Abstract
Multidrug resistance (MDR) contributes to the failure of chemotherapy and high mortality in non-small cell lung cancer (NSCLC). We aim to identify MDR genes that predict tumor response to chemotherapy. 199 NSCLC fresh tissue samples were tested for chemosensitivity by MTT assay. cDNA microarray was done with 5 samples with highest resistance and 6 samples with highest sensitivity. Expression of ABCC3 mRNA and protein was detected by real-time PCR and immunohistochemisty, respectively. The association between gene expression and overall survival (OS) was examined using Cox proportional hazard regression. 44 genes were upregulated and 168 downregulated in the chemotherapy-resistant group. ABCC3 was one of the most up-regulated genes in the resistant group. ABCC3-positive expression correlated with lymph node involvement, advanced TNM stage, more malignant histological type, multiple-resistance to anti-cancer drugs and reduced OS. ABCC3 expression may serve as a marker for MDR and predictor for poor clinical outcome of NSCLC.
Similar content being viewed by others
Introduction
Lung cancer is the leading cause of cancer morbidity and mortality worldwide. Non-small cell lung cancer (NSCLC) accounts for 80–85% of lung cancer with a 5-year survival rate of 15%1. Although surgery is regarded as the best possible treatment for NSCLC, only 20–25% of tumors are suitable for potentially curative resection2 and despite complete surgical resection, the recurrence rate remains high (30–70%). Adjuvant chemotherapy may reduce recurrences by eradicating the subclinical and micro-metastatic disease2. However, the heterogeneity of NSCLC tumors and resistance to multiple drugs results in a poor response to chemotherapy and reduced survival2. Multidrug resistance (MDR) is a multifactorial process and a major obstacle to treatment for NSCLC patients2. Identification of biomarkers for predicting an individual's resistance to chemotherapy and for rationally selecting the most efficacious therapeutic agents is dependent upon understanding the molecular alterations that contribute to MDR in lung cancer.
In vitro anticancer drug sensitivity tests can identify biomarkers to predict chemosensitivity, improve chemotherapeutic response and design individualized chemotherapy3,4,5. Such tests as MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), CD-DST (collagen gel droplet-embedded culture drug sensitivity test) and HDRA (histoculture drug response assay) have been developed for a variety of malignancies. MTT chemosensitivity test might help to predict tumor response in ovarian cancer and identify the optimum adjuvant chemotherapy for esophageal squamous cell carcinoma. HDRA might be feasible for predicting the efficacy and selecting the proper anticancer drug for lung cancer6. CD-DST test for “new generation” anticancer drugs by using surgically resected specimens of NSCLC patients may predict tumor response to postoperative chemotherapy7. We applied MTT assay using resected NSCLC tumor tissues to stratify patients into chemotherapy-sensitive and resistant groups. We then analyzed the gene expression profile of the resistant and sensitive group by cDNA Microarray to identify genes associated with MDR. 212 genes with differential expression were identified including 44 upregulated and 168 down regulated in the chemotherapy-resistant group compared to the sensitive group. ABCC3 was one of the most up-regulated genes in the resistant group.
ABCC3 is a member of the ATP-binding cassette (ABC) transporters family. ABC transporters are highly expressed in tumor cells where they actively efflux a broad spectrum of anticancer drugs and thus contribute to MDR8,9,10,11. ABCC3 is a member of the multidrug resistance-associated protein (MRP) subfamily which is involved in MDR. ABCC3 expression has been shown to be higher in NSCLC than that in SCLC12 and ABCC3 expression correlated with decreased sensitivity of lung cancer cells to anticancer drugs (vincristine, etoposide and cisplatin)13 and especially to methotrexate and doxorubicin in NSCLC14,15. ABCC3 is also overexpressed in Anip973/NVB cells consistent with involvement of ABCC3 in NVB-resistance in lung cancer16. However, the mechanism underlying the role of ABCC3 in MDR to taxanes (paclitaxel and docetaxel), gemcitabine and cisplatin in NSCLC remains largely unknown. To assess whether ABCC3 expression has predictive value in chemosensitivity and prognosis in NSCLC, we examined the expression of ABCC3 in patients with resected NSCLC tumors and evaluated its association with tumor characteristics and sensitivity to chemotherapy. Our goal was to examine the specific genetic traits of tumors as a means of predicting their chemosensitivity to “new generation” anticancer drugs. The results, if confirmed, would possibly identify biomarkers useful to predict clinical outcome of NSCLC.
