Abstract
Renal dysfunction (RD) often characterizes the worse course of patients with advanced heart failure (AHF). Many prognosis assessments are hindered by researcher biases, redundant predictors, and lack of clinical applicability. In this study, we enroll 1736 AHF/RD patients, including data from Henan Province Clinical Research Center for Cardiovascular Diseases (which encompasses 11 hospital subcenters), and Beth Israel Deaconess Medical Center. We developed an AI hybrid modeling framework, assembling 12 learners with different feature selection paradigms to expand modeling schemes. The optimized strategy is identified from 132 potential schemes to establish an explainable survival assessment system: AIHFLevel. The conditional inference survival tree determines a probability threshold for prognostic stratification. The evaluation confirmed the system’s robustness in discrimination, calibration, generalization, and clinical implications. AIHFLevel outperforms existing models, clinical features, and biomarkers. We also launch an open and user-friendly website www.hf-ai-survival.com, empowering healthcare professionals with enhanced tools for continuous risk monitoring and precise risk profiling.
Similar content being viewed by others
Introduction
Heart failure (HF) is a life-threatening condition marking the final common pathway for many cardiac diseases1. Most HF cases progress into an advanced stage ultimately, characterized by persistent symptoms despite maximal therapy2,3. The prevalence of advanced HF (AHF) is increasing typically following a pattern of gradual deterioration interspersed with episodes of acute worsening, leading to sudden death4. Prognostic stratification is important for timely referral to an appropriate center, to properly convey expectations to patients and families, and to plan treatment and follow-up strategies1. Despite many prognostic parameters and tools, accurately assessing outcomes for AHF remains complex5.
Chronic kidney disease (CKD) has consistently been recognized as a prevalent comorbidity in HF5,6, and when present, carried the highest population attributable risk for all-cause mortality (ACM) and AHF hospitalization among all comorbidities7,8,9,10,11. The complex interaction between HF and renal dysfunction (RD) accelerated disease progression, driven by neurohormonal and inflammatory activation, elevated venous pressure, and hypoperfusion8,10,12. Clinically, AHF patients with RD may encounter additional harm because such patients often receive lower doses of drugs, and diagnostic tests using contrast media are avoided13,14. Recent studies have highlighted that patients with comorbid AHF and RD are often not optimally treated with evidence-based medical therapies, even when their eGFR levels would not contraindicate such treatments due to kidney dysfunction. There is a pressing need for further efforts to mitigate such risk15. While emerging biomarkers like natriuretic peptides provided some post-discharge prognostic value, they are not specific enough for AHF or RD cases, constraining their clinical utility16,17,18. The efficacy of management and prognostication guidance using serial natriuretic peptide measurements remained unestablished19. Additionally, heterogeneity within HF and RD populations reduced the efficacy of a single biomarker5,6,20. Therefore, comprehensive survival predictions should be prioritized in this high-risk population for clear prognostication and targeted interventions15,21,22,23.
The high dimensionality and interactivity of clinical data rendered hypothesis-driven traditional statistical methods less effective. Currently, the predictive capability of conventional prognostic models for HF is generally limited24,25. Artificial intelligence (AI) is increasingly employed to establish prognostic tools for predicting death, readmission, or composite endpoints; however, no algorithm can perfectly assess HF outcomes26. Despite many options available, researchers’ preferences and knowledge limitations can lead to suboptimal use of modeling algorithms, resulting in reduced predictive power27. Algorithm selection should be based on various objectives, and conditions, aligned with known rules28. Additionally, current risk models, often burdened with redundant or costly predictors, prove impractical for daily clinical practice and inaccessible in primary care29. The real-world application of predictive models requires balancing accuracy, interpretability, and complexity. Despite the potency, AI’s ‘black-box’ nature posed challenges in providing clear interpretations and transparency to clinicians, a key factor limiting the clinical implementation of many HF prognostic models24,30,31. Moreover, the lack of extensive external validation and calibration in many HF prognostic models restricted their generalizability across diverse clinical settings and populations32.
In this study, we aimed to develop and validate an explainable predictive system for survival assessment of patients with AHF and RD in multicenter retrospective longitudinal cohorts. Our AI hybrid modeling framework maximized the use of various algorithms, reducing the impact of researcher bias. We also integrated the system with clinical interpretability and prognostic stratification, optimizing patient management strategies. This system (AIHFLevel) has been translated into a convenient web application to facilitate its utility for clinicians, available at ‘www.hf-ai-survival.com’.
Results
Population characteristics overview
The graphic abstract of the study is summarized in (Fig. 1a). At the CRCCD center, we retrospectively enrolled 712 patients with AHF and RD between September 2018 and December 2020 as our in-house cohort. An independent external cohort of 1024 patients from June 2001 to October 2012 was sourced from the BIDMC center. The demographic and clinical characteristics of the CRCCD cohort were depicted in (Fig. 1b). In-depth baseline data for both cohorts, including comorbidity status, blood routine, coagulation, renal and liver functions, cardiac assessments, echocardiography results, medication history, were detailed in (Supplementary Tables 1 and 2).
a Overview of the study methodology. Image was created with a licensed version of bioRender.com. b Heatmap providing a snapshot of the general baseline demographic and clinical profile in the Henan Province Clinical Research Center for Cardiovascular Diseases (CRCCD) in-house cohort. This visual representation includes data on all-cause mortality (ACM), gender distribution, age, smoking habits (current smokers and previous smokers), drinking status (any consumption of alcohol in the previous six months), chronic kidney disease (CKD) staging, NYHA classification for cardiac function, comorbidities such as arrhythmia and coronary artery disease (CAD), history of percutaneous coronary intervention (PCI), and medication usage (including digoxin, aspirin, diuretics, valsartan, clopidogrel, and nifedipine). c Kinetics plots revealed evolving baseline profiles among AHF and RD patients, tracing the intricate paths of disease progression. This analysis, covering NYHA cardiac function grades I to IV, CKD stages I to V, and HF subtypes (HFpEF, HFmrEF, HFrEF), distinguished eight unique patterns of progression for each path. The clustering visualized these patterns through a gradient of membership values, represented in a spectrum from warm colors (dark red for high membership) to cool colors (sky blue for low membership). d Kaplan–Meier estimates provide a nuanced view of conditional survival up to 30 months post-diagnosis for patients with AHF and RD, stratified by initial survival increments of 0–18 months. The matrix format, where each column signifies the elapsed months since the initial diagnosis and each row delineates the cumulative survival probability from that juncture, offers an insightful prognosis tool. For instance, a patient who has already achieved a 6-month survival post-diagnosis has an 89% chance of reaching the 20-month mark and an 85% likelihood of attaining a 30-month survival threshold. e Univariate Cox proportional hazards analysis, leveraging 93 variables derived from medical health record data from the CRCCD cohort (n = 712), discerned 50 variables significantly associated with ACM survival. Statistic test: two-sided Wald test. Data were presented as hazard ratio (HR) with 95% confidence interval (CI).
Within the CRCCD cohort, the average age was 59 years (SD = 16), with a distribution of 39.0% females (n = 277) and 61.0% males (n = 435). 49.0% of these patients (n = 346) had comorbid CAD, and 32.0% (n = 228) presented with arrhythmia. A majority exhibited compromised cardiac function, with 52.0% (n = 368) classified as NYHA IV and 33.0% (n = 235) as NYHA III. Through the trajectory analysis, we explored the dynamic baseline profiles, revealing significant non-linear shifts in disease progression across NYHA classes I-IV, CKD stages I-V, and HF subtypes (HFpEF, HFmrEF, HFrEF), underscoring the complex nature of AHF&RD evolution (Fig. 1c). The dynamic soft clustering approach distinguished eight distinct progression patterns for each trajectory (Fig. 1c, Supplementary Tables 3-5). Our findings indicated that the trajectory of AHF&RD development does not follow a straightforward, linear journey; rather, it is marked by significant, nonlinear shifts that underscored the intricate and dynamic nature of disease evolution. Moreover, the occurrence rates for ACM and MACE were 21% (n = 151) and 33% (n = 233), respectively, with mean times to ACM and MACE at 19 months (SD = 8) and 17 months (SD = 8). The survival probability increased per half year already survived relative to the total survival time. The Kaplan–Meier estimates for conditional survival indicated that the probability of 30-month survival directly after diagnosis of AHF with RD increased from 76% to 85%, 92%, and 95% per additional half year survived (Fig. 1d). In the BIDMC cohort, the average age was significantly higher at 71 years (SD = 14), with a near-balanced gender ratio (46.2% female (n = 473)). Incidences of CAD and arrhythmias comorbidities were reported in 49.2% (n = 504) and 60.6% (n = 621) of patients. 55.2% (n = 565) of patients experienced ACM during the follow-up period, with an average time to event of 19 months (SD = 20). Notably, heart failure subtypes were distributed as 58.2% (n = 414) HFpEF, 23.5% (n = 167) HFmrEF, and 18.3% (n = 131) HFrEF in the CRCCD cohort, compared to 66.9% (n = 685) HFpEF, 11.5% (n = 118) HFmrEF, and 21.6% (n = 221) HFrEF in BIDMC cohort. This distribution pattern corroborated previous reports that CKD and worsening renal function both appear more common in HFpEF as compared to in HFmrEF and HFrEF perhaps due to shared pathophysiological mechanisms6,8,15,33,34.
Survival assessment system AIHFLevel
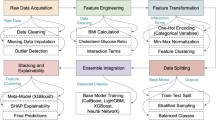
The workflow of our AI hybrid framework was succinctly illustrated in (Fig. 2a), with detailed elaboration provided in (Supplementary Fig. 9). We commenced with 93 candidate predictors derived from electronic health records (EHR), narrowing them down through univariate Cox proportional hazards regression and Log-rank tests to 50 and 63 significant predictors, respectively (Supplementary Tables 6 and 7, Fig. 1d, Supplementary Figs. 1 and 2). A core set of 46 predictors, showing consistent significance in both tests, were defined as candidate survival features for inclusion in the modeling framework. 12 AI algorithms were performed on 46 candidate survival features to fit models, yielding 132 distinct modeling schemes (Fig. 2b). Each scheme’s performance was rigorously evaluated using a tripartite strategy: 10 repeated 10-fold cross-validation, Monte-Carlo cross-validation (MCCV, 100 iterations with a 0.7 sampling ratio), and bootstrap analysis (1000 iterations with a 0.7 sampling ratio) (Supplementary Table 8). This evaluation process identified the integration of Surv.gbm and Surv.Xgboost, enhanced by the Filter & Wrapper Hybrid Method, as the optimal modeling scheme for populations with AHF and RD. This scheme achieved the highest average C-index of 0.821, demonstrating superior discriminative power over alternative models (Fig. 2b).
a AI modeling hybrid framework overview. The image was created with a licensed version of bioRender.com. b Schematic illustration of optimal scheme identification. The diagram delineates the comprehensive evaluation process of 132 distinct modeling schemes through an array of lollipop plots, including 10 repeated 10-fold cross-validation, 100 iterations of Monte-Carlo cross-validation (MCCV) with a 70% sampling ratio, and a thorough bootstrap analysis comprising 1000 iterations. c Analysis of non-linear relationship between AIHFLevel and ACM risk using restricted cubic spline regression based on Replication cohort (Poverall <0.0001, and Pnon-linear < 0.0001). Non-linear patterns indicating a ‘fast-to-low’ increase in ACM risk associated with rising AIHFLevel. Univariate Cox regression analysis highlighted the AIHFLevel as a significant clinical predictor for ACM, with a hazard ratio (HR) of 1.615 (95% CI = 1.417–1.863). Statistic test: two-sided Wald test: P < 0.0001. Line chart displaying the estimated logarithm HRs represented by blue lines, along with 95% CIs indicated by shading. d Cumulative Kaplan-Meier estimates the delineating time to the survival difference for ACM stratified by AIHFLevel within the Replication cohort. e Time-dependent ROC analysis for predicting ACM within the Replication cohort. AUCs at 6-, 12-, 24-, and 30-months demonstrating strong predictive accuracy: 0.902, 0.932, 0.932, 0.903. f Calibration curves depicting the predicted versus observed probabilities of ACM as evaluated by AIHFLevel within the Replication cohort. g Decision Curve Analysis (DCA) illustrating net benefit curves of AIHFLevel for predicting ACM within the Replication cohort. h Analysis of non-linear relationship between AIHFLevel and ACM risk using restricted cubic spline regression within Meta cohort (Poverall <0.0001, and Pnon-linear < 0.0001). Univariate Cox regression analysis highlighted the AIHFLevel as a significant clinical predictor for ACM, with an HR of 1.878 (95% CI = 1.770–1.992). Statistic test: two-sided Wald test: P < 0.0001. Line chart displaying the estimated logarithm HRs represented by blue lines, along with 95% CIs indicated by shading. i Cumulative Kaplan-Meier estimates delineating time to the first adjudicated occurrence of ACM stratified by AIHFLevel within Meta cohort. j Time-dependent ROC analysis for predicting ACM within Meta cohort. AUCs at 6-, 12-, 24-, and 30-months confirming predictive excellence: 0.925, 0.947, 0.965, and 0.960. k Calibration curves depicting the predicted versus observed probabilities of ACM within the Meta cohort. l DCA illustrating net benefit curves of AIHFLevel for predicting ACM within Meta cohort.
