 329–338 (2023).
Helgason, H. et al. Evaluation of large-scale proteomics for prediction of cardiovascular events. JAMA 330, 725–735 (2023).
Gadd, D. A. et al. Blood protein assessment of leading incident diseases and mortality in the UK Biobank. Nature Aging 4, 939–948 (2024).
Avram, R. Revolutionizing cardiovascular risk prediction in patients with chronic kidney disease: machine learning and large-scale proteomic risk prediction model lead the way. Eur. Heart J. 44, 2111–2113 (2023).
Williams, S. A. et al. A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk. Sci. Transl. Med. 14, eabj9625 (2022).
Nurmohamed, N. S. et al. Proteomics and lipidomics in atherosclerotic cardiovascular disease risk prediction. Eur. Heart J. 44, 1594–1607 (2023).
Barrett, J. C. et al. Metabolomic and genomic prediction of common diseases in 700,217 participants in three national biobanks. Nat. Communi. 15, 10092 (2024).
Nightingale Health Biobank Collaborative Group. Metabolomic and genomic prediction of common diseases in 700,217 participants in three national biobanks. Nat. Communi. 15, 10092 (2024).
Schuermans, A. et al. Integrative proteomic analyses across common cardiac diseases yield new mechanistic insights and enhanced prediction. medRxiv 19, 2023.12.19.23300218 (2023).
Hastie, T. & Tibshirani, R. Generalized additive models: some applications. J. Am. Stat. Assoc. 82, 398 (1987).
Lou Y., Caruana, R. & Gehrke, J. Intelligible models for classification and regression. In Proc. 18th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. (IEEE, 2013).
Grinsztajn, L., Oyallon, E. & Varoquaux, G. Why do tree-based models still outperform deep learning on typical tabular data? Adv. Neural Inform. Process. Syst. 35, 507–520 (2022).
Lundberg, S. M. & Lee, S. I. A unified approach to interpreting model predictions. Advances in Neural Information Processing Systems, 4768–4777 (IEEE, 2017).
UK Biobank. https://biobank.ndph.ox.ac.uk/ukb/help.cgi?cd=value_type (2007).
Rubin, D. Inference and missing data. Biometrika 63, 581–592 (1976).
Stekhoven, D. J. & Buhlmann, P. MissForest–non-parametric missing value imputation for mixed-type data. Bioinformatics 28, 112–118 (2012).
Chen, Z., Tan S., Chajewska, U., Rudin C. & Caruna, R. Missing values and imputation in healthcare data: can interpretable machine learning help? In Proc. Conference on Health, Inference, and Learning (eds Bobak J. M., Tasmie S., Andrew B., Joyce C. H.) 86–99 (PMLR, 2023).
Nori, H., Jenkins, S., Koch, P., & Caruana, R. Interpreml: a unified framework for machine learning interpretability. arXiv https://arxiv.org/abs/1909.09223 (2019).
Snoek, J., Larochelle, H. & Adams, R. P. Practical Bayesian optimization of machine learning algorithms. Adv. Neural Inform. Process. Syst. 25, 1–9 (2012).
Bergstra J., Yamins, D. & Cox, D. Making a science of model search: hyperparameter optimization in hundreds of dimensions for vision architectures. In: Proc. of the 30th International Conference on Machine Learning (ICML 2013)) 115–123 (PMLR, 2013).
Goff, D. C. Jr. et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association task force on practice guidelines. Circulation 129, S49–S73 (2014).
Khan, S. S. et al. Development and validation of the American Heart Association’s PREVENT equations. Circulation 149, 430–449 (2024).
Hippisley-Cox, J., Coupland, C. & Brindle, P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. BMJ 357, j2099 (2017).
Graham, I. M., Di Angelantonio, E. & Huculeci, R. European Society of Cardiology’s cardiovascular risk C. New way to “SCORE” risk: updates on the ESC scoring system and incorporation into ESC cardiovascular prevention guidelines. Curr. Cardiol. Rep. 24, 1679–1684 (2022).
Carter, A. R. et al. Cross-sectional analysis of educational inequalities in primary prevention statin use in UK Biobank. Heart 108, 536–542 (2022).
Thompson, D. J. et al. UK Biobank release and systematic evaluation of optimised polygenic risk scores for 53 diseases and quantitative traits. medRxiv https://doi.org/10.1101/2022.06.16.22276246 (2022).
Ke, G. et al. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inform. Process. Syst. 30, 1–9 (2017).
Cao, Z., Jia, Y., Zhu, B. BNP and NT-proBNP as diagnostic biomarkers for cardiac dysfunction in both clinical and forensic medicine. Int. J. Mol. Sci. 20, 1820 (2019).