Results
MTT assay
MTT assay was successfully performed using primary cancer cells from tissues of 199 patients; 69 were highly sensitive to all the 5 drugs and were categorized as the highly sensitive group; 44 were resistant to all the 5 drugs and were categorized as resistant group; and 86 were partially sensitive to anticancer drugs and categorized as the moderate-sensitive group. Samples for 11 patients (6 patients with highest sensitivity and 5 patients with the highest resistance to all the 5 drugs) were selected for gene expression microarray. Resistance index of the 11 patients is shown in Supplemental Table 1.
Gene expression microarray
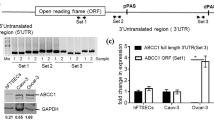
To identify potential predictors for chemosensitivity, cDNA microarray was used to analyze the gene expression profiling of the resistant and sensitive group. Fold change measurements were analyzed by SAM. The transcripts with fold change >2 (upregulated) or <1/2 (downregulated) and |Score(d)| ≥ 2 were considered to have significantly differential expression between the two groups. The differentially expressed elements were identified by the two-sided statistical tolerance interval (95%) with a cut-off of false discovery rate at 10%. Two hundred-twelve (212) genes were differentially expressed between the resistant and sensitive group. Compared to sensitive group, 44 were upregulated and 168 downregulated in chemotherapy-resistant group. Those genes spanned distinct functional families involved in biological processes including cell apoptosis, cell cycle, cell adhesion, signal transduction and ABC transporter (Supplemental Table 2 and 3). Hierarchical clustering was performed on the 11 arrays based on the differentially expressed genes (Fig. 1).
ABCC3 expression by real time-PCR in NSCLC
The upregulated genes with potential predictive value for chemosensitivity in NSCLC were evaluated by real time-PCR (date was not shown). ABCC3 was one of the most differentially expressed in the resistant group. We performed quantitative RT-PCR for ABCC3 in the 199 NSCLS tumor tissues. ABCC3 mRNA level was higher in the resistant group compared to the sensitive group (mean ± SD: 0.4587 ± 0.1324 vs. 0.1324 ± 0.0866, P = 0.001) (Fig. 2).
ABCC3 expression by IHC in NSCLC and its correlation with clinicopathological characteristics ABCC3 immunoreactivity was present in the cell membrane and cytoplasm of NSCLC tumor (Fig. 3). ABCC3-positive expression was observed in 63 cases including 23 with squamous cell carcinoma (SCC), 38 with adenocarcinoma (ADC) and 2 with other types (Table 1). ABCC3 expression correlated with pN status (P < 0.001), pathological TNM stage (P < 0.001) and histological type (P = 0.04).
Prognostic significance of ABCC3 expression
ABCC3-positive expression correlated with shorter OS of NSCLC patients (P log-rank < 0.001) (Fig. 4). In univariate analysis, positive pN status, advanced TNM stage and ABCC3-positive expression correlated with poor prognosis (P < 0.001). In multivariate regression model, ABCC3 expression remained a significant prognostic factor independent of pN status, pTNM stage and histological type (P < 0.001) (Table 2).
Expression of ABCC3 and in vitro drug sensitivity
Among the 199 samples, 69 (34.6%) were sensitive, 44 (22.1%) were resistant and 86 (44.2%) were partially sensitive to the 5 anticancer drugs. 15 (21.7%), 20 (23.2%) and 26 (59.1%) samples of the sensitive, partial sensitive and resistant group, respectively, were ABCC3 positive. A significant association was observed between ABCC3-positive expression and reduced sensitivity to the 5 anticancer drugs using the MTT assay (P < 0.001), suggesting that ABCC3-positive expression is predictive of chemoresistance to the 5 anticancer drugs (Supplemental Table 4).
Discussion
In this study, we performed in vitro chemosensitivity tests using “new generation” anticancer drugs for 199 NSCLC patients. We observed 212 genes were significantly altered in the resistant group compared to the sensitive group. These genes are involved in various biological processes such as cell apoptosis, cell cycle, cell adhesion, signal transduction and ABC transporter, which have been shown to predict chemosensitivity in NSCLC. The resistant group with ABCC3-positive expression had poor OS.