Consequently, the AIHFLevel system was developed using the Discovery cohort, guided by this optimal scheme. AIHFLevel utilized a set of 12 readily accessible predictors: age (year), arrhythmia comorbidity (Yes or No), CAD comorbidity (Yes or No), CKD stage (I, II, III, IV, V), lymphocyte percentage (%), mean corpuscular hemoglobin concentration (MCHC, g/L), eGFR (ml/(min/1.73 m3)), serum creatinine (Cr, μmol/L), serum total bilirubin (TBIL, μmol/L), serum cardiac troponin I (CTnI, ng/ml), left ventricular ejection fraction (EF, %), stroke volume (SV, %) (Supplementary Table 9).
Uniformly prognostic implication and predictive performance of AIHFLevel
To explore potential non-linear relationships between AIHFLevel scores and hazard ratio (HR) for ACM, we initially estimated the associations with restricted cubic spline analysis. In the Replication and Discovery cohorts, we consistently observed a pattern of non-linear associations along with the increase of AIHFLevel: ‘fast-to-low increase’ of risk for ACM (Poverall <0.0001, and Pnon-linear < 0.0001) (Fig. 2c, Supplementary Fig. 3a). Univariate Cox regression analysis underscored AIHFLevel’s significance as a clinical predictor for ACM (Replication cohort: HR = 1.615, P < 0.0001, 95%CI = 1.417–1.863; Discovery cohort: HR = 2.245, P < 0.0001, 95%CI = 2.039–2.472) (Fig. 2c, Supplementary Fig. 3a). To assess AIHFLevel’s prognostic efficacy, subjects were stratified into high and low AIHFLevel groups based on the median score. Kaplan–Meier curve for ACM indicated significantly shorter survival for the high AIHFLevel group in both Replication and Discovery cohorts (Log-rank test, P < 0.0001) (Fig. 2d, Supplementary Fig. 3b). Discriminatory power of AIHFLevel was quantified through ROC analysis, with AUCs at 6-, 12-, 24-, and 30-months demonstrating strong predictive accuracy: 0.902, 0.932, 0.932, 0.903 in the Replication cohort and 0.931, 0.952, 0.973, 0.976 in the Discovery cohort, respectively (Fig. 2e, Supplementary Fig. 3c). Calibration analyses for 6-, 12-, and 24-month survival predictions showed high concordance with observed outcomes in Replication and Discovery cohorts (Fig. 2f, Supplementary Fig. 3d). Additionally, Decision Curve Analysis (DCA) affirmed AIHFLevel’s significant clinical utility and net benefit at 6-, 12-, and 24-month survival intervals, validating its value in clinical decision-making (Fig. 2f, Supplementary Fig. 3e). Comparable excellence was observed in Meta cohort, where a robust positive correlation between AIHFLevel scores and ACM risk was evident (Poverall < 0.0001, and Pnon-linear < 0.0001) (HR = 1.878, P < 0.0001, 95%CI = 1.770–1.992)) (Fig. 2h). The high AIHFLevel group exhibited a significantly greater incidence of long-term ACM (P < 0.0001) (Fig. 2i), with AUCs at 6-, 12-, 24-, and 30-months confirming predictive excellence: 0.925, 0.947, 0.965, and 0.960, respectively (Fig. 2j). Calibration and DCA plots further corroborated AIHFLevel’s predictive accuracy and clinical benefit, reinforcing its applicability across varying prognostic thresholds (Fig. 2j, l).
Robustness and superior performance of AIHFLevel
In clinical practice, prognostication and stratification for management traditionally relied on a range of clinicopathological characteristics, such as serum cardiac troponin levels, natriuretic peptides, pharmacotherapy, renal function, age, and comorbidity status1. To assess their predictive efficacy, we evaluated these readily accessible conventional clinical traits derived from the EHR system. Our findings revealed that AIHFLevel consistently outperformed in predictive accuracy. This superiority was quantitatively confirmed through C-index and Integrated Brier Score (IBS) across Discovery, Replication, and Meta cohorts, suggesting AIHFLevel’s potential for integration into clinical workflows (Fig. 3a, b, Supplementary Fig. 4a, and Supplementary Table 10).
a Comparative predictive efficacy of AIHFLevel against collected 93 readily accessible clinical traits in the Replication cohort (n = 214). AIHFLevel exhibits notably higher predictive accuracy, demonstrated by superior C-index values and IBS. The statistical significance of differences was determined using the compareC R package, employing a one-shot, nonparametric approach. C-index was presented with 95% confidence interval (CI). Statistic tests: two-sided z-score test. b Comparative predictive efficacy of AIHFLevel against collected 93 readily accessible clinical traits in the Meta cohort (n = 712). C-index was presented with 95% CI. Statistic tests: two-sided z-score test. c Comparative predictive efficacy of AIHFLevel against established risk and prognostic models within the Replication cohort (top, n = 214) and Meta cohort (bottom, n = 712). The risk scores for each model were computed based on their predefined features and coefficients as outlined in their original publications. The analysis, visualized through bar graphs for the C-index and line graphs for the IBS. C-index was presented with 95% CI. We employed a one-shot, nonparametric statistical comparison, using two-sided z-tests to ascertain significance. d Multivariate Cox regression analysis of AIHFLevel for ACM within Replication cohort (left, n = 214) and Meta cohort (right, n = 712). Upon adjusting for significant clinical traits, AIHFLevel consistently demonstrated independent prognostic value. Statistic test: two-sided Wald test. Dot plots illustrated the adjusted hazard ratios with the horizontal line indicating the 95% CI for each variable. Bar graphs highlighted the -log10(adjusted P-values) to denote statistical significance levels. e Subgroup analysis estimating the clinical prognostic value of AIHFLevel across different pre-specified subgroups. The length of the horizontal line represented the 95% confidence interval for each subgroup, with a vertical dotted line indicating the hazard ratio of all patients. Statistic test: two-sided Wald test. The vertical solid line denoted HR = 1. f SHAP summary dot plot, stacked vertically to show density. On the X-axis, the contribution of each predictor to the system’s output was quantified by the SHAP value. The probability of survival decreased with increasing the SHAP value of a predictor. A positive SHAP value indicated an increased risk of ACM, while a negative value suggests a protective effect. Each dot represents an individual patient’s SHAP value for a specific predictor. The color coding of dots reflected the actual predictor values for patients, with red indicating higher values and blue indicating lower values. This color gradient demonstrates the relationship between predictor values and their effect on ACM risk. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Given the development of numerous objective risk markers and composite prognostic scores for heart failure, we expanded our analysis to include a variety of published risk markers and models, including systemic immune-inflammation index (SII), neutrophil-lymphocyte ratio (NLR), neutrophils to leukocyte‐neutrophil count (dNLR), lymphocyte-monocyte ratio (LMR), platelet-lymphocyte ratio (PLR), albumin-to-fibrinogen ratio (AFR), triglyceride‐glucose (TyG)‐index, MAGGIC-HF, PREDICT-HF, BCN Bio-HF, REMATCH-HF, and 3C-HF score35,36,37,38,39.
Initially, we computed the risk scores for each model based on their predefined features and coefficients as outlined in their original publications, testing their predictive performance using C-index and IBS. We also added a comparative model by refitting the variables used in AIHFLevel using the cox regression method to create a prognostic model for comparison with AIHFLevel. AIHFLevel exhibited superior accuracy compared to other models (Fig. 3c, Supplementary Fig. 4b, Supplementary Table 11). While some model might perform well within their original dataset, they faltered across other cohorts, likely due to a lack of generalizability and potential overfitting. Building upon this preliminary analysis, we conducted a secondary, in-depth evaluation to underscore the AIHFLevel’s robustness and superior predictive capacity. This involved a refitting process, where AIHFLevel and other models were retrained using only their respective predictors—without incorporating their original coefficients—in a unified proportional hazards regression framework applied to the Discovery cohort. This methodological refinement aimed at facilitating an equitable, standardized comparison, thus validating the robustness of AIHFLevel under a stringent evaluative setting. The outcomes of this recalibration underscored AIHFLevel’s optimal performance, surpassing that of comparator models across the Discovery, Replication, and Meta cohorts (Supplementary Fig. 5). This iterative validation reaffirmed AIHFLevel’s superiority over conventional models, and illuminated the resilience of its predictive capacity, demonstrating a potent tool in prognostic assessment. In addition, we performed the Surv.Xgboost algorithm to refit each model again with their respective predictors based on the Discovery cohort, revealing that AIHFLevel still maintained superior accuracy across all cohorts (Supplementary Fig. 6). The consistent outperformance under equivalent algorithmic conditions also indicated the inherent robustness and predictive reliability of AIHFLevel’s predicters. Such findings demonstrated the efficacy of our AI hybrid modeling framework in identifying a set of potent predictors. 12-predictor AIHFLevel not only forecasted the prognosis of AHF&RD patients with remarkable accuracy but also achieved this with a streamlined feature set, significantly boosting its clinical utility and readiness for broader implementation.
Clinical interpretability underlying AIHFLevel
After adjustment for available clinical traits with significant prognostic value, multivariate Cox regression analysis confirmed the independent prognostic significance of AIHFLevel for ACM across the Replication, Meta, and Discovery cohorts (Fig. 3d, Supplementary Fig. 4c). Stratification analysis further revealed the consistent prognostic value of AIHFLevel across different pre-specified subgroups, delineated by dataset (Discovery cohort, Replication cohort, Meta cohort), age (>65 or ≤65 years), cardiac function grade (NYHA I/II versus NYHA III/IV), heart failure (HF) subtype (HFpEF, HFmrEF, HFrEF), chronic kidney disease (CKD) stage (I/II versus III, IV/V), and history of percutaneous coronary intervention (PCI) (Yes versus No) (Fig. 3e).
In the real-world deployment of AI predictive models, balancing accuracy, interpretability, and complexity is significant40. Such models often act as ‘black boxes’ to clinicians, masking the reasoning behind their predictions. Beyond prognostic outcomes, the significant risks associated with various predictors demand attention. Understanding these risks is essential for guiding clinical decision-making and managing reversible risk factors. Therefore, Shapley Additive exPlanation (SHAP) approach was leveraged to interpret the AIHFLevel system’s outputs, the influence of each predictor on the system’s predictions. Our explainable analysis provided two types of explanations: global explanation of the system at the feature level and local explanation at the individual level. Global explanation described the overall functionality of the model. The global explanation, as depicted in the SHAP summary plot (Fig. 3f), analyzed the 12 predictors by their contribution to AIHFLevel’s decision-making process, arranging them in a descending order based on average SHAP values. GFR, age, cTNI, creatinine, and EF emerged as the most significant features, suggesting that AIHFLevel’s adeptness at pinpointing key physiological indicators—specifically, renal and cardiac function statuses—for refined prognostic assessments. Moreover, the web-based AIHFLevel system incorporated local explanation, analyzing how a certain prediction was made for a new specific individual by incorporating the individualized input data. The elaboration of these functionalities will be described in the following sections.