Thompson, A. et al. Lipoprotein-associated phospholipase A(2) and risk of coronary disease, stroke, and mortality: collaborative analysis of 32 prospective studies. Lancet 375, 1536–1544 (2010).
Traylor, M. et al. A novel MMP12 locus is associated with large artery atherosclerotic stroke using a genome-wide age-at-onset informed approach. PLoS Genet 10, e1004469 (2014).
Adela, R. & Banerjee, S. K. GDF-15 as a target and biomarker for diabetes and cardiovascular diseases: a translational prospective. J. Diabetes Res. 2015, 490842 (2015).
Wang, D. et al. GDF15: emerging biology and therapeutic applications for obesity and cardiometabolic disease. Nat. Rev. Endocrinol. 17, 592–607 (2021).
Wollert, K. C., Kempf, T. & Wallentin, L. Growth differentiation factor 15 as a biomarker in cardiovascular disease. Clin. Chem. 63, 140–151 (2017).
Carrasco-Zanini, J. et al. Proteomic signatures improve risk prediction for common and rare diseases. Nat. Med. 30, 2489–2498 (2024).
Sheng, W., Ji, G. & Zhang, L. Role of macrophage scavenger receptor MSR1 in the progression of non-alcoholic steatohepatitis. Front. Immunol. 13, 1050984 (2022).
Gudgeon, J., Marin-Rubio, J. L. & Trost, M. The role of macrophage scavenger receptor 1 (MSR1) in inflammatory disorders and cancer. Front. Immunol. 13, 1012002 (2022).
Ma, M., Hou, C. & Liu, J. Effect of PCSK9 on atherosclerotic cardiovascular diseases and its mechanisms: Focus on immune regulation. Front. Cardiovasc. Med. 10, 1148486 (2023).
Sabatine, M. S. et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376, 1713–1722 (2017).
Goncalves, I. et al. Elevated plasma levels of MMP-12 are associated with atherosclerotic burden and symptomatic cardiovascular disease in subjects with type 2 diabetes. Arterioscler. Thromb. Vasc. Biol. 35, 1723–1731 (2015).
Newby, A. C. Metalloproteinase production from macrophages—a perfect storm leading to atherosclerotic plaque rupture and myocardial infarction. Exp. Physiol. 101, 1327–1337 (2016).
Han, W. K., Bailly, V., Abichandani, R., Thadhani, R. & Bonventre, J. V. Kidney injury molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 62, 237–244 (2002).
Fry, A. et al. Comparison of sociodemographic and health-related characteristics of UK biobank participants with those of the general population. Am. J. Epidemiol. 186, 1026–1034 (2017).
Climente-Gonzalez, H., Oh, M., Chajewska, U. & Mukherjee, S. novonordisk-research/ML-Proteomics-CVD-Prediction.). 1.0.0 edn. Zenodo. https://github.com/novonordisk-research/ML-Proteomics-CVD-Prediction (2025).
Acknowledgements
We thank Dr. Alice Rose Carter for her help in implementing the QRISK3 score. This research has been conducted using the UK Biobank Resource under Application Numbers 53639 and 65851.
Author information
Authors and Affiliations
Contributions
The high-level experiment was conceptualized by H.C.-G., M.O., U.C., W.G., G.F., C.L., and J.M.M.H. Preprocessing of proteomics and clinical data was carried out by H.C.-G., M.O., W.G., and S.H. Baseline models were established by H.C.-G. and M.O. The EBM models were developed by U.C. The LightGBM models were primarily developed by S.M. The model evaluation pipeline was implemented by R.H. Biological interpretation of the results was provided by M.T. The cross-team coordination was carried out by P.P.D.V. and E.V. All authors contributed to data analysis and interpretation. The initial draft of the paper was prepared by C.L., H.C.-G., M.O., and U.C. H.C.-G., M.O., U.C., R.H., S.M., W.G., M.T., S.H., G.F., J.G., P.P.D.V., E.V., N.K., L.D., R.A., C.L., and J.M.M.H. provided feedback on the manuscript. Requests for materials and other inquiries should be directed to C.L. and H.C.-G. All authors have given approval for the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing interests: H.C.-G., W.G., M.T., S.H., G.F., N.K., and J.M.M.H. are employees of Novo Nordisk Research Centre Oxford. M.O., U.C., R.H., S.M., P.P.D.V., L.D., R.A. and C.L. are employees of Microsoft Corporation. E.V. and J.G. are employees of Novo Nordisk A/S.
Peer review
Peer review information
Communications Medicine thanks Ming Wai Yeung and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Climente-González, H., Oh, M., Chajewska, U. et al. Interpretable machine learning leverages proteomics to improve cardiovascular disease risk prediction and biomarker identification. Commun Med 5, 170 (2025). https://doi.org/10.1038/s43856-025-00872-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-025-00872-0