Multiple transporters constitute the major mechanisms of MDR8,9,10,11. ABC transporters confer resistance to many anticancer agents. We identified ABCC3 which has been associated with the prognosis of different type of tumors, as one of the most upregulated genes in the chemoresistant group. In pancreatic cancer ABCC3 mRNA has been shown to be upregulated and associated with tumor grading17. ABCC3 overexpression in glioblastoma multiforme (GBM) also correlates with a higher risk of death and may be a potential therapeutic target for GBM18. In breast cancer amplification of ABCC3 occurs in primary breast tumors, predominantly in HER2-amplified and luminal tumors19. In our study, 31.6% of the NSCLC samples expressed ABCC3 and its expression correlated with advanced pN status and pathological TNM stage and the patients with ABCC3-positive expression had 2.63 times higher risk for death.
ABCC3 was highly expressed in tumor cells but not in type II alveolar pneumocytes20. Moreover, the number of ABCC3-positive cells was significantly increased compared to the precursor lesion of lung adenocarcinoma (atypical adenomatous hyperplasia, AAH) to localized non-mucinous bronchioloalveolar carcinoma (LNMBAC). Together, these data suggest that ABCC3 is a candidate novel tumor marker for histological diagnosis and functions as a driving factor in carcinogenesis. We found ABCC3 expression was higher in ADC than SCC. Recent studies have shown that gene expression patterns of lung ADC differ substantially from SCC and each has different etiology, carcinogenesis and clinicopathological behavior21,22. These observations suggest that high expression of ABCC3 may confer a more malignant phenotype in lung ADC patients. ABCC3 overexpression has been implicated in acquired MDR in cancer cell lines. ABCC3 also contributes to resistance to doxorubicin in breast cancer23, to 5-fluorouracil in pancreatic cancer cells24 and resistance to adriamycin in hepatocellular carcinoma cells, which however, can be effectively reversed by tetramethylpyrazine25. Our results are consistentwith ABCC3 expression being predictive of resistance to paclitaxel, docetaxel, gemcitabine, vinorelbine and cisplatin in NSCLC.
The limitation in our study includes the fact that the results of MTT were not compared with the optimal regimen. The current study did not include the functional study of drug-resistant related genes that have been screened, thus further studies are needed to explore the detailed process of multiple-drug resistance and to provide more evidence of how ABCC3 is involved in MDR in vitro. Although we collected samples from large amounts of NSCLC patients, only 11 samples were eventually selected for gene expression microarray, which also limited the extent of application. Our observations need to be validated in a multiple-centered study involving patients with a large sample size. However, our observations support the notion that chemotherapy combined with targeted therapy may improve the efficacy in patient with MDR tumors.
Most previous studies have screened drug-resistant genes from cell lines in vitro, while our study validated and predicted drug resistance by using primary cells of human cancer tissues. We conducted drug resistance experiments, identified drug-resistant genes directly from clinical specimens of NSCLC patients and validated the observations in vivo, which accurately reflected the original drug sensitivity vs. resistance in patients. We have employed a new method to popularize the MDR genes screening, which has clinical implications for individualized therapy for a variety of tumors.
In summary, MTT drug response assay in surgically resected NSCLC may have clinical utility for predicting tumor response to chemotherapy and to prospectively identify drug sensitive and resistant subpopulations. ABCC3-positive expression correlated with enhanced chemoresistance and reduced OS in NSCLC patients. For patients with resectable stage IB to IIIA NSCLC, cisplatin-based regimen (in combination with Paclitaxel, Docetaxel, Gemcitabine, Vinorelbine, etc.) is the majorchoice for postoperative adjuvant therapy. However, some patients develop multiple-drug resistance, which leads to failures in therapy and a poor prognosis. By using in vitro MTT assay, we could evaluate an individual patient's sensitivity vs. resistance to conventional anticancer drugs before the treatment is employed. Based on the results, an unnecessary cytotoxicity/side effects caused by an insensitive drug could be avoided. In our study, we found the correlation of ABCC3 expression with multiple-drug resistance and reduced overall survival, confirming that in vitro MTT assay is reliable and feasible in NSCLC.
Therefore, we propose ABCC3 as a novel marker to predict tumor response to chemotherapy and postoperative clinical outcome of NSCLC patients, which will be helpful to stratify patient subpopulation for an efficient therapy and realize the concept of personalized medicine. Future studies will investigate whether the inhibition of ABCC3 increases the sensitivity to chemotherapeutic agents and provides new effective targeted therapy of NSCLC.
Methods
Patients and tissue specimens
Each patient had signed informed consent for medical record review and tissue sample donation before surgery. This study was approved by the institutional review board at Harbin Medical University and conducted according to all current ethical guidelines. Fresh tumor tissues from 205 patients with NSCLC were obtained at the Department of Thoracic Surgery of the Affiliated Tumor Hospital of Harbin Medical University between May 2008 and August 2009. The patients were follow-up until June, 2012.