Prognostic stratification underlying AIHFLevel
We further performed a conditional inference survival tree to explore heterogeneity in trends for prognosis among AHF&RD patients, stratifying individuals based on the AIHFLevel values (Fig. 4a). This approach delineated three distinct prognostic states: (Low-Risk: Defined by AIHFLevel ≤ 0.435; Intermediate-Risk: AIHFLevel between 0.435 and 1.548; High-Risk: AIHFLevel > 1.548) (Fig. 4b). Kaplan–Meier analysis revealed significant differences in ACM incidence across three prognostic states (P < 0.0001), indicating the stratification’s efficacy (Fig. 4c). Further validation with unsupervised t-SNE on AIHFLevel 12-predictor profiles visualized the risk heterogeneity among AHF&RD patients. This dimensional reduction technique effectively segregated samples into discernible clusters within a two-dimensional space, suggesting robust discrimination capabilities (Fig. 4d). TrinROC analysis confirmed AIHFLevel’s discriminatory power for the prognostic states. In the Replication cohort, the trinormal ROC test statistic reached 1063.675 (P < 0.0001), with a Volume Under the ROC Surface (VUS) statistical test statistic of 14.759 (P < 0.0001) and a trinormal VUS of 0.876. Similarly, in the Meta cohort, the trinormal ROC test statistic achieved 8682.598 (P < 0.0001), with a VUS statistical test statistic of 29.331 (P < 0.0001) and a trinormal VUS of 0.867 (Fig. 4e). The baseline characteristics also varied in accordance with the prognostic stratification (Fig. 4f). The high-risk state was predominantly associated with female or elderly patients. CAD comorbidity was more prevalent in the high-risk state. Additionally, lower LVEF and HFrEF were highly associated with high-risk, where patients were more likely to present with deteriorated cardiac and renal function (Fig. 4g).
a Conditional Inference Survival Tree Analysis: Identifies prognostic heterogeneity within AHF&RD patients, categorizing them into three prognostic states based on AIHFLevel values: Low-Risk (AIHFLevel ≤ 0.435), Intermediate-Risk (AIHFLevel between 0.435 and 1.548), and High-Risk (AIHFLevel > 1.548). Image was created with licensed version of bioRender.com.b Distribution of AIHFLevel values among low-risk, intermediate-risk, and high-risk groups in the Replication cohort (Top, P < 0.0001, n = 214) and Meta cohort (Bottom, P < 0.0001, n = 712). Statistic tests: two-sided wilcoxon test. c Kaplan-Meier curves showing the survival difference for ACM across three prognostic states, with log-rank test results indicating statistical significance. d Unsupervised t-SNE analysis spatially segregates samples by prognostic states into two dimensions, showcasing effective discrimination. e Trinormal snapshot of ROC surface showing AIHFLevel’s discriminatory efficacy among the three prognostic states. f Heatmap depicting the baseline characteristics distribution within Meta cohort, including gender, alcohol status, smoking status, age, occurrence of ACM, coronary artery disease (CAD), hypertension (HTN), arrhythmia comorbidities, NYHA cardiac function, chronic kidney disease (CKD) stage, HF subtype, nifedipine/diltiazem (CCB) medication history. g Pie chart showing the two-sided Chi-squared test of general baseline factors for prognostic stratification within Meta cohort. h Critical transition signal analysis in the CRCCD cohort (n = 712). Top left: Schematic diagram illustrating a phase transition during disease progression. Top right: Bar plot showing DNB-Composite Index (CI) per module across three prognostic states. The Biomodule (module with the highest CI) of each state indicated the CT levels per state. Intermediate-risk exhibited the most significant CT signal. Bottom left: Distribution of DNB-scores in each state. SDi, the average standard deviation of all variables inside the biomodule; PCCi, the average Pearson’s correlation coefficient (PCC) of absolute value for variable-pairs inside the biomodule; PCCo, the average PCC absolute value for feature-pairs between the biomodule and others (absolute value). The CI is expected to increase abruptly and significantly before the CT to the deteriorated phase and can serve as an early warning signal. Bottom right: Comparison between observed (red dot) and simulated (box plot) CI validated the robustness of CT and tipping-point captured by DNB model. i Global shift of our prognostic stratification based on the CRCCD cohort (n = 712; low-risk: n = 427, intermediate-risk: n = 177, high-risk: n = 427). Euclidean expression distances were calculated between high-risk samples and remaining samples (blue), within samples of high-risk (green), and within remaining samples (grey). Inset summarized the average distances between pairs of samples as a percentage of the average distance between high-risk and remaining samples. In the boxplots, centre line indicates median, bounds of box indicate 25th and 75th percentiles, and whiskers indicate minimum and maximum.
Based on nonlinear dynamic theory, complex diseases typically traverse through distinct phases: ‘Before-deterioration’ (a relatively stable phase with gradual change), ‘Pre-disease’ (a critical state or tipping point which is the limit of the stable state just before the transition to the deteriorated state), and ‘Deteriorated phase’ (typically irreversible to the Before-deterioration phase)41. Our trajectory analysis has revealed that AHF&RD progression indeed does not follow a gradual path, instead undergoing nonlinear and drastic transitions at certain points (Fig. 1c). Identifying the tipping point just at such critical transition (CT) is crucial for timely intervention. Thus, we employed a tipping-point theory-based model, the enhanced dynamic network biomarker (DNB), to convert static snapshots of three AIHFLevel-derived prognostic states into a dynamic movie, exploring malignant phase transition in disease progression (Fig. 4h, Critical transition signal analysis in Methods). Following quantifying each module per cross-section through the DNB model, we captured a potent CT signal preceding disease deterioration at the intermediate-risk phase, where targeted interventions should be implemented to potentially alter the progression trajectory (Fig. 4h, Supplementary Tables 12 and 13). We then calculated Euclidean distance to investigate the global variance between and within prognostic states. The divergence between high-risk individuals and the other individuals or within the high-risk individuals was significantly larger than the distance within intermediate-risk & low-risk samples (Fig. 4i). These findings further validated the clinical relevance and the rationality of our prognostic stratification, suggesting its potential in guiding early preventive strategies.
Sensitivity analysis
To enhance the robustness assessment of AIHFLevel, we expanded the evaluation to include major adverse cardiovascular events (MACE) as an alternative outcome based on the Meta cohort. Initially, stratifying patients into high and low AIHFLevel groups based on median values, the Kaplan–Meier curves revealed a significantly higher rate of MACE in the high AIHFLevel group (P < 0.0001) (Fig. 5a). Significant risk differences for MACE were also observed across the three prognostic states (P < 0.0001) (Fig. 5b) Calibration curves indicated similar predicted and actual probabilities for MACE (Fig. 5c). DCA analysis further demonstrated the predictive potential for MACE (Fig. 5d). Stratification analysis demonstrated the AIHFLevel’s consistent prognostic value for MACE across different subgroups (Fig. 5e). ROC analysis confirmed AIHFLevel’s discriminative ability for MACE, and AUCs at 6-, 12-, 24-, and 30-month were respectively 0.825, 0.848, 0.861, 0.846 (Fig. 5f). Notably, AIHFLevel demonstrated enhanced accuracy in predicting ACM over MACE (MACE: C-index = 0.798, IBS = 0.115; ACM: C-index = 0.913, IBS = 0.0595) (Fig. 5g). Designed to estimate individual survival distributions, AIHFLevel was initially trained with ACM as its primary endpoint. However, when evaluated against MACE as an alternative endpoint, it still demonstrated reliable predictive performance. The ability to accurately assess outcomes across different clinical endpoints indicated AIHFLevel’s comprehensive predictive capabilities and robustness in clinical prognostication.
a Cumulative Kaplan–Meier estimates of the time to the first adjudicated occurrence of MACE within the Meta cohort, with log-rank test results indicating statistical significance. b Kaplan-Meier curve showing the incidence difference for MACE across three prognostic states, with log-rank test results indicating statistical significance. c Calibration curve depicting the predicted versus observed probabilities of MACE at 6, 12, and 24 months, as evaluated by AIHFLevel within Meta cohort (n = 712). d DCA illustrating net benefit curves of AIHFLevel for predicting MACE at 6, 12, and 24 months within Meta cohort (n = 712). The X-axis represented the threshold probability for critical care outcomes, while the Y-axis quantified the net benefit. e Subgroup analysis estimating AIHFLevel’s prognostic value for MACE across different subgroups. Statistic test: two-sided Wald test. The length of the horizontal line represented the 95% confidence interval for each subgroup, with a vertical dotted line indicating the hazard ratio of all patients. The vertical solid line denoted HR = 1. HR > 1 indicated AIHFLevel as a risk prognostic factor. f Time-dependent ROC analysis for predicting MACE within Meta cohort (n = 712). AUCs at 6-, 12-, 24-, and 30-months demonstrating strong predictive accuracy: 0.825, 0.848, 0.861, 0.846. g Comparative AIHFLevel’s efficacy in assessing ACM and MACE within the Meta cohort (n = 712) using C-index and IBS. C-index was presented with 95% confidence interval (CI).
Extrapolation of AIHFLevel to the heterogeneous populations
AIHFLevel has been rigorously evaluated within our designated testing datasets (Replication and Meta cohorts); however, external validation remains imperative. Models can correspond erroneously or be fitted to peculiarities in training datasets so well that it losed generalizability to heterogeneous data unseen by the training process. Recent studies suggested that these limitations can potentially be addressed by validation on different data modalities (i.e. dataset from diverse hospital systems technology platforms, even regions and ethnic backgrounds) for predictive analytics42. Therefore, we have incorporated an independent external cohort of 1024 patients, spanning June 2001 to October 2012, from the BIDMC center. The imperative for future AI systems lies in their broad applicability across various healthcare settings and geographic locales26. The significant population heterogeneity observed between CRCCD and BIDMC centers provided a robust test bed for assessing the AIHFLevel’s generalizability (Supplementary Table 14).
Scaled Schoenfeld residual analysis showed AIHFLevel’s proportional hazard assumption was met over time (P = 0.7647), confirming its time-invariance (Fig. 6a). We also noted a nonlinear relationship between AIHFLevel and the risk for ACM, characterized by a ‘fast-to-low’ increase in risk (Poverall <0.0001, Pnon-linear < 0.0001) (Fig. 6b). Univariate Cox regression analysis further solidified AIHFLevel’s role as a significant prognostic factor (HR = 1.956, P < 0.0001, 95% CI = 1.838-2.082) (Fig. 6b). Kaplan–Meier survival curves illustrated that higher AIHFLevel correlated with significantly lower survival rates (P < 0.0001) (Fig. 6c). ROC analysis also indicated an enhanced accuracy of AIHFLevel: AUCs for assessing ACM at 1-, 2-, 3-, 4- year were 0.788, 0.816, 0.824, 0.846, respectively (Fig. 6d). Calibration and decision curve analyses further confirmed AIHFLevel’s robust clinical utility across diverse patient populations (Fig. 6e, f). Based on established stratification criteria, patients were categorized into three prognostic states (Fig. 6g), and three states also presented significant risk differences (Fig. 6h). The AIHFLevel’s stratification efficacy was validated using both t-SNE and trinROC methods, indicating significant discriminatory power (trinormal ROC test statistic = 6938.615, P < 0.0001; VUS = 0.894, P < 0.0001) (Fig. 6i, j). Multivariate Cox regression, adjusting for potential confounders, demonstrated AIHFLevel’s independent prognostic value (Fig. 6k). Comparatively, AIHFLevel outperformed traditional clinical indicators and other models according to C-index and IBS, showcasing its superior predictive performance (Fig. 6l, m, Supplementary Tables 15 and 16). These validations underscore the robust extrapolation and generalizability of AIHFLevel, indicating the potential as a clinical translation tool.
a Proportional hazard assumption of Cox regression for AIHFLevel demonstrated no significant correlation between Schoenfeld residuals and time. Statistical tests: two-sided Schoenfeld residuals test. b Analysis of the non-linear relationship between AIHFLevel and ACM risk using restricted cubic spline regression within the external BIDMC cohort (Poverall <0.0001, and Pnon-linear < 0.0001). Non-linear patterns indicating a ‘fast-to-low’ increase in ACM risk associated with rising AIHFLevel. Univariate Cox regression analysis highlighted the AIHFLevel as a significant clinical predictor for ACM, with a HR of 1.956 (95% CI = 1.838-2.082). Statistic test: two-sided Wald test. Line chart displaying the estimated logarithm HRs represented by blue lines, along with 95% CIs indicated by shading. c Cumulative Kaplan-Meier estimates delineating time to the survival difference for ACM stratified by AIHFLevel. d Time-dependent ROC analysis for predicting ACM. AUCs at 1-, 2-, 3-, 4-year demonstrating strong predictive accuracy: 0.788, 0.816, 0.824, 0.846. e Calibration curves depicting the predicted versus observed probabilities of ACM as evaluated by AIHFLevel. f DCA illustrating net benefit curves of AIHFLevel for predicting ACM. g Distribution of AIHFLevel across three prognostic states in the BIDMC cohort (P < 0.0001, n = 1024). Statistic tests: two-sided wilcoxon test, as determined by established stratification criteria. Centre line indicates median, bounds of box indicate 25th and 75th percentiles, and whiskers indicate minimum and maximum. h Kaplan-Meier curves showing the survival difference for ACM across three prognostic states. i Trinormal snapshot of ROC surface demonstrating the discriminatory power of AIHFLevel on three prognostic states. j t-SNE dimension reduction analysis spatially segregated samples by prognostic states into two dimensions, showcasing effective and stable discrimination. k Multivariate Cox regression of AIHFLevel for ACM risk in the BIDMC (n = 1024). Upon adjusting for potential confounders, AIHFLevel demonstrated independent prognostic value. Dot plots illustrated the adjusted hazard ratios with the horizontal line indicating the 95% confidence interval for each variable. Bar graphs highlighted the -log10(adjusted P-values) to denote statistical significance levels. Statistic test: two-sided Wald test. l Comparative predictive efficacy of AIHFLevel against clinical traits in the BIDMC (n = 1024). C-index was presented with 95% CI. Statistic tests: two-sided z-score test. m Comparative predictive efficacy of AIHFLevel against established risk and prognostic models in the BIDMC (n = 1024). The analysis, visualized through bar graphs for the C-index and line graphs for the IBS, using two-sided z-score test to ascertain significance: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Convenient web application for clinical utility
Based on the Django web framework, the AIHFLevel system has been deployed in a user-friendly application (https://www.hf-ai-survival.com), designed to enhance the utility in clinical scenarios (Fig. 7a). The system incorporated a range of functionalities for the analysis of individual patients, including prognostic stratification, calculation of long-term survival probabilities, all-cause mortality predictions at indicated time points, local interpretation, and the predictor contribution.
a The user interface of the application website showcases a comprehensive suite of functionalities, including prognostic stratification, calculation of long-term survival probabilities, all-cause mortality predictions at indicated time points, local interpretation, and predictor contribution. The left panel displayed the initial screen where users input the actual values for the 12 predictors and the desired time point for any new individual patient. The right panel presented the interface for displaying the results of the survival assessment, providing a clear and concise overview of the prognostic outcomes. b Illustration of the practical application through an exemplary case. Users input data through 13 queries: age (72 years), arrhythmia comorbidity (No), CAD comorbidity (Yes), CKD stage (IV), Cr (101.3 μmol/L), LVEF (51%), eGFR (23.490 ml/(min/1.73 m3)), Lymphocyte (23.8%), MCHC (328.00 g/L), SV (42%), CTnI (0.35 ng/ml), TBIL (2.79 ng/ml), and the time point of interest (600-day). The image was created with a licensed version of bioRender.com. c Long-term survival curve. The y-axis represents the percentage value of survival probability, while the x-axis represents any future time point. The arrow and dotted line indicate the calculation of all-cause mortality at the specified time point of interest. d Histogram depicting the predicted survival probability values for this patient at several important future time points (180, 365, 730, 900 days, and the specified time point) from the current moment. Data were presented with 95% CI. e Radar graph indicating contributions of each predictor to the survival assessment. f The X-axis quantifies each predictor’s contribution to the system’s output using SHAP values, with the probability of survival decreasing as the SHAP value increases. A positive SHAP value indicates an increased risk of ACM, while a negative value suggests a protective effect. For this case, the values of eGFR, SV, CTnI, age, and Cr were pushing the decision towards a worse prognosis, while Cr, LVEF, and MCHC were influencing the decision toward a favorable prognosis.
To intuitively demonstrate the practical application, we presented an exemplary case. Users simply input data through 13 queries: age (72 years), arrhythmia comorbidity (No), CAD comorbidity (Yes), CKD stage (IV), Cr (101.3 μmol/L), LVEF (51%), eGFR (23.490 ml/(min/1.73 m3)), Lymphocyte (23.8%), MCHC (328.00 g/L), SV (42%), CTnI (0.35 ng/ml), TBIL (2.79 ng/ml), and the time point of interest (600-day) (Fig. 7b). Upon entry, the system automatically performed conditional inference survival tree, categorizing this individual as high-risk state. Subsequently, it generated a long-term survival curve, calculating a 45.5% probability of all-cause mortality at 600-day (Fig. 7c). The bar graph offered an intuitive view of the survival profile over time, with predicted survival probabilities at 180-, 365-, 730-, 900-, 600-day being 78.6%, 61.5%, 48.3%, 45.5%, and 54.5%, respectively (Fig. 7d). Furthermore, the radar graph indicated the predictor contribution to the survival assessment (Fig. 7e). As observed, the values of eGFR, SV, CTnI, age, and Cr were pushing the decision towards the worse prognosis, while Cr, LVEF, and MCHC were pushing the decision towards the favorable prognosis (Fig. 7f). This suggested that aligning most predictors values closer to their normal ranges, such as eGFR and SV, through targeted interventions or management strategies could mitigate the risk of ACM for this individual, despite the overall prediction pushed this case into the high-risk prognosis. Overall, this system could help physicians select the optimal treatments and offer personalized recommendations to improve outcomes based on the output risk stratification and interpretability of personal information.
Discussion
Accurate prognostication is significantly important in AHF to identify the ideal time for referral to an appropriate center and to plan therapy and follow-up strategies43,44. Predicting the outcome of HF is complex and difficult. Despite the development of many risk scores reported in recent decades25,45, some prognostic tools were not specifically designed for AHF populations where a majority of patients also suffer from RD. A small number of researchers have translated their mode into a practical prognostic tool rather than just for algorithm development. In addition, some tools were developed and validated in selected clinical trial populations or at a single center with geographic and ethnic limitations1. To bridge this gap, we identified predictors among profiles comprising almost 100 variables, including biological data, biomarkers, histories, and imaging data using our original workflow, to discover novel predictors that were not hypothesis-driven without presumptions. we adopted an original approach to identify and validate a robust AI-powered survival assessment system AIHFLevel based on cohorts from multicenter to predict ACM for patients with AHF and RD. This system was further deployed into a web-based application for clinicians and patients to have a better understanding, forming an improved management plan.
AI techniques have been investigated by many groups as tools to improve survival predictions in HF; however, obstacles still remain before AI progresses to clinical practice46,47,48. For instance, appropriate initiatives are required to maximize accuracy while avoiding overfitting and deciding how many and which clinical parameters need to be included in the prediction so that gathering this information on a new patient is not overly expensive or burdensome49. Moreover, despite the importance of selecting an optimal modeling algorithm, researchers might mostly choose the algorithms based on their preferences and knowledge limitations, leading to limited predictive power31. Some studies don’t elaborate on the rationale and reasons for selecting such algorithms, which makes it difficult to find the best modeling approach to fit one dataset. However, some researchers advocate for hybrid learning models, such as ensemble-based approaches, which combine multiple models to enhance predictive performance50. Mienye and colleagues demonstrated that ensemble-based learning can outperform single models in predicting heart failure events51. Therefore, Our study innovates beyond existing risk prediction frameworks by developing an ensemble hybrid model framework—AI hybrid modeling framework. Through this process, we minimized redundant information and established a system based on 12 predictors, named AIHFLevel. Our modeling framework functions by utilizing a collection of diverse and independent models, which allowed AIHFLevel to reduce generalization error and enhance predictive reliability.
Our study not only applied existing risk prediction learning algorithms but innovatively developed an ensemble hybrid model framework, potentially marking a significant advancement in the field of risk prediction for AHF with RD comorbidity. We draw upon established concepts in AI modeling, notably hybrid paradigms that incorporate ensemble-based approaches52,53. Herein, instead of choosing the best individual learning algorithm, our methodology focuses on constructing ensemble modeling schemes to improve survival assessment accuracy, especially for the comorbidity of AHF with RD. We employed 12 different AI learning algorithms and broadened potential model schemes by integrating three distinctive feature engineering approaches: Embedded, Filter, and Wrapper. This integration reduces generalization errors and ensures robust generalizability across various clinical scenarios. The novel of our framework methodology included: (1) Integration of multiple learning algorithms: We incorporate a diverse array of advanced algorithms, each contributing uniquely to the ensemble. This integration enhances robustness and reduces the likelihood of model overfitting. Included are various forms of regularization and sophisticated machine learning techniques such as Support Vector Machines, Random Forests, and Gradient Boosting Machines. (2) Advanced feature engineering techniques: Our model employs a combinational family of regularization, sequential forward floating selection (SFFS), and Correlation-Adjusted Survival Score (CARS). This allowed for a comprehensive exploration of potential predictive variables and enhances the model’s ability to handle high-dimensional data effectively. (3) Ensemble-based modeling: Leveraging the strengths of ensemble learning, our framework boosts predictive accuracy by strategically combining models to mitigate individual weaknesses and capitalize on their collective strengths, thus ensuring more reliable and consistent predictions. (4) Comprehensive assessment strategy: A systematic and unbiased assessment strategy included 10 repeated 10-fold cross-validation, Monte-Carlo cross-validation, and bootstrap analysis, which enhanced the model’s generalizability and reliability in diverse clinical settings and populations.
In general, incorporating an AI model into clinical practice requires satisfying multiple criteria, particularly the need for algorithms to be tested across multiple cohorts and to demonstrate universal applicability across various healthcare settings, systems, and geographic locations. Diverse databases may contain poorly classified or reported variables and employ different technologies and platforms, highlighting the importance of validation using independent cohorts54. Moreover, the timing of endpoint event observation largely depends on the surveillance schedule, and the follow-up protocol can influence patient survival, thereby affecting the model’s performance in different settings55. Hence, validation across different data modalities (i.e. datasets from diverse hospital systems technology platforms, even regions, and ethnic backgrounds) for predictive analytics is essential42. In this study, the disparity in follow-up, attrition, detection, and selection schedules between CRCCD Center and Beth Israel Deaconess Medical BIDMC Center provided a robust test bed for assessing the AIHFLevel’s generalizability56. Despite the challenges, AIHFLevel has been confirmed as a robust tool for risk stratification and estimation, as evidenced by rigorous assessments of various criteria, including discrimination, calibration, performance, generalization, and clinical utility in the Discovery, Replication, Meta, and External heterogeneous cohorts.