Histopathologic subtypes were determined according to the WHO classification. The International Staging System was used for pathological staging. All tumors were staged according to the pathological tumor/node/metastasis (pTNM) classification of the American Joint Committee on Cancer (7th edition). All tumors were histologically reviewed by pathologists. Tumor tissue collected at the time of curative surgical resection was placed immediately into normal saline. Then a pathologist conducted a representative tumor biopsy and stored the tissue in RPMI-1640 for primary culture of NSCLC cells and subsequent MTT assay within 30 minutes. Long-term storage of the tissues was at −80°C. At the time of surgery and tissue collection, none of the patients had received neoadjuvant chemotherapy or radiation therapy.
Primary culture of NSCLC cells
Each fresh specimen was homogenized and digested in 0.25% trypsin. The dispersed cancer cells were filtered through an 80 μm nylon mesh, washed twice in 500 μL RNase free PBS, centrifuged at 1000 rpm for 5 min and then incubated in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% (v/w) penicillin/streptomycin at 37°C in a humidified atmosphere of 5% CO2. The samples of 6 patients were contaminated and excluded from the consequent experiments. In total, the culture of primary cells was successful in 199 samples.
MTT drug response assay
The NSCLC cells from each patient were seeded at a density of 1 × 105 per well in 96-wellplates with 190 μL culture medium for 24 h. Then the cells were treated with 10 μL gradient concentrations (10×, 5×, 2.5×, 1.25×, 0.625×, 0.312×, 0.156 × ppc) of each of the drugs paclitaxel, docetaxel, gemcitabine, vinorelbine and cisplatin for 72 h. “ppc” represents peak plasma concentration (the highest level of drug that occurs in the plasma portion of blood after administration of the drug). The tumor cells treated with PBS served as controls. Then 20 μL tetrazolium salts (MTT, Sigma, 5 mg/mL) were added to each well and incubated for 4 h. The salts were cleaved into a colored formazan product by metabolically active cells and 100 μL DMSO was added.
After checking for complete solubilization of the purple formazan crystals, the optical density (OD) was measured at 490 nm wavelength. The inhibition rate (IR) was calculated as the formula: IR = (ODcontrol group − ODexperimental group)/ODcontrol group × 100%26,27,28. IR ≥ 50% was defined as highly sensitive; IR 30% ~ 50% as moderately sensitive; and IR ≤ 30% as resistant. The data were presented as mean ± standard deviation (SD) from four replicate wells per microtiter plate. The samples with the highest sensitivity or resistance to all the 5 drugs were selected for gene expression microarray.
Gene expression microarray
Extraction and purification of total RNA was performed using TRIZOL reagent (Invitrogen, Carlsbad, CA). RNA purity and integrity was controlled by BioAnalyser 2100 (Agilent technologies, CA, USA). Gene expression profiling was performed using Human Genome U133 Plus2.0 Affymetrix (Santa Clara, USA), which included 47,400 transcripts and represented 38,500 human genes. Target preparation for Affymetrix Genechips™ was performed according tothe manufacturer's instruction. 4 μg of total RNA was reverse-transcribed and amplified by using the RNA amplification kit (Ambion, TX, USA). Hybridization, washing, staining and scanning of chips were carried out according to standard protocols (Affymetrix, Santa Clara, CA). The mean ± SD of replicated spots was calculated for each geneby using the Acuity 4.0 software (Molecular Devices, LLC, USA).
Microarray validation
Real time-PCR
The mRNA level of each gene was measured by real-time-PCR using TaqMan method and quantified using an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, USA). GAPDH was used as the internal control. All experiments were performed in triplicate. Relative mRNA abundance was calculated as 2−ΔCt [ΔCt = Ct (ABCC3) − Ct(GAPDH)].
Immunohistochemistry (IHC)
IHC assay was performed using the labeled streptavidin-biotin (LSAB) method. In brief, tissue sections (4 μm) were cut from the tissue blocks. To retrieve antigenicity, tissue slides were immersed in 1 mM EDTA (pH 8.0) and bathed in a steamer at 100°C for 15 min. To block the endogenous peroxidase activity, the slides were incubated in methanol containing 0.3% H2O2 for 15 min. The slides were washed with PBS and incubated with a specific mouse monoclonal primary antibody for human ABCC3 (1:800 dilution, Abcam, Cambridge, UK) at 4°C overnight. After washing with PBS, the slides were incubated with streptavidin-biotinylated anti-mouse IgG (Dako, Carpinteria, CA, USA) as the secondary antibody for 10 min at room temperature. The slides were washed with PBS for 3 times and stained using diaminoben-zidine tetrahydrochloride (DAB), counterstained using hematoxylin, dehydratedand placed on coverslips. The negative controls were performed by omitting the primary antibody and replaced with PBS. Adenocarcinoma of intestine was used as a positive control, which was provided with kit and confirmed to express this protein. The positive staining was observed in the membrane of a cell.