In the clinical setting, clinicopathological and baseline characteristics such as age, cardiac troponin, medical history, echocardiographic left ventricular parameters, and comorbidity status, determine clinical management and prognosis57,58. Here, we compared the AIHFLevel with almost 100 clinical variables and molecular characteristics collected from AHF patients. According to the index of concordance and IBS, AIHFLevel outperformed other features in all cohorts, suggesting a potential alternative to assess prognosis and drive personalized management of patients in the clinic. It has been reported that chronic low-grade inflammation in AHF with RD activates a harmful immune response and leads to further cardiac and renal impairment59. The inflammation degree in CKD and AHF has been proven to be able to predict ACM60. More importantly, the AIHFLevel demonstrated higher accuracy than classic biomarkers, especially inflammatory indicators. Moreover, the conventional Cox proportional hazards regression model and some other published risk models such as 3C-HF of Senni M, REMATCH-HF of Lietz K, etc are often overestimated single cohorts, resulting in the reduced accuracy in another cohort35,36,37,38,39, due to the poor generalizability from overfitting. In contrast, AIHFLevel has undergone optimization through a comprehensive algorithm network with a superior extrapolation possibility and significant advantages over other models. Following the initial comparison, we advanced to a secondary, in-depth analysis to further validate AIHFLevel’s robustness. This phase involved a strategic refitting process where AIHFLevel and the comparative models were retrained using only their respective predictors, without incorporating their original coefficients, through the Cox proportional hazards regression. This recalibration ensured a fair and uniform comparison, thus rigorously testing AIHFLevel’s robustness in a more stringent evaluative setting. The results demonstrated that both AIHFLevel and its refitted version outperformed the comparator models across cohorts, reaffirming AIHFLevel’s superiority and showcasing its resilience and inherent predictive strength. Building upon these analyses, we employed the Surv.Xgboost algorithm—a key component in AIHFLevel’s development—to refit each comparative model using only their respective predictors. This approach established a uniform framework for assessing the inherent strength of each model’s selected features in a consistent algorithmic environment. The consistent superiority of AIHFLevel across various algorithmic frameworks underscores the inherent robustness and predictive reliability of its selected predictors. This performance highlights the effectiveness of our AI hybrid modeling framework in identifying a set of robust predictors. The 12-predictor AIHFLevel not only accurately forecasts the prognosis of AHF&RD patients but also does so with a streamlined feature set, greatly enhancing its clinical utility and potential for broader application. While the AIHFLevel system has demonstrated robust predictive accuracy across various validation scenarios, a notable limitation of our study is the inability to conduct direct comparisons of the underlying modeling methods with every model evaluated. Due to the diverse nature of existing models, each employing potentially unique feature engineering techniques and specific modeling parameters, it is challenging to replicate the exact conditions under which these models were originally developed. As a result, our comparisons are primarily based on performance metrics rather than methodological nuances, which might lead to variations in how these models would perform under identical conditions. This limitation underscores the need for a standardized framework for model comparison in future studies, to ensure that the performance metrics are directly comparable and reflective of methodological rigor.
Some burdens are specific to the application of predictive models in clinical translation. In particular, the majority of current risk estimations using dozens of redundant features from various types of dimensions are inconvenient and almost impossible to implement in daily clinical practice35,36,37,38,39,61,62,63,64. By a hybrid feature selection paradigm containing Filter & Wrapper & Embedded techniques, our framework ultimately locked to 12 features for risk prediction Objective data for these variables are accessible, making it readily available in the current clinical setting. Sometimes, an accurate yet complex model did not necessarily translate into a practical clinical tool. The adoption of ML systems in medical decision support could be hindered by a nebulous associated with intrinsic mechanisms difficult to understand65. This study presents a significant advantage in utilizing the SHAP approach to demystify the “black-box” nature of AI models. AIHFLevel system could explain the AI model via a global explanation that described the overall functionality of a model and a local explanation that detailed how a certain prediction is made for an individual patient by inputting the individualized data. Furthermore, our prediction model is conveniently accessible through a web-based tool developed using the Django framework, enabling broader dissemination among clinicians. AIHFLevel’s direct integration of interpretability highlights the critical role of factors such as age, GFR, cTNI, and serum creatinine in influencing long-term survival outcomes for patients with AHF and RD. In contrast to previous studies that primarily focused on the importance of variables at a group or historical level without offering actionable insights for individual patients, our system provides a personalized visualization of predictors35,36,37,38,39,61,62,63,64. This feature allows users to see how each factor impacts the survival outcome at an individual level, suggesting that aligning most risk predictor values closer to their normal ranges through targeted interventions or management strategies could reduce the risk of ACM for the patient, even if the overall prediction categorizes the case as high-risk.
Several limitations should be noted. Although the use of a large and heterogeneous patient population, was an observational study, future validation should be conducted in a prospective multicenter cohort. Moreover, further randomized and controlled studies were required to figure out whether individualized and prompt therapeutic measures according to the AIHFLevel system could improve patient outcomes. Even though the system can inform the contribution of 12 clinical indexes to survival for each case, echocardiography might not be available for all AHF patients with RD, especially in primary care clinics. Additionally, regarding therapeutic use, a prognostic factor fails to substitute for the need for predictive elements. Put differently, if the system shows an overall poor prognosis score for a patient, the physician should conduct additional booster sessions for her or him. This is the reason we integrated interpretability. If age, for example, is the first feature that explains the worst survival probability, then the decision to intensify therapy can be very different from a case when comorbidity status, or pathology results as the main factors. The system gives back the final word and determination to the physician.
In conclusion, we employed an AI hybrid modeling framework to develop and validate AIHFLevel, a web-based system designed for evaluating the long-term survival profiles of patients with AHF and RD. The system integrated outcome prediction, clinical interpretability and prognostic stratification, and outperformed other clinical traits and composite risk models. Through 13 straightforward queries, the system empowered users to understand the influence of each predictor on every single individual survival outcome, thereby enabling the optimization of management strategies and targeted interventions in clinical practice.
Methods
Data source
This study was conducted to the ethical guidelines of the Declaration of Helsinki. The research was based on two medical centers. The in-house cohort data was accessed from the electronic health records (EHR) system of the Henan Province Clinical Research Center for Cardiovascular Diseases (CRCCD, Zhengzhou, Henan, China) from September 2018 to December 2020. Situated in the North China Plain, CRCCD’s organizational structure extends across the most densely populated areas of Central China. A total of 13 institutions, all of which are CRCCD members, participated in the study. The participating institutions comprised 11 independent tertiary hospitals (The First Affiliated Hospital of Zhengzhou University center hub, The First Affiliated Hospital of Henan University of Science and Technology Subcenter, Henan Provincial Chest Hospital Subcenter, Nanyang Central Hospital Subcenter, The First Affiliated Hospital of Xinxiang Medical University Subcenter, Xinyang Central Hospital Subcenter, Xuchang Central Hospital Subcenter, The Second Affiliated Hospital of Zhengzhou University Subcenter, Zhengzhou Central Hospital Subcenter, The Seventh People’s Hospital of Zhengzhou Subcenter, Zhoukou Central Hospital Central), one Professional Council, and one Academic Steering Committee responsible for supervision of the study design and operations. CRCCD used an online electronic data capture system (DCS) designed to ensure accurate data collection. Patient hospitalization records were registered in the DCS by the doctors in charge at each institution. Patient-identifiable information was dissociated and anonymized. Data were automatically checked for missing or contradictory entries and values out of the normal range. Additional editing and checks for duplicated records were performed by clinical research coordinators at the general office of the registry. This study was conducted to the ethical guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Committee of Henan Province Clinical Research Center for Cardiovascular Diseases (No.2021-KY-0720). Informed consent was obtained from each patient for their data to be used for research purposes.
The external cohort data was accessed from a single-center available database called the Medical Information Mart for Intensive Care III using the pgAdmin PostgreSQL (version 9.6) Structured Query Language. This database was approved by the Institutional Review Boards of Beth Israel Deaconess Medical Center (BIDMC, Boston, Massachusetts, USA) and the Massachusetts Institute of Technology, included information on 46,520 patients who were admitted to the hospital of BIDMC in Boston, Massachusetts from June 2001 to October 201256. The use of de-identified patient health information is not considered human subjects research, thus eliminating the need for individual patient consent due to the anonymity of the data. We completed the National Institutes of Health Course in the United States, passing the exam of human protection research participants (No.9971167). Moreover, access to the data was approved after completing the Collaborative Institutional Training Initiative (CITI) program “Data or Specimens Only Research” by author Ge Zhang (certification number: 41407001).
We reported our study in line with the TRIPOD (Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis) recommendations (Supplementary Table 17)66.
Study population
The inclusion criteria were as follows:
-
(1)
Over 18 years old.
-
(2)
Diagnosis with AHF using the International Classification of Disease 9th revision (ICD9) after the first admission, referring to the 2021 ESC advanced HF diagnostic criteria8.
At least two of the following three criteria must be met despite the treatment:
-
(a)
Severe and persistent HF symptoms [New York Heart Association (NYHA) class III or IV].
-
(b)
Severe cardiac dysfunction was defined by at least one of the following:
-
(i)
ejection fraction less than or equal to 30%;
-
(ii)
isolated right ventricular (RV) failure (e.g., arrhythmogenic right ventricular cardiomyopathy (ARVC));
-
(iii)
inoperable severe valvular anomalies;
-
(iv)
inoperable severe congenital anomalies;
-
(v)
the persistence of elevated BNP or NT-proBNP values, or severe left ventricular (LV) diastolic dysfunction or structural abnormalities defined by HFpEF (according to the definitions of HFpEF: objective evidence of cardiac structural, functional and serological abnormalities consistent with the presence of left ventricular diastolic dysfunction; NT-proBNP >125 (SR) or >365 (AF) pg/mL; BNP > 35 (SR) or >105 (AF) pg/mL; AF= atrial fibrillation, SR= sinus rhythm).
-
(i)
-
(c)
Low output paroxysmal HF requiring positive inotropes and vasoactive drugs; or pulmonary or systemic congestion episodes requiring high intravenous dose diuretics; or hospitalization in the past 12 months; or malignant arrhythmia resulting in >1 unscheduled visit.
-
(a)
Episodes of pulmonary or systemic congestion requiring high-dose i.v. diuretics (or diuretic combinations) or episodes of low output requiring inotropes or vasoactive drugs or malignant arrhythmias causing >1 unplanned visit or hospitalization in the last 12 months.
(d) Severe impairment of exercise capacity with inability to exercise or low 6-minute walk test (6MWT) distance (<300 m) or pVO2 < 12 mL/kg/min or <50% predicted value, estimated to be of cardiac origin.
Detailed information on AHF operationalized identification in the study was provided in (Supplementary Table 18).
(3) Diagnosis of renal insufficiency, characterized by an estimated glomerular filtration rate (eGFR) of less than 90 ml/(min·1.73 m²) and abnormalities in at least one of the following: blood creatinine or blood urea nitrogen levels.
Exclusion criteria:
-
(1)
Initial onset of acute heart failure.
-
(2)
Diagnosis of primary renal disease.
-
(3)
Diagnosis of an infectious disease or malignant tumor.
-
(4)
Receiving renal dialysis treatment.
-
(5)
Incomplete clinical case data: with missing >20% of individual data on relevant covariates necessary for research.
-
(6)
Hospitalization time <2 days.
Given that this study was a hypothesis-driven exploratory study based on multicenter retrospective longitudinal cohorts, no attempt was made to estimate the necessary sample size for the study67. Instead, all eligible patients from both centers were included to maximize statistical power.
At the CRCCD center, through the DCS system, we retrospectively enrolled 1256 patients from September 2018 to December 2020 following the inclusion criteria. Of those, 457 patients were excluded due to meeting the exclusion criteria or not signing the consent. Additionally, 87 patients were lost to follow-up during the follow-up period, and 712 patients were finally included and formed an in-house cohort. From the BIDMC center, using the PostgreSQL tool, we identified 1024 eligible patients between June 2001 and October 2012 as an independent external cohort, applying the same inclusion and exclusion criteria as the in-house cohort.
Data collection
We retrieved the patient data within the initial 24 hours following admission. Additionally, variables with over 20% missing values were excluded in the following analyses to minimize the bias resulting from missing data. Finally, 93 easily accessible variables were listed as candidates for constructing the survival assessment system subsequently.
The variables included: (1) general information (gender, age, NYHA class, smoking status, alcohol status, chronic kidney disease (CKD) stage, HF subtype); (2) comorbidity state (coronary artery disease (CAD), hypertension (HTN), arrhythmia, diabetes (DM), hyperlipidemia (HL), cerebro-vascular disease (CeVD), peripheral vascular disease (PVD), thyroid disorder (TD), chronic obstructive pulmonary disease (COPD)); (3) blood routine result (white blood cell count (WBC), red blood cell count (RBC), hemoglobin (Hb), platelet count (Plt), neutrophils count (NE), lymphocytes count (LY), monocytes count (MO), eosinophils count (EO), basophils count (BA), lymphocytes percentage (LY_Per), monocytes percentage (MO_Per), eosinophils percentage (EO_Per), basophils percentage (BA_Per), basophils neutrophils (NE_Per), haematocrit (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), red cell distribution width (RDW), mean platelet volume (MPV), plateletcrit (PCT), platelet distribution width (PDW)); (4) coagulation function result (thrombin time (TT), prothrombin time activity percentage (PTA), international Normalized Ratio (INR), activated partial thromboplastin time (APTT), fibrinogen (Fg), prothrombin time (PT)); (5) renal function result (blood urea nitrogen (BUN), serum creatinine (Cr), serum uric acid (UA), BUN/Cr ratio (BUN_Cr), glomerular filtration rate (GFR)); (6) liver function (total protein (TP), albumin (ALB), globulins (GLO), total bilirubin (TBIL), direct bilirubin (DBIL), indirect bilirubin (IBIL), total cholesterol (TCHO), triglycerides (TRIG), high density lipoprotein cholesterol (HDL), low-density lipoprotein cholesterol (LDL)); (7) cardiac function biomarker (NT-proBNP (BNP), cardiac troponin I (TN), creatine kinase (CK), creatine kinase MB Isownzyme (CKMB), C-reactive protein (CRP)); (8) (glucose (GLU), glycated haemoglobin (HbA1c)); (9) Echocardiography result (right ventricle internal diameter (RVID), interventricular septal thickness (IVS), left ventricle internal diameter (LVID), left ventricular posterior wall thickness (LVPWT), aortic valve annulus diameter (AOAD), left atrium diameter (LAD), pulmonary artery valve diameter (PAVD), right atrium supero-inferior diameter (RALRD), right atrium supero-inferior diameter (RASID), early diastolic mitral wave velocity (MVE), late diastolic mitral wave velocity ratio (MVA), left ventricular ejection fraction (EF), stroke volume (SV), left ventricular fraction shortening (FS)); (10) medication history (aspirin, ticagrelor, clopidogrel, sacubatril/valsartan, ACEI_ARB, beta.blocker,diuretics, statin, nitrates, digoxin, nifedipine/diltiazem (CCB), pantoprazole/omeprazole/esomeprazole (PPI)); (11) percutaneous coronary intervention history (PCI history).
Missing data handling
The variables with over 20% missing values were discarded as described previously. The TRIPOD guidelines suggested applying multiple imputations when missing data were present, as complete case analyses can lead to reduced sample size, biased estimates, and loss of Information66. After enrolling patients based on the inclusion and exclusion criteria, no missing data were found in the BIDMC cohort, while the extent of missingness in the CRCCD cohort is detailed in (Supplementary Table 19). Initially, our data was presumed to be Missing At Random (MAR), aligning with the assumptions of prior studies68. Little’s test was subsequently applied to affirm that the data was not Missing Completely At Random (MCAR) (Chi-square = 1483.688, Degrees of Freedom = 1104, P = 1.26e−13) as noted in (Supplementary Table 19)69. Therefore, multiple imputation was performed through the method of chained equations procedure70, ensuring a statistically robust approach to addressing missing data in our study.
Multiple imputation with chained equations (MICE) specified a multivariate model through conditional distributions, where each variable with missing data is imputed conditionally on all other variables. In our dataset with p (p = 93) variables and k (k = 20) variables missing data (with p-k complete cases), the procedure was as follows:
-
(1)
For each of the k variables with missing data, an imputation model was specified.
(a) numeric data: bayesian linear regression imputation.
(b) factor data with 2 levels: logistic regression imputation.
(c) factor data with >2 unordered levels: polytomous regression imputation.
(d) factor data with >2 ordered levels: proportional odds model.
-
(2)
Imputation values are drawn from the overall distribution of observed values for each variable with missing data. These initial imputed values are refined in subsequent steps.
-
(3)
For the first variable with missing data:
-
(a)
Regression is performed based on other variables (including the complete data of the first variable; and other observed or imputed variables).
-
(b)
Regression parameters: extract estimated regression coefficients and the variance-covariance matrix of the regression model from the previous regression (and for linear regression models fitted for continuous variables, the estimated variance of the residuals).
-
(c)
Parameters are randomly perturbed to reflect the uncertainty in data generation.
The conditional distribution for each sample with missing data in the first variable is determined using the perturbed regression coefficients.
-
(d)
Imputation values are then selected for each missing datum based on the conditional distribution.
-
(4)
Steps 3 and 4 are repeated in a cycle, imputing each variable with missing data.
-
(5)
The process of steps 3 and 4 is repeated 20 iterations. The final imputed values form the first completed imputed dataset.
-
(6)
Steps 3 to 5 are repeated 5 times to generate 5 imputed datasets.
-
(7)
Construction of the multiple imputed datasets is completed and now M datasets are obtained (assuming M = 5). These datasets are then treated as complete for statistical analysis. The results were pooled into one dataset by applying Rubin’s rules71.
In addressing missing data for continuous and categorical variables, our analysis revealed a uniform pattern in the data distribution before and after interpolation, as detailed in (Supplementary Fig. 7) and (Supplementary Fig. 8). This consistency was validated using the Wilcoxon rank-sum test for continuous variables and the chi-squared test for categorical variables, ensuring the integrity and reliability of our data analysis.
Study outcome
For the CRCCD cohort, collection of the follow-up information was conducted by extracting case system records, contacting patients, and relatives, and referring physicians through telephone or mail. For the BIDMC cohort, we gathered information from the publicly available single-center database, Medical Information Mart for Intensive Care III, utilizing the pgAdmin PostgreSQL (version 9.6) Structured Query Language. The primary endpoint event was ACM during in-hospital and out-hospital deaths. The secondary endpoint was a major adverse cardiovascular event (MACE). MACE included one or several of the following conditions: readmitted for symptomatic HF, nonfatal acute coronary syndrome, nonfatal ischemic stroke, new-onset cardiac arrhythmia, the use of mechanical circulatory support and implementation of heart transplantation, and cardiac death. Each chart was reviewed separately to determine the presence of MACE, and the American College of Cardiology/American Heart Association definitions provided for clinical trials were used to identify MACE72.
AI hybrid modeling framework generated survival assessment system (AIHFLevel)
There are many modeling algorithms to analyze data and assess outcomes currently. Predictions provided by different algorithms may vary depending on the characteristics of the dataset. It remained challenging to identify the appropriate learner tailored to the application’s requirements. However, some researchers have suggested that the use of hybrid learning models, e.g. ensemble-based approaches that is, a combination of two models, and enhancing existing learning models can be helpful. Mienye and colleagues provided evidence that ensemble-based learning may perform better than the single model in the prediction of HF events51. Ensemble modeling operates by employing a collection of diverse and independent models to make predictions, thereby reducing the generalization error and improving the reliability of the predictive outcomes53. Central to our framework is the pursuit of the suitable learner. The graphic illustration was presented in (Fig. 2a, Supplementary Fig. 9)
-
(1)
Data application: The CRCCD in-house cohort was used to evaluate the viability of various algorithms and identify the optimal modeling schemes, while the BIDMC external cohort served to validate the system’s generalizability. ACM serves as an outcome to train our system.
-
(2)
46 candidate survival features: We incorporated 93 readily accessible variables, as detailed earlier. In the CRCCD cohort, univariate Cox proportional hazards (PH) regression and Log-rank test were conducted. 46 variables were identified as candidate survival features, demonstrating statistical significance (P < 0.05) in both tests.
-
(3)
12 modeling algorithms: L1-regularized PH regression (L1.regularization), L2-regularized PH regression (L2.regularization), Elastic-Net-regularized PH regression (ENet.regularization), Survival random forest SRC learner (Surv.RFsrc), Survival support vector machine (Surv.SVM), survival tree (Surv.rpart), Cox model with likelihood-based boosting (Surv.Coxboost), Boosted generalized linear survival learner (Surv.glmboost), Extreme gradient boosting survival learner (Surv.Xgboost), Survival gradient boosting machine learner (Surv.gbm), Survival fully parametric learner (Surv.Fully-parametric), Accelerated oblique random survival forest learner (Surv.aorsf). Details of their hyperparameters are presented in (Supplementary Table 20).
-
(4)
132 modeling schemes:
-
(a)
Single Algorithm Model Fitting:12 algorithms were performed on 46 candidate survival features to fit models, generating 15 schemes: L1.regularization (lambda.min), L1.regularization (lambda.1se), L2.regularization (lambda.min), L2.regularization (lambda.1se), ENet.regularization (lambda.min), ENet.regularization (lambda.1se), Surv.RFsrc, Surv.SVM, Surv.rpart, Surv.Coxboost, Surv.glmboost, Surv.Xgboost, Surv.gbm, Surv.Fully-parametric, Surv.aorsf. For regularization algorithms (L1, L2, and ENet), we utilized ‘lambda.min’ and ‘lambda.1se’ as λ regularization parameters to expand the range of possible schemes.
-
(b)
Model Fitting using a Combinatorial Family of Algorithms: Feature selection on 46 candidate survival features was initially executed with each algorithm, followed by fitting the selected features with the remaining algorithms.
-
(i)
Embedded Method: For L1.regularization and ENet.regularization, we directly leveraged their built-in selection mechanisms based on L1 and elastic-net penalties. 5-fold cross-validation served as the resampling strategy for evaluating feature subset performance. The other 9 algorithms, excluding regularization methods, were then applied to fit models on the selected features, generating 18 schemes.
-
(ii)
Filter & Wrapper Hybrid Method: This method combined the rapid training capability of the external technique Filter (feature ranking) and the intensive computation of the Wrapper (using the model as a black box to evaluate feature subset according to their predictive power)73. Apart from the regularization methods, each of the remaining 9 algorithms executed the following steps:
-
(i)
First, the filter method was adopted to rank the features in descending order according to the Correlation-Adjusted Survival Score (CARS)74. Next, sequential forward floating selection (SFFS) as a wrapper method was used to select features75. Features were added in feature sets in sequence, and feature retention was based on the performance of each algorithm at every step. The 5-fold cross-validation assisted the performance assessment. If the performance with a new feature set did not increase, the new feature was removed from the feature set. Supplementary Fig. 10 illustrated the main procedure of the Filter & Wrapper Hybrid Method. Finally, the remaining 11 algorithms were used to fit models on the selected features, generating 99 schemes.
-
(5)
Comprehensive Assessment:
In the CRCCD in-house cohort, we implemented 132 modeling schemes, each generating a separate model. The evaluation of these models’ effectiveness and relevance was conducted via a rigorous comprehensive assessment strategy. This included 10 repeated 10-fold cross-validation, Monte-Carlo cross-validation (MCCV, 100 iterations with a 0.7 sampling ratio), and bootstrap analysis (1000 iterations with a 0.7 sampling ratio).
The concordance index (C-index), calculated across three strategies, served as the pivotal performance metric. The modeling scheme with the highest average C-index was recognized as the most effective for AHF&RD patients.
This comprehensive assessment strategy offered significant advantages:
-
(a)
Comprehensive Coverage: Integrating varied validation techniques ensured a more thorough and resilient evaluation. This multifaceted approach countered the risks of overfitting, bias inherent in single-method validation, and selection bias by researchers.
-
(b)
Diverse Scenario Analysis: Using MCCV and bootstrap analysis allowed for an extensive examination of the models’ performance across diverse datasets and hypothetical clinical scenarios, thus ensuring that the model is not overly tailored to a specific dataset or condition. This enhances the model’s generalizability, making it more applicable and reliable in different clinical settings and populations.
-
(c)
Precision in Model Selection: Applying the average C-index across diverse validation strategies refined the selection of the most effective model for AHF&RD, ensuring a robust choice backed by consensus performance metrics.
-
(6)
Validation and Extrapolation:
In the CRCCD in-house cohort, we divided the participants into a Discovery cohort (70%, n = 498) for model fitting using the optimal scheme, which led to the modeling of our prediction system AIHFLevel. A Replication cohort (30%, n = 214) was then employed for the initial validation of AIHFLevel. Further validation and generalization processes were conducted on the complete CRCCD Meta cohort and the external BIDMC cohort.
External validation across diverse data modalities, such as patients from BIDMC with varying hospital systems, technological platforms, regional, and ethnic backgrounds, is crucial for predictive analytics. This approach can effectively address the issue of model overfitting on internal data, ensuring broader applicability and generalizability of the predictive system.