Scoring
The staining results were interpreted independently and blindly by two pathologists. For each slide, three to five randomly selected fields were evaluated. For each field, the percentage of DAB-positive-tumor cells was calculated as: [(number of DAB-positive tumor cells/total number of tumor cells) × 100]. The relative staining intensity was defined as negative for <5%; weak (+) for 5–25%; moderate (++) for 25–50% and strong (+++) for >50% of the tumor cells stained positive for ABCC3.
Statistical analysis
The association between ABCC3 expression and tumor characteristics (tumor size, lymph node, pathological TNM stage, histological classification) or sensitivity to chemotherapy was evaluated by χ2 test and Cox regression. Kaplan-Meier plot was used to estimate median survival time (MST). Overall survival (OS) time was calculated from the date of diagnosis to the date of death or last follow-up. The association between ABCC3 expression and survival time was analyzed by log-rank test. The effect of ABCC3 expression on OS was estimated using hazard ratio (HR) and 95% CI was estimated using Cox proportional hazard regression model, with adjustment for other clinical factors. A P-value <0.05 was considered statistically significant. All analyses were performed using SPSS 18.0 (SPSS Inc., Chicago, IL).
References
Jemal, A., Siegel, R., Xu, J. & Ward, E. Cancer statistics, 2010. CA Cancer J clin 60, 277–300, 20073 (2010).
Group, N. M.-A. C. et al. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet 375, 1267–1277 (2010).
Bertelsen, C. A., Sondak, V. K., Mann, B. D., Korn, E. L. & Kern, D. H. Chemosensitivity testing of human solid tumors. A review of 1582 assays with 258 clinical correlations. Cancer 53, 1240–1245 (1984).
Shaw, G. L. et al. Correlation of in vitro drug sensitivity testing results with response to chemotherapy and survival: comparison of non-small cell lung cancer and small cell lung cancer. J Cell Biochem Suppl. 24, 173–185 (1996).
Kobayashi, H. Collagen gel droplet culture method to examine in vitro chemosensitivity. Methods Mol Med. 110, 59–67 (2005).
Yoshimasu, T. et al. Data acquisition for the histoculture drug response assay in lung cancer. The J Thorac Cardiovasc Surg. 133, 303–308 (2007).
Higashiyama, M. et al. Prediction of chemotherapeutic effect on postoperative recurrence by in vitro anticancer drug sensitivity testing in non-small cell lung cancer patients. Lung cancer 68, 472–477 (2010).
Deeley, R. G., Westlake, C. & Cole, S. P. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiological Rev 86, 849–899 (2006).
Kruh, G. D., Belinsky, M. G., Gallo, J. M. & Lee, K. Physiological and pharmacological functions of Mrp2, Mrp3 and Mrp4 as determined from recent studies on gene-disrupted mice. Cancer metastasis Rev 26, 5–14 (2007).
Marquez, B. & Van Bambeke, F. ABC multidrug transporters: target for modulation of drug pharmacokinetics and drug-drug interactions. Curr Drug Targets 12, 600–620 (2011).
Borst, P., Zelcer, N. & van de Wetering, K. MRP2 and 3 in health and disease. Cancer Lett 234, 51–61 (2006).
Young, L. C. et al. Expression of multidrug resistance protein-related genes in lung cancer: correlation with drug response. Clin Cancer Res. 5, 673–680 (1999).
Young, L. C., Campling, B. G., Cole, S. P., Deeley, R. G. & Gerlach, J. H. Multidrug resistance proteins MRP3, MRP1 and MRP2 in lung cancer: correlation of protein levels with drug response and messenger RNA levels. Clin Cancer Res. 7, 1798–1804 (2001).
Kool, M. et al. MRP3, an organic anion transporter able to transport anti-cancer drugs. Proc Natl Acad Sci U S A 96, 6914–6919 (1999).