Discrimination was quantified by AUC and C-index for specific time points and global time assessment. Integrated Brier score (IBS) was used as an overall summative measure of predictive performance, and calibration was evaluated through calibration plots. The decision curve analysis (DCA) served to determine whether the clinical value of the AIHFLevel system increased the net benefit over a realistic range of threshold probabilities.
Prognostic stratification
We employed an unbiased non-parametric conditional inference survival tree, integrating tree-structured regression models. This method utilized a recursive partitioning algorithm to naturally categorize samples based on survival time and covariates76.
-
(1)
The inference tree was built based on AIHFLevel system, demonstrating the potent capability to separate survivors and non-survivors.
-
(2)
We performed a binary split on the features used by AIHFLevel, determining the optimal cut point based on log-rank statistics. This approach allowed for precise stratification.
-
(3)
Stopping rules were based on Bonferroni-adjusted p-values to determine tree size. The minimum criterion for node split was defined as p < 0.001. The outcomes of this modeling are represented in a single tree (High risk, Intermediate risk, Low risk). Additionally, Kaplan-Meier (KM) curves were constructed for each subgroup identified through the survival tree methodology.
The features used by AIHFLevel were processed using the t-distributed stochastic neighbor-embedding (t-SNE) technique for dimensional reduction, visualization, and validation of prognostic stratification. To assess the discriminatory power of the AIHFLevel system across three risk states, we applied the trinormal-based ROC test and Volume Under the ROC Surface (VUS) based statistical tests, alongside the trinormal snapshot of the ROC surface77,78.
To further substantiate the robustness of our prognostic stratification, we quantified the global difference between pairs of EHR profiles using the Euclidean distance approach79.
The Euclidean distance:
Where xi and yi are the value of AIHFLevel system’s features i over two profiles (High-risk subgroup and remaining samples) with p and q samples respectively (x1, x2,…, xp), (y1, y2,…, yq). n is the number of features present in the profile.
Critical transition signal analysis
In complex disease progression, transitions could occur abruptly and nonlinearly, not always following a slow, linear trajectory41. A critical transition signal (CTS) or tipping point represented a sharp shift from one state to another. According to this concept, the disease progression can be segmented into three stages: ‘Before-deterioration state’, a relatively stable state where the disease undergoes gradual and slow change, ‘Pre-disease state’, which is the limit of the normal state just before the transition to the disease state, and the ‘Deteriorated state’, which is another relatively stable state and is usually irreversible to the ‘Pre-disease state’. Identifying the critical state or tipping point just before this transition is crucial for early prevention. We optimized a mathematical model, dynamic network biomarker (DNB) method, detecting early-warning signals or CTS of deterioration of the disease.
Let X denote the p × n matrix of the value levels of P EHR variables (g1, g2,… gp) in rows and n samples (s1, s2,… sn) in columns. When the n = n1 +…+ nR samples are classified into R distinct states (based on identified prognostic stratification: High risk, Intermediate risk, Low risk), we can group the columns of X to have X = [X1|···|XR], where Xr denotes the p × nr submatrix for samples in a state r ∈ (1, 2,…, R) and nr denotes the number of samples observed in the r-th state.
Let PCC refer to the Pearson correlation coefficient between any two variables (gi and gj). Let |.| take the absolute value.
-
(1)
Network partition: Repeat the following steps for each state r,
-
(a)
Calculate all pairwise PCC_r(gi,gj).
-
(b)
(b) Network partition by greedily optimizing ‘modularity’ in network communities (FDR of PCC_r(gi,gj) <0.05).
Module refers to a cluster of network nodes (e.g. EHR variables) highly linked (by correlation).
-
(2)
Identify CTS: Repeat the following steps for each module m,
-
(a)
PCCi(m) = Avg(|PCC(g,g)|), where (g.)∈ m.
-
(b)
PCCo(m) = Avg(|PCC(gi,gj)|), where (gi.)∈ m, (gj.)∉ m
-
(c)
SDi(m) = Avg(SD(g)), where (g.)∈ m, SD refers to the standard deviation.
-
(d)
CI(m) = PCCi(m)*SDi(m)/PCCo(m), CI refers to the composite index.
-
(e)
The biomodule (module with the highest CI(m) score) of each state indicated the CTS levels in each state. The DNB scores (PCCi, PCCo, SDi, CI) of biomodules represented the DNB scores of each state.
The CI is expected to increase abruptly and significantly before the critical transition to the disease state and can serve as an early warning signal.
-
(3)
CTS validation: To ensure robustness and reliability, we finally estimated the expected DNB scores from Xr by bootstrapping size biomodule variables from the HER background 1000 times.
System interpretability
To reasonably explain the decision-making process and adapt treatment strategies, the physicians require an understanding of how the AIHFLevel system relies on the system’s features or any comorbidity of the specific subject. The Shapley Additive exPlanation (SHAP) value was imputed to tackle the transparency issue, estimating each feature’s contribution based on cooperative game theory80. On an individual scale, we further visualized the features of any new subject participating in the prediction and how they impacted the future survival outcome.
Subgroup and sensitivity analyses
To validate the robustness of our result, sensitivity analyses were performed:
-
(1)
Dual-Outcome Evaluation: Our system was designed to estimate the survival distribution for a given individual, with ACM serving as an outcome to train our system. In addition to confirming the system’s robustness in predicting ACM, we also conducted an evaluation to assess its performance with MACE as an alternative outcome. This approach allowed us to comprehensively gauge the system’s predictive capabilities for different clinical endpoints.
-
(2)
Subgroup analysis: We conducted analyses among five pre-specified subgroups [Age (>65 or ≤65 years), Cardiac function grade (NYHA I/II or NYHA III/IV), HF subtype (HFpEF, HFmrEF, HFrEF), CKD stage (stage I/II, III, IV/V), PCI history (Yes or No)] to evaluate the prognostic value of AIHFLevel on long-term ACM and MACE among AHF patients with RD.
Statistical analysis
Data processing, statistical analysis, and plotting were conducted in R software (version 4.3.2). Continuous variables were statistically compared through Wilcoxon-rank-sum test or Student’s t-test, while categorical variables were analyzed by Chi-square test or Fisher’s exact test. The assumption of proportional hazards was verified using Schoenfeld residuals and log-log inspection. The restricted cubic spline regression (3 knots) was applied to investigate the possibly nonlinear relationships between AIHFLevel and prognosis. Non-linearity was assessed via a likelihood ratio test comparing models with linear terms against those with both linear and cubic spline terms. The fuzzy c-means soft clustering and dynamic baseline data pattern analysis were performed using Mfuzz software, focusing on cardiac function grade, CKD stage, and HF subtype subgroups. Comparative analysis of Integrated Brier Score (IBS) and Concordance Index (C-index) was conducted using the survcomp package, while the survminer package was utilized for optimal cut-off point determination. Cox regression analysis, logrank test, visualization of ROC and calibration curves, and trinormal snapshot of ROC surface were implemented by the survival, survminer, pROC, timeROC, rms, and trinROC packages. Model fitting was conducted using the mlr3, mlr3learners, mlr3verse, mlr3tuning, mlr3proba, and mlr3extralearners packages. Statistical significance was set at a two-sided P-value < 0.05. Error bars represent 95% confidence intervals.
Online system deployment
To enhance the model’s practicality in clinical settings, the AIHFLevel system has been further encapsulated into a user-friendly web application (https://www.hf-ai-survival.com), that can work on any new case, providing a more intuitive and understandable way to interpret the working principle. For any subject with AHF&RD, using the answers to 13 easy questions, the application assesses the future survival from diagnosis and the contribution of each index to the outcome. The web server was powered by Django, a high-level Python Web framework (version 4.1.3) (https://djangoproject.com).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The in-house data in this study was accessed from the electronic health records system of the Henan Province Clinical Research Center for Cardiovascular Diseases. The in-house individual-level data is protected and cannot be shared openly due to data privacy laws, ethical restrictions, and confidentiality agreements. For access to additional information required to reanalyze the data supporting the findings, please contact corresponding author ([email protected]) with a detailed request and may be required to sign a data use agreement to ensure the protection of participant confidentiality. Requests will be evaluated by the Institutional Ethics Committee of Henan Province Clinical Research Center for Cardiovascular Diseases, and a response will be provided within 30 business days. The external data was accessed from the Beth Israel Deaconess Medical Center Resource, which is available at Multi-parameter Intelligent Monitoring III database. Source data are provided with this paper.
Code availability
Essential original scripts for implementing AIHFLevel system in this paper is available through GitHub website (https://github.com/DrZoggg/AIHFLevel).
References
Crespo-Leiro, M. G. et al. Advanced heart failure: a position statement of the Heart Failure Association of the European Society of Cardiology. Eur. J. Heart Fail 20, 1505–1535 (2018).
Fang, J. C. et al. Advanced (stage D) heart failure: a statement from the Heart Failure Society of America Guidelines Committee. J. Card. Fail 21, 519–534 (2015).
Truby, L. K. & Rogers, J. G. Advanced heart failure: epidemiology, diagnosis, and therapeutic approaches. JACC Heart Fail 8, 523–536 (2020).
Xanthakis, V. et al. Prevalence, neurohormonal correlates, and prognosis of heart failure stages in the community. JACC Heart Fail 4, 808–815 (2016).
McDonagh, T. A. et al. Focused Update of the 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 44, 3627–3639 (2023). 2023.
Löfman, I., Szummer, K., Dahlström, U., Jernberg, T. & Lund, L. H. Associations with and prognostic impact of chronic kidney disease in heart failure with preserved, mid-range, and reduced ejection fraction. Eur. J. Heart Fail 19, 1606–1614 (2017).
Beldhuis, I. E. et al. Evidence-based medical therapy in patients with heart failure with reduced ejection fraction and chronic kidney disease. Circulation 145, 693–712 (2022).
McDonagh, T. A. et al. ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 42, 3599–3726 (2021). 2021.
Schefold, J. C., Filippatos, G., Hasenfuss, G., Anker, S. D. & von Haehling, S. Heart failure and kidney dysfunction: epidemiology, mechanisms and management. Nat. Rev. Nephrol. 12, 610–623 (2016).
Iorio, A. et al. Prevalence and prognostic impact of non-cardiac co-morbidities in heart failure outpatients with preserved and reduced ejection fraction: a community-based study. Eur. J. Heart Fail 20, 1257–1266 (2018).
Beldhuis, I. E. et al. Efficacy and safety of Spironolactone in patients with HFpEF and chronic kidney disease. JACC Heart Fail. 7, 25–32 (2019).
Krishnathasan, K. et al. Advanced heart failure in adult congenital heart disease: the role of renal dysfunction in management and outcomes. Eur. J. Prev. Cardiol. 30, 1335–1342 (2023).
McAlister, F. A. et al. Renal dysfunction in patients with heart failure with preserved versus reduced ejection fraction: impact of the new Chronic Kidney Disease-Epidemiology Collaboration Group formula. Circ. Heart Fail 5, 309–314 (2012).
Unger, E. D. et al. Association of chronic kidney disease with abnormal cardiac mechanics and adverse outcomes in patients with heart failure and preserved ejection fraction. Eur. J. Heart Fail 18, 103–112 (2016).
Patel, R. B. et al. Kidney function and outcomes in patients hospitalized with heart failure. J. Am. Coll. Cardiol. 78, 330–343 (2021).
Myhre, P. L. et al. Influence of NT-proBNP on efficacy of Dapagliflozin in heart failure with mildly reduced or preserved ejection fraction. JACC Heart Fail. 10, 902–913 (2022).
Wang, F., Kaushal, R. & Khullar, D. Should health care demand interpretable artificial intelligence or accept “Black Box” medicine? Ann. Intern. Med 172, 59–60 (2020).
Myhre, P. L. et al. Association of natriuretic peptides with cardiovascular prognosis in heart failure with preserved ejection fraction: secondary analysis of the TOPCAT Randomized Clinical Trial. JAMA Cardiol. 3, 1000–1005 (2018).
Tsutsui, H. et al. Natriuretic peptides: role in the diagnosis and management of heart failure: A scientific statement from the Heart Failure Association of the European Society of Cardiology, Heart Failure Society of America and Japanese Heart Failure Society. Eur. J. Heart Fail 25, 616–631 (2023).
Reddy, Y. N. V., Carter, R. E., Obokata, M., Redfield, M. M. & Borlaug, B. A. A simple, evidence-based approach to help guide diagnosis of heart failure with preserved ejection fraction. Circulation 138, 861–870 (2018).
George, L. K. et al. Heart failure increases the risk of adverse renal outcomes in patients with normal kidney function. Circ. Heart Fail 10, e003825 (2017).
Mark, P. B. et al. Major cardiovascular events and subsequent risk of kidney failure with replacement therapy: a CKD Prognosis Consortium study. Eur. Heart J. 44, 1157–1166 (2023).
Bansal, N. et al. Burden and outcomes of heart failure hospitalizations in adults with chronic kidney disease. J. Am. Coll. Cardiol. 73, 2691–2700 (2019).
Gautam, N. et al. Contemporary applications of machine learning for device therapy in heart failure. JACC Heart Fail. 10, 603–622 (2022).
Olsen, C. R., Mentz, R. J., Anstrom, K. J., Page, D. & Patel, P. A. Clinical applications of machine learning in the diagnosis, classification, and prediction of heart failure. Am. Heart J. 229, 1–17 (2020).
Kresoja, K.-P., Unterhuber, M., Wachter, R., Thiele, H. & Lurz, P. A cardiologist’s guide to machine learning in cardiovascular disease prognosis prediction. Basic Res Cardiol. 118, 10 (2023).
Eloranta, S. & Boman, M. Predictive models for clinical decision making: Deep dives in practical machine learning. J. Intern. Med. 292, 278–295 (2022).
Kee, O. T. et al. Cardiovascular complications in a diabetes prediction model using machine learning: a systematic review. Cardiovasc. Diabetol. 22, 13 (2023).
Jeong, K., Mallard, A. R., Coombe, L. & Ward, J. Artificial intelligence and prediction of cardiometabolic disease: Systematic review of model performance and potential benefits in indigenous populations. Artif. Intell. Med. 139, 102534 (2023).
Rajkomar, A. et al. Scalable and accurate deep learning with electronic health records. NPJ Digit. Med. 1, 18 (2018).
Volovici, V., Syn, N. L., Ercole, A., Zhao, J. J. & Liu, N. Steps to avoid overuse and misuse of machine learning in clinical research. Nat. Med. 28, 1996–1999 (2022).
Lewis, E. F. Machine learning and social determinants of health-an opportunity to move beyond race for inpatient risk prediction in patients with heart failure. JAMA Cardiol. 7, 854–855 (2022).
Löfman, I. et al. Incidence of, associations with and prognostic impact of worsening renal function in heart failure with different ejection fraction categories. Am. J. Cardiol. 124, 1575–1583 (2019).
Casado, J. et al. Clinical characteristics and prognostic influence of renal dysfunction in heart failure patients with preserved ejection fraction. Eur. J. Intern. Med. 24, 677–683 (2013).
Pocock, S. J. et al. Predicting survival in heart failure: a risk score based on 39 372 patients from 30 studies. Eur. Heart J. 34, 1404–1413 (2013).
Simpson, J. et al. Prognostic models derived in PARADIGM-HF and validated in ATMOSPHERE and the Swedish Heart Failure Registry to predict mortality and morbidity in chronic heart failure. JAMA Cardiol. 5, 432–441 (2020).
Lupón, J. et al. Development of a novel heart failure risk tool: the barcelona bio-heart failure risk calculator (BCN bio-HF calculator). PLoS One 9, e85466 (2014).
Lietz, K. et al. Outcomes of left ventricular assist device implantation as destination therapy in the post-REMATCH era: implications for patient selection. Circulation 116, 497–505 (2007).
Senni, M. et al. Predicting heart failure outcome from cardiac and comorbid conditions: the 3C-HF score. Int J. Cardiol. 163, 206–211 (2013).
Cleland, J. G. F., Li, C. & Jones, Y. Artificial intelligence needs clinical intelligence to succeed. JACC Heart Fail. 8, 588–591 (2020).
Chen, L., Liu, R., Liu, Z.-P., Li, M. & Aihara, K. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci. Rep. 2, 342 (2012).
Khan, M. S. et al. Artificial intelligence and heart failure: A state-of-the-art review. Eur. J. Heart Fail 25, 1507–1525 (2023).
Zhang, G. et al. Smooth muscle cell fate decisions decipher a high-resolution heterogeneity within atherosclerosis molecular subtypes. J. Transl. Med. 20, 568 (2022).
Zhang, G. et al. Atherosclerotic plaque vulnerability quantification system for clinical and biological interpretability. iScience 26, 107587 (2023).
Zhang, G. et al. Uncovering the genetic links of SARS-CoV-2 infections on heart failure co-morbidity by a systems biology approach. ESC Heart Fail 9, 2937–2954 (2022).
Samad, M. D. et al. Predicting survival from large echocardiography and electronic health record datasets: optimization with machine learning. JACC Cardiovasc. Imaging 12, 681–689 (2019).
Kwon, J.-M., Kim, K.-H., Jeon, K.-H. & Park, J. Deep learning for predicting in-hospital mortality among heart disease patients based on echocardiography. Echocardiography 36, 213–218 (2019).
Hearn, J. et al. Neural networks for prognostication of patients with heart failure. Circ. Heart Fail 11, e005193 (2018).
Liu, Y., Chen, P.-H. C., Krause, J. & Peng, L. How to read articles that use machine learning: Users’ guides to the medical literature. JAMA 322, 1806–1816 (2019).
Wynants, L. et al. A simulation study of sample size demonstrated the importance of the number of events per variable to develop prediction models in clustered data. J. Clin. Epidemiol. 68, 1406–1414 (2015).
Mienye, I. D., Sun, Y. & Wang, Z. An improved ensemble learning approach for the prediction of heart disease risk. Inform. Med. Unlocked 20, 100402 (2020).
Kunapuli, G. Ensemble methods for machine learning. Simon and Schuster (2023).
Kim, Y., Heider, P. & Meystre, S. Ensemble-based methods to improve de-identification of electronic health record narratives. AMIA Annu. Symp. Proc. 2018, 663–672 (2018).
Moons, K. G. et al. Risk prediction models: II. External validation, model updating, and impact assessment. Heart 98, 691–698 (2012).
Li, T., Jiang, S. & Yang, Y. Database selection and heterogeneity-more details. More Credibil. JAMA Oncol. 4, 1295 (2018).
Johnson, A. E. W. et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3, 160035 (2016).
Heidenreich, P. A. et al. AHA/ACC/HFSA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 145, e895–e1032 (2022). 2022.
Zhang, L. et al. Differential expression profiles of plasma exosomal microRNAs in dilated cardiomyopathy with chronic heart failure. J. Cell Mol. Med. 27, 1988–2003 (2023).
Machowska, A., Carrero, J. J., Lindholm, B. & Stenvinkel, P. Therapeutics targeting persistent inflammation in chronic kidney disease. Transl. Res. 167, 204–213 (2016).
Colombo, P. C. et al. Inflammatory activation: cardiac, renal, and cardio-renal interactions in patients with the cardiorenal syndrome. Heart Fail Rev. 17, 177–190 (2012).
Xanthopoulos, A. et al. Larissa heart failure risk score: a proposed simple score for risk stratification in chronic heart failure. Eur. J. Heart Fail 20, 614–616 (2018).
Carluccio, E. et al. The ‘Echo Heart Failure Score’: an echocardiographic risk prediction score of mortality in systolic heart failure. Eur. J. Heart Fail 15, 868–876 (2013).
Canepa, M. et al. Performance of prognostic risk scores in chronic heart failure patients enrolled in the european society of cardiology heart failure long-term registry. JACC Heart Fail 6, 452–462 (2018).
Freitas, P., Ferreira, A. M. & Aguiar, C. Comparison of prognostic scores in chronic heart failure. JACC Heart Fail 6, 887–888 (2018).
Sansone, M., Fusco, R., Pepino, A. & Sansone, C. Electrocardiogram pattern recognition and analysis based on artificial neural networks and support vector machines: a review. J. Health Eng. 4, 465–504 (2013).
Moons, K. G. M. et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann. Intern Med 162, W1–W73 (2015).
Greener, J. G., Kandathil, S. M., Moffat, L. & Jones, D. T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 23, 40–55 (2022).
O’Connor, C. et al. Clinical factors related to morbidity and mortality in high-risk heart failure patients: the GUIDE-IT predictive model and risk score. Eur. J. Heart Fail 21, 770–778 (2019).
Little, R. J. A. A test of missing completely at random for multivariate data with missing values. J. Am. Stat. Assoc. 83, 1198–1202 (1988).
Azur, M. J., Stuart, E. A., Frangakis, C. & Leaf, P. J. Multiple imputation by chained equations: what is it and how does it work? Int J. Methods Psychiatr. Res 20, 40–49 (2011).
Rubin, D. B. Statistical matching using file concatenation with adjusted weights and multiple imputations. J. Bus. Econ. Stat. 4, 87–94 (1986).
Hicks, K. A. et al. ACC/AHA key data elements and definitions for cardiovascular endpoint events in clinical trials: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Data Standards (Writing Committee to Develop Cardiovascular Endpoints Data Standards). J. Am. Coll. Cardiol. 66, 403–469 (2015). 2014.
Chandrashekar, G. & Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 40, 16–28 (2014).
Bommert, A., Welchowski, T., Schmid, M. & Rahnenführer, J. Benchmark of filter methods for feature selection in high-dimensional gene expression survival data. Brief. Bioinform. 23, bbab354 (2022).
Pudil, P., Novovičová, J. & Kittler, J. Floating search methods in feature selection. Pattern. rn Recognit. Lett. 15, 1119–1125 (1994).
Hothorn, T., Hornik, K. & Zeileis, A. Unbiased recursive partitioning: a conditional inference framework. J. Comput. Graph. Stat. 15, 651–674 (2006).
Noll, S., Furrer, R., Reiser, B. & Nakas, C. T. Inference in receiver operating characteristic surface analysis via a trinormal model-based testing approach. Stat 8, e249 (2019).
Xiong, C. et al. A parametric comparison of diagnostic accuracy with three ordinal diagnostic groups. Biom. J. 49, 682–693 (2007).
Hu, J. et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 31, 522–529 (2013).
Lundberg, S. & Lee, S. I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 30, 4765–4774 (2017).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82222007 to J.T., 82170281 to J.T., and U2004203 to J.Z.), the Henan Thousand Talents Program (No. ZYQR201912131) to J.Z., Henan Province Science and Technology Research Joint Project (No. 222301420054) to J.T., Henan Province Key R&D Program (No.241111313300) to J.Z., Funding for Scientific Research and Innovation Team of The First Affiliated Hospital of Zhengzhou University (ZYCXTD2023008 to J.Z. and QNCXTD2023001 to J.T.), Central Plains Youth Top Talent to J.T. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. Figs. 1a, 2a, 4a, 7b, created with licensed version of BioRender.com (https://www.biorender.com/), released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. We are very grateful for their contributions.
Author information
Authors and Affiliations
Contributions
J.T., K.C., J.Z., G.Z. conceived, designed, and/or supervised the project. G.Z. performed data analysis, manuscript writing, and figure preparation. G.Z. and Z.T. implemented the development and validation of the system. Z.W., Z.Q., S.X. and K.L. contributed to the collection and process of study data. C.S., D.L., Z.Z., Y.X., S.X., K.L., S.Z., R.W. T.L. and Y.Z. assisted with data analysis and/or figure preparation. All authors discussed the results and supervised the manuscript. All authors had full access to all the data in the study and accepted the responsibility to submit it for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jacob Joseph and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, G., Wang, Z., Tong, Z. et al. AI hybrid survival assessment for advanced heart failure patients with renal dysfunction. Nat Commun 15, 6756 (2024). https://doi.org/10.1038/s41467-024-50415-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-50415-9
This article is cited by
-
A physics-informed and data-driven framework for robotic welding in manufacturing
Nature Communications (2025)
-
Association of platelet-to-lymphocyte ratio with 1-year all-cause mortality in ICU patients with heart failure
Scientific Reports (2024)