Lockhart, A. C., Tirona, R. G. & Kim, R. B. Pharmacogenetics of ATP-binding cassette transporters in cancer and chemotherapy. Mol Cancer Ther. 2, 685–698 (2003).
Hong, X. et al. Multidrug resistance-associated protein 3 and Bcl-2 contribute to multidrug resistance by vinorelbine in lung adenocarcinoma. Int J Mol Med. 28, 953–960 (2011).
Konig, J. et al. Expression and localization of human multidrug resistance protein (ABCC) family members in pancreatic carcinoma. Int J Cancer. 115, 359–367 (2005).
Kuan, C. T. et al. MRP3: a molecular target for human glioblastoma multiforme immunotherapy. BMC cancer 10, 468 (2010).
O'Brien, C. et al. Functional genomics identifies ABCC3 as a mediator of taxane resistance in HER2-amplified breast cancer. Cancer Res. 68, 5380–5389 (2008).
Hanada, S. et al. Expression profile of early lung adenocarcinoma: identification of MRP3 as a molecular marker for early progression. J Pathol. 216, 75–82 (2008).
Daraselia, N. et al. Molecular signature and pathway analysis of human primary squamous and adenocarcinoma lung cancers. Am J Cancer Res. 2, 93–103 (2012).
Petty, R. D., Nicolson, M. C., Kerr, K. M., Collie-Duguid, E. & Murray, G. I. Gene expression profiling in non-small cell lung cancer: from molecular mechanisms to clinical application. Clin Cancer Res. 10, 3237–3248 (2004).
Liu, Y., Peng, H. & Zhang, J. T. Expression profiling of ABC transporters in a drug-resistant breast cancer cell line using AmpArray. Mol Pharmacol. 68, 430–438 (2005).
Hagmann, W., Jesnowski, R., Faissner, R., Guo, C. & Lohr, J. M. ATP-binding cassette C transporters in human pancreatic carcinoma cell lines. Upregulation in 5-fluorouracil-resistant cells. Pancreatology. 9, 136–144 (2009).
Wang, X. B. et al. Inhibition of tetramethylpyrazine on P-gp, MRP2, MRP3 and MRP5 in multidrug resistant human hepatocellular carcinoma cells. Oncol Rep. 23, 211–215 (2010).
Keiko Yagi, K. K., Kunio & Seiki Relationship of P-Glycoprotein and p53 Protein to Chemosensitivity in Colorectal Cancer. Int J Clin Oncol. 2, 81–91 (1997).
Chen, Z., Liu, G., Chen, M., Xu, B., Peng, Y., Chen, M. & Wu, M. Screen anticancer drug in vitro using resonance light scattering technique. Talanta. 77, 1365–1369 (2009).
Kuang, Y. H., Chen, X., Su, J., Wu, L. S., Liao, L. Q., Li, D., Chen, Z. S. & Kanekura, T. RNA interference targeting the CD147 induces apoptosis of multi drug resistantcancer cells related to XIAP depletion. Cancer Lett. 276, 189–95 (2009).
Acknowledgements
This work is supported by National Natural Science of China grants (30772540 and 8177224 to LC, 81301991 to YZ) and Science and Technology of Heilongjiang Province 2grant (GC07C349, to LC).
Author information
Authors and Affiliations
Contributions
L.C. and X.D. conceived and directed the project. Y.Z. and H.L. designed experiments. Y.Z., H.L., A.Y., Y.Y., Q.M., L.S., C.L. and H.P. carried out experiments, conducted data analysis and interpreted the results. Y.Z., H.L., L.C. and X.D. wrote and edited the paper. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
supplementary table1-4
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Zhao, Y., Lu, H., Yan, A. et al. ABCC3 as a marker for multidrug resistance in non-small cell lung cancer. Sci Rep 3, 3120 (2013). https://doi.org/10.1038/srep03120
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03120
This article is cited by
-
Cell density modulates chemoresistance in breast cancer cells through differential expression of ABC transporters
Molecular Biology Reports (2023)
-
Nanobodies targeting ABCC3 for immunotargeted applications in glioblastoma
Scientific Reports (2022)
-
Drug resistance: from bacteria to cancer
Molecular Biomedicine (2021)
-
Analysis at the single-cell level indicates an important role of heterogeneous global DNA methylation status on the progression of lung adenocarcinoma
Scientific Reports (2021)
-
Co-delivery of cisplatin and siRNA through hybrid nanocarrier platform for masking resistance to chemotherapy in lung cancer
Drug Delivery and Translational Research (2